Abstract
Cell loss contributes to the pathogenesis of many inherited and acquired human diseases. We have developed a system to conditionally ablate cells of any lineage and developmental stage in the mouse by regulated expression of the diphtheria toxin A (DTA) gene by using tetracycline-responsive promoters. As an example of this approach, we targeted expression of DTA to the hearts of adult mice to model structural abnormalities commonly observed in human cardiomyopathies. Induction of DTA expression resulted in cell loss, fibrosis, and chamber dilatation. As in many human cardiomyopathies, transgenic mice developed spontaneous arrhythmias in vivo, and programmed electrical stimulation of isolated-perfused transgenic hearts demonstrated a strikingly high incidence of spontaneous and inducible ventricular tachycardia. Affected mice showed marked perturbations of cardiac gap junction channel expression and localization, including a subset with disorganized epicardial activation patterns as revealed by optical action potential mapping. These studies provide important insights into mechanisms of arrhythmogenesis and suggest that conditional lineage ablation may have wide applicability for studies of disease pathogenesis.
Cell loss is commonly observed in diseased tissues, arising either as a consequence of necrosis or programmed cell death (1–3). If of sufficient magnitude, cell loss by either pathway may impair the ability of the tissue to adequately meet its functional demands, leading to clinically overt disease. Moreover, in tissues with highly organized structure, the loss of relatively few but strategically placed cells also may result in disease. In the central nervous system, for example, loss of specific dopaminergic striatal neurons may result in Parkinson’s disease (4), whereas in the heart, focal cell loss and scar formation may lead to conduction disturbances, either by blocking the specialized conduction pathway (5) or by promoting slow impulse conduction and facilitating reentry (6, 7). Thus, a versatile system to specifically ablate cells of any lineage and developmental stage would be of substantial benefit to model human diseases of various etiologies. Moreover, if the cell death signal could be reversibly controlled, not only could disease progression be modeled, but the processes of repair and/or regeneration could be examined as well.
In this report, we describe studies conditionally expressing the diphtheria toxin A (DTA) gene (8) in transgenic mice, using a binary approach with tetracycline (TCN)-responsive promoters (9). As a first example of the power of this system, we wished to explore how cell loss and ventricular remodeling contribute to cardiac rhythm disturbances. We reversibly targeted expression of DTA to cardiac myocytes to model a spectrum of pathological features commonly observed in arrhythmogenic human cardiomyopathies. These hearts displayed a remarkably high incidence of spontaneous and inducible ventricular tachycardia. Moreover, we found that expression of the connexin43 (Cx43) gap junction channel protein was significantly dysregulated, and in some instances, associated with perturbed conduction properties, which likely contributed to the arrhythmogenic milieu.
METHODS
Generation and Maintenance of Transgenic Mice.
The myosin heavy chain (MHC)α-tTA transgenic strain has been described previously (10). The transgene consists of the rat α MHC promoter driving expression of the tetracycline-controlled transactivator (9). To generate the tTA-responsive diphtheria toxin A (DTA) response transgene (tet-DTA), the DTA-coding region was placed downstream of heptamerized tetO sequences fused to a cytomegalovirus minimal promoter (8, 9). Transgenic mice were generated according to standard techniques in a CBAxB6F2 background, and founders were identified by Southern blot analysis. For most experiments, mating pairs from the two lines were supplemented with tetracycline hydrochloride (1 mg/ml) in the drinking water and antibiotic treatment was continued during gestation and maintained in the progeny until selected postnatal time points (11). Subsequently, offspring were genotyped by PCR, using primer pairs (5′-TCAGACCGAGATTTCTCCATCCC-3′) and (5′-GAATTCAGGCTCGCCTGCAGTTGG-3′) for the MHCα-tTA transgene and (5′-GGCGTGTACGGTGGGAGG-3′) and (5′-GGCATTATCCACTTTTAGTGC-3′) for the tet-DTA transgene. For both primer-pairs, reactions were heated to 94°C for an initial 3 min and then amplified by denaturing at 94°C for 30 sec, annealing at 60°C for 30 sec, and extending at 72°C for 1 min, for a total of 35 cycles. PCR products were visualized by ethidium bromide staining after agarose gel electrophoresis.
Telemetry Recordings.
Mice were anesthetized with inhaled methoxyfluorane. The peritoneum was exposed with a midline incision. A TA10EA-F20 implant (Datasciences, Minneapolis) was secured within the peritoneal cavity, and leads were implanted s.c. corresponding to position II (12, 13). Mice were allowed to recover for 2 days after surgery. Subsequently, the digital signal was transmitted from the implant to an RPC-1 receiver and BCM consolidation matrix and converted to analogue with an RO8 D/A converter (Datasciences, Minneapolis). This signal was then fed into the ECG100A amplifier and MP100WS acquisition system (Biopac Systems, Santa Barbara, CA) for data collection and analysis.
Langendorff Preparations.
Mice were heparinized and anesthetized. The hearts were quickly removed and rinsed in a Tyrode’s solution containing (in mM): NaCl, 130, NaHCO3 24, NaH2PO4 1.2, MgCl2 1.0, glucose 5.6, KCl 4.0, and CaCl2 1.8 and equilibrated with a 95% O2/5% CO2 gas mixture. Hearts were then rapidly cannulated and perfused in a retrograde fashion via an aortic cannula with warm (37–39°C) oxygenated Tyrode’s solution (1–2 ml/min). Once connected to the Langendorff perfusion system, the hearts were placed in a custom built perfusion chamber where warm oxygenated Tyrode’s solution was continuously superfused at 37–39°C. This chamber also served as a volume conductor for recording in vitro electrocardiograms. Hearts were stabilized against and imaged through the glass front of the chamber. To ensure that the hearts were in sinus rhythm and contracting forcefully and rhythmically, they were allowed to equilibrate for 20 min in the perfusion apparatus while the electrocardiogram was monitored.
High Resolution Optical Action Potential Mapping of the Mouse Heart.
A voltage sensitive dye (di-4-ANEPPS, Molecular Probes) was used to record electrical activity (14). Fluorescence associated with the electrical activity was recorded in the absence of mechanical artifacts by continuously perfusing diacetyl monoxime (15 mM). The video imaging used was similar to that previously described (15). Briefly, light from a 150-watt xenon arc lamp (OptiQuip, Highland Mills, NY) was made quasimonochromatic by the use of an interference filter (520 nm) together with a heat filter. A fiber optic cable directed the excitation light source toward the epicardial surface of the heart. The emitted light was collected and passed through an emission filter (645 nm) and projected onto a CCD video camera. Isochrone maps were generated from filtered video imaging data by analyzing each pixel’s value over time. Two different cameras were used: Dalsa (Waterloo, Ontario), 1484 frames/sec or Cohu (San Diego, CA), 240 frames/sec. Video images of the epicardial surface were acquired with an A/D frame grabber (Bitflow, Woburn, MA or Epix, Buffalo Grove, IL). To reveal the signal, the background fluorescence was subtracted from each frame. No temporal or spatial filtering was used in the data analysis. To increase the temporal resolution of the Cohu camera, we implemented a synchronous acquisition system (16). With this system, the frame rate of the camera is synchronized with each stimulus delivered to the tissue. During video acquisition, the stimulus was delayed by 0, 1, 2, and 3 ms with respect to the start of each frame of the movie. The four parts of the movies were identified, averaged, and shuffled to reveal an activation sequence with a 1-ms resolution. Acquisition with the Cohu system was limited to signals that were regular.
Induction of Ventricular Arrhythmia.
Susceptibility to induction of ventricular arrhythmias in control and transgenic hearts was determined by applying 1-sec burst pacing at 1.5 times diastolic threshold at rates near the 1:1 capture limit (BCL ≈60 ms). This protocol was repeated 20 times on both the anterior and posterior surfaces of the heart. Hearts were classified as inducible if arrhythmias (defined as more than five ectopic ventricular activations) occurred spontaneously, occurred during our normal pacing protocol, or occurred during the burst pacing protocol.
Histology.
At the completion of the optical-mapping protocol, hearts were immediately fixed in 10% formalin, sectioned in the short axis, and stained with hematoxylin and eosin. The histology was graded as normal (0), mild (+), moderate (++), or severe (+++) by an expert cardiac pathologist (S.M.F.) who was blinded to the genotype of the mouse, length of tetracycline treatment, or outcome of the optical-mapping study.
Immunohistochemistry.
Paraffin sections were deparaffinized and boiled in citrate buffer to increase antigen availability, as previously described (17). Sections were then blocked in 10% goat serum and incubated overnight at 4°C with rabbit anti-Cx43 polyclonal antisera (18) at 1:100 dilution, followed by incubation with a goat-anti rabbit Cy3 conjugated secondary antibody. Sections were initially visualized with an Axiophot microscope (Zeiss) equipped with epifluorescence and then viewed with a Bio-Rad MRC600 laser-scanning confocal microscope for quantification.
Western Blot Analysis.
Lysates were prepared by sonicating ventricular tissue in water supplemented with protease inhibitors. Samples were then solubilized in Laemmli buffer for 10 min at 25°C and proteins (10 μg) separated on duplicate 10% SDS/PAGE. One gel was directly stained with Coomassie Blue. The second gel was transferred to nitrocellulose and incubated with the rabbit anti-Cx43 antisera at 1:5000 dilution, followed by a protein A-peroxidase conjugated secondary antibody. Immunoreactive bands were visualized by enhanced chemiluminescence and autoradiographic signals were quantified by densitometry.
RESULTS
Generation of tet-DTA Transgenic Mice.
In the absence of transactivator, the DTA target transgene should be transcriptionally silent. However, generation of viable, healthy mice carrying such a transgene was difficult; only 6 of 300 (1.75%) live births carried the transgene. Given the toxicity of DTA, in which a single molecule is considered sufficient to inhibit protein synthesis and cause cell death, it is likely that transcriptional leakiness in at least one essential lineage during embryogenesis reduced the efficiency of transgenic recovery, which typically is ≈10–40% of live births in the facility used to generate these mice. Lines were established from selected tet-DTA founders. Histological analysis of a panel of tissues from one such line, including brain, kidney, spleen, liver, heart, skeletal muscle, eye, skin, pancreas, and intestines, showed no evidence of pathology attributable to abnormal cell death; this line was selected for further studies.
Transactivation of DTA in Utero is Embryonic Lethal.
We examined whether the DTA target gene could be successfully transactivated in the heart by breeding a line of tet-DTA transgenic mice with a second strain we had previously characterized, MHCα-tTA, which expresses the tetracycline-controlled transactivator exclusively in cardiac myocytes (10, 11). Offspring were genotyped 3 weeks after birth. In the absence of maternal tetracycline supplementation during gestation and in the perinatal period, no (0/18) binary transgenic mice harboring both the MHCα-tTA and the tet-DTA transgenes were recovered, deviating significantly from the expected Mendelian distribution of 25%. In marked contrast, when similar matings were performed in the presence of suppressive doses of gestational and perinatal tetracycline, the distribution of genotypes normalized, with recovery of 22% (5/23) binary pups. These data strongly suggested that the DTA target transgene was successfully transactivated in the heart in a tetracycline-repressible manner. Genotype analysis at earlier time points showed that death in binary transgenic progeny occurred between day 10.5 after conception and fetal day 19 (not shown), although the precise time interval and histopathology were not further delineated in this study.
Transactivation of DTA in Adult Mice.
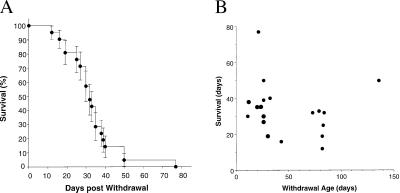
In contrast to the embryonic lethality observed with persistent expression of the target transgene in utero, we predicted that induction of DTA expression in postnatal mice would lead to progressive cardiac injury. Therefore, we tested this hypothesis by withdrawing tetracycline treatment in binary mice at varying ages after birth and following their subsequent clinical course. Withdrawal of the antibiotic was uniformly fatal. The median survival after tetracycline withdrawal was 32 days, with the earliest death occurring 12 days after drug withdrawal and the latest death recorded 77 days after stopping the antibiotic (Fig. 1A). We found no relationship between the age of the mice when tetracycline was withdrawn and the length of survival (Fig. 1B), suggesting that transactivation of DTA was equally effective in neonatal and adult hearts and consistent with our earlier observations using reporter genes (10). Death was typically sudden, with no ante-mortem indications of illness.
Figure 1.
(A) Kaplan–Meier curve. Matings were carried out between single transgenic MHCαtTA and tet-DTA strains. Tetracycline was administered throughout gestation and withdrawn at varying times after birth. Survival of double-transgenic offspring from the time of tetracycline withdrawal was determined. Median survival was 32 days (n = 19 binary mice). (B) Survival as a function of withdrawal age. No relationship between age at the time of tetracycline withdrawal and subsequent length of survival was observed (r2 = 0.02).
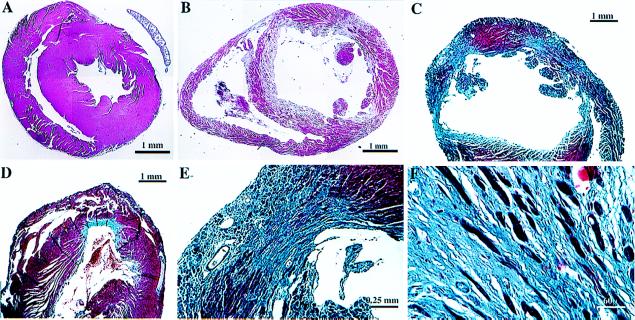
We recovered hearts from several binary transgenic mice, which had died suddenly, as well as others, which were killed and examined microscopically. All of the specimens showed evidence of myocyte loss and patchy fibrosis in the ventricles, although the extent of these changes varied substantially amongst individual specimens, reflecting the extent of DTA expression (Fig. 2). In some instances, prominent ventricular dilatation was observed. Tissue destruction and mural thrombus formation was prominent in atrial tissue as well (not shown), consistent with the expected spatial profile of MHCα-driven transgenes.
Figure 2.
Histological analysis. Hearts were fixed in formalin and paraffin sections prepared and stained with either hematoxylin and eosin (A and B) or trichrome (C–F) to highlight the interstitial fibrosis. Representative low-power views from a control heart (A) and individual binary transgenic hearts (B–D). Note the chamber dilatation (B and C) and marked variability in the extent of fibrosis (C and D) in binary hearts. Higher power views demonstrate the presence of scant residual myocytes in the most severe lesions (E and F). Scales are indicated. A and B are montages.
Spontaneous and Inducible Cardiac Arrhythmias.
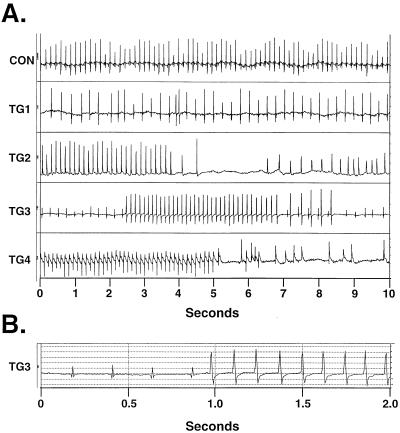
The structural heterogeneity in these hearts, coupled with the clinical picture of sudden death, suggested that cardiac arrhythmias might be prominent in the binary transgenic mice. Therefore, indwelling telemetry devices were implanted to record cardiac rhythm in unrestrained conscious mice. A variety of arrhythmias were observed in DTA-expressing transgenic mice, including atrial fibrillation, pauses, and complex ventricular ectopy, including runs of ventricular tachycardia (Fig. 3). In contrast, no comparable arrhythmias were detected in nontransgenic controls.
Figure 3.
Telemetric recordings. (A) Representative rhythm strips from nontransgenic (control) and binary transgenic mice (TG1–TG4). Abnormalities including atrial fibrillation (TG1), pauses (TG2), and runs of ventricular tachycardia (TG3 and TG4) were observed. (B) Replotting of TG3 trace at faster chart speed reveals altered QRS morphology of ectopic ventricular beats.
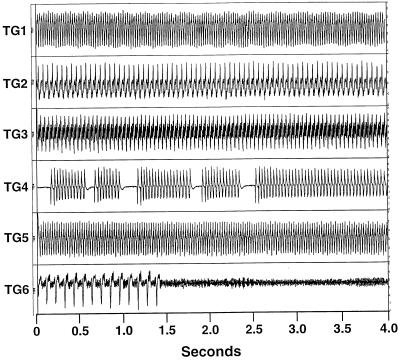
We wished to more quantitatively examine the relationship between cardiac structure, conduction properties and ventricular arrhythmogenesis. Therefore, isolated-perfused hearts from a cohort of binary transgenic mice and age-matched controls were examined 1 mo after withdrawal of tetracycline. Transgenic hearts had a markedly increased propensity to develop reentrant ventricular tachycardia. In contrast to the relative resistance of control hearts to the induction of ventricular arrhythmias (1/9), the majority (12/15) of transgenic hearts displayed either spontaneous or inducible ventricular tachycardia (P < 0.01 by χ2 analysis). Both nonsustained and sustained runs of ventricular tachycardia were observed (Fig. 4), with some episodes lasting several minutes at cycle lengths as short as 25 msec (≈2,400 bpm).
Figure 4.
Ventricular tachycardia in isolated-perfused hearts. Representative tracings of ventricular arrhythmias in binary transgenic mice. Recurrent bursts and nonsustained and sustained ventricular tachycardia were observed in individual hearts.
Epicardial Activation Patterns in Myopathic Hearts.
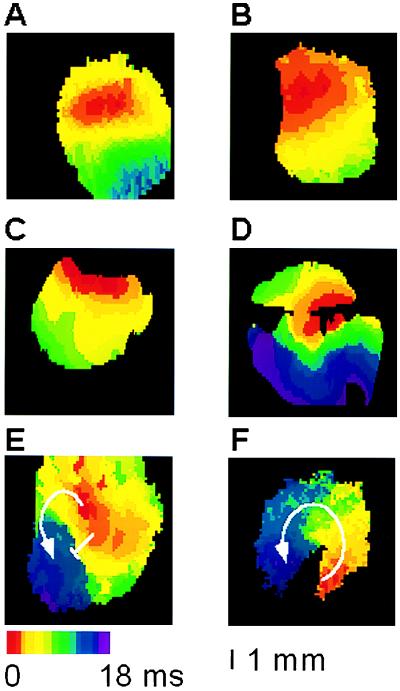
By using an optical action potential mapping system, we evaluated the patterns of epicardial activation in these same hearts during ventricular pacing. In all nontransgenic controls, activation proceeded smoothly from the pacing site with the expected anisotropic profile (Fig. 5A). Most binary transgenic hearts showed preserved conduction maps (Fig. 5B), but a subset displayed markedly disturbed activation profiles, with either disorganized conduction or evidence of block during pacing (Fig. 5C–E). In several instances, not only could the baseline epicardial activation map be obtained, but the camera was appropriately positioned to directly image the induced reentrant ventricular tachycardia, including one heart in which the area of slow conduction observed during pacing clearly served as an obstacle for the reentrant arrhythmia (Fig. 5 E and F).
Figure 5.
Optical action potential analysis. Isolated-perfused hearts were studied as described. Representative images showing normal anisotropic conduction observed in all control hearts (A) and most binary transgenics (B). In three transgenic mice, varying degrees of disorganized epicardial activation were observed (C–E). An area of slow conduction observed during pacing (E) served as an obstacle for inducible ventricular tachycardia (F). Hearts were imaged from either the anterior (A, B, and D) or posterior (C, E, and F) surfaces.
Histopathology of Arrhythmogenic Hearts.
Hearts were examined at the conclusion of the mapping studies and the severity of pathology graded (Table 1). In all control hearts, no significant abnormalities were detected. In contrast, a spectrum of histological abnormalities was observed in transgenic hearts, ranging from mild patchy fibrosis to diffuse severe cell loss and fibrosis (see Fig. 2B–D). Whereas the likelihood of developing ventricular tachycardia increased with worsening pathology, surprisingly, the majority of even mildly diseased transgenic hearts were arrhythmogenic, including one sample with markedly disorganized conduction despite relatively well-preserved structure (Fig. 5E).
Table 1.
Isolated-perfused heart analysis
| Group | Histology, n | Ventricular tachycardia
|
Disorganized conduction | ||
|---|---|---|---|---|---|
| Spontaneous | Induced | Total (%) | |||
| Control | Normal (9) | 0 | 1 | 1 (11%) | 0 |
| Transgenic | Mild (5) | 1 | 2 | 3 (60%) | 1 |
| Moderate (5) | 1 | 3 | 4 (80%) | 0 | |
| Severe (5) | 2 | 3 | 5 (100%) | 2 | |
This table summarizes the results of the isolated-perfused heart analysis. Control (n = 9) and binary transgenic (n = 15) hearts were optically imaged during the pacing protocol. The occurrence of either spontaneous or inducible ventricular tachycardia was then determined. At the end of the study, hearts were fixed in formalin and sectioned, and the extent of pathology was graded by an observer who was blinded as to genotype status or results of the optical mapping analysis.
Cx43 Expression in Arrhythmogenic Hearts.
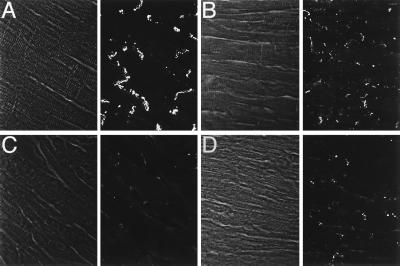
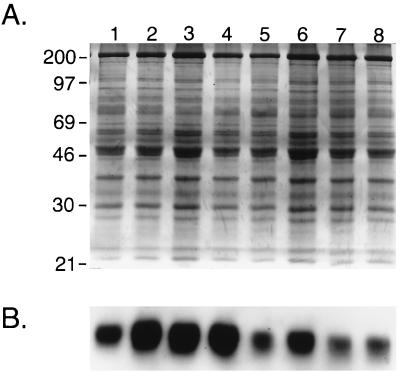
To understand potential factors contributing to the altered epicardial activation, especially in the mildly damaged hearts in which little fibrosis was evident, we examined the expression of Cx43, the major gap junction channel protein in the ventricle, using confocal microscopy (17, 18). Localization of Cx43 immunoreactivity into discrete appositional plaques at intercalated discs was substantially reduced in transgenic hearts (Fig. 6). Although this remodeling of gap junctions was most profound in the severely damaged hearts, spatial dysregulation of Cx43 was quite apparent even in minimally damaged hearts. Moreover, the reduction in junctional Cx43 staining was not restricted to focal areas of cell loss and fibrosis but was observed heterogeneously throughout the entire ventricular myocardium, including areas which appeared relatively normal by routine staining. The loss of Cx43 from appositional sites was confirmed by direct Western blot analysis, in which a 60% reduction (P < 0.05) in Cx43 content in the transgenic ventricles was observed, as illustrated in Fig. 7. This reduction did not reflect simply the loss of myocytes, inasmuch as the expression of sarcomeric myosin heavy chain was unchanged (not shown).
Figure 6.
Cx43 immunostaining. Representative confocal images of Cx43 expression in control (A) and binary transgenic hearts with mild (B), moderate (C), or severe (D) disease. Paired images show phase and immunofluorescence. In the moderately and severely diseased hearts, regions at a distance from focal fibrosis are shown. Images were acquired for equivalent times so that signal intensities could be compared directly. Substantial reduction in the intensity of junctional staining is evident in all transgenic hearts. (Original magnification was 60×.)
Figure 7.
Western blot analysis. Companion gels either stained with Coomassie Blue to visualize total proteins loaded (A) or further processed for immunoblotting to visualize Cx43 expression (B). The distribution of total proteins in control (lanes 1–4) and transgenic (lanes 5–8) hearts was similar, but Cx43 abundance in binary transgenic hearts was significantly reduced (n = 4 for each group; P < 0.05). This experiment was repeated in triplicate.
DISCUSSION
Cell loss is observed in many disease states, arising in response to a broad range of stimuli including environmental toxins, metabolic insults such as ischemia, as well as inherited or somatic genetic mutations. Accordingly, a versatile system to specifically ablate cells of any lineage and developmental stage would be of substantial benefit to model human diseases of various etiologies. Moreover, if the cell death signal could be reversibly controlled, not only could disease progression be modeled, but the processes of repair and/or regeneration could be examined as well. Previously, methods to selectively kill cells by genetic means have been used primarily for studies of lineage specification during development (19–23), to some extent reflecting the constraints of available transcriptional regulatory elements, few of which are both cell-type specific and amenable to temporal control. Here, by incorporating a binary approach with tetracycline-responsive promoters, cell death may be both spatially and temporally controlled.
In this report, we have focused our attention on the effects of conditional DTA expression in the heart to gain insight into the relationship between myocardial disease and arrhythmogenesis. Myocyte loss induces an exceedingly complex response in the heart, with both anatomic and functional sequelae. Structural heterogeneity arises by depletion of myocytes, especially when interstitial fibrosis accompanies the pathologic process (24). Moreover, the residual myocyte population is subjected to increased hemodynamic load, leading to ventricular remodeling, compensatory hypertrophy, and marked alterations in myocardial gene expression and function, including those genes regulating cardiac excitability (25–31). Both processes may contribute to the arrhythmogenic substrate and understanding the relative contribution of each is an important goal for rational design of anti-arrhythmic therapies.
We had initially expected that only the most diseased hearts would be arrhythmogenic, and we were surprised to find that the majority of transgenic hearts, including those with only mild cell loss and interstitial fibrosis, supported reentrant ventricular tachycardia. Even more unexpected was the demonstration of perturbed conduction profiles in minimally damaged hearts. These observations suggested that factors other than disruption of gross myocardial architecture must contribute to the arrhythmogenic substrate. Cardiac propagation is considered stochastic at the microscopic level and alterations in gap junction distribution and myocardial microstructure are postulated to contribute to the genesis of cardiac arrhythmias (32, 33). Remodeling of gap junctions has been demonstrated in patients with ischemic heart disease, cardiac hypertrophy, and Chagasic cardiomyopathy (34–38). Despite limited experimental data, remodeling of gap junctions is nonetheless considered an important factor in the high frequency of arrhythmias in these settings (reviewed in ref. 39). Our data lend strong experimental support to the hypothesis that dysregulation of gap junctions and altered impulse conduction contribute to the arrhythmogenic substrate, even in hearts that microscopically appear to have rather modest structural disease.
Interestingly, in heterozygous mice with a targeted mutation of the Cx43 gene (40), despite evidence for slowed epicardial activation (41), no ventricular tachycardia is induced with the protocol described here (G.M., unpublished observations). These observations suggest that heterogeneous remodeling of gap junction channels may be required to create an arrhythmogenic substrate, rather than a uniform reduction of Cx43 content. We previously observed that transactivation of target genes in the heart using the MHCα-tTA mice is somewhat nonhomogeneous on a cell by cell basis (10). Thus, heterogeneity of DTA-induced cell loss and injury may contribute both to the arrhythmogenic milieu and the variability in length of survival of individual mice after tetracycline withdrawal. Whether or not significant alterations in ion channel gene expression and function, as has been observed in a variety of experimental and human cardiomyopathies (28, 31), also contribute to the arrhythmogenic substrate remains to be determined.
Although numerous arrhythmia models in larger mammals have been described (42–46), none offer the obvious advantages of mouse molecular genetics to identify additional factors that influence the susceptibility to arrhythmogenesis. Furthermore, arrhythmias in mice engineered to model human diseases with single gene defects, such as familial hypertrophic cardiomyopathy (47), may differ mechanistically from those arising in the more clinically prevalent acquired forms of heart disease.
The adaptation of existing techniques to study murine physiology has improved substantially in the past few years, driven by the mouse’s suitability for genetic manipulation, (reviewed in ref. 48). For example, methodology for in vivo cardiac electrophysiology testing was recently described that successfully revealed conduction disturbances and inducible ectopy in the alpha MHC Arg-403ΔGln knockin model of familial hypertrophic cardiomyopathy (49, 50). To this battery of studies, we now add optical action potential mapping and arrhythmia induction of the isolated-perfused heart. The mouse is relatively resistant to cardiac arrhythmogenesis and thus induction of even short runs of ventricular tachycardia has been considered significant (50). Accordingly, we defined ventricular tachycardia as greater than five ectopic ventricular activations. Although the DTA and murine familial hypertrophic cardiomyopathies differ substantially in design, the nature of arrhythmias observed in the present model appear significantly more severe and compared with in vivo testing, the protocol we described in this report shows excellent sensitivity and specificity for arrhythmia induction. Thus, it is likely that the two approaches will provide complementary information in terms of electrophysiological perturbations and clinical sequelae.
In summary, conditional lineage ablation may have broad applicability for studies of disease pathogenesis and progression. For example, the role of individual cellular components of complex tissues may be examined by specific deletion of appropriate lineages in the adult context. Moreover, the capacity to repress DTA-dependent cell death by resumption of tetracycline therapy should allow for studies of tissue repair and/or regeneration. Finally, conditional expression of DTA may be of therapeutic value to ablate cells with uncontrolled proliferation or latent infections (51).
Acknowledgments
We thank Rendi Cheng for expert technical assistance. This study was supported in part by grants from the American Heart Association, New York City Affiliate and National Center (to G.I.F.) and the National Institutes of Health (to G.I.F. and J.J.). G.I.F. is an Established Investigator of the American Heart Association.
ABBREVIATIONS
- TCN
tetracycline, tTA, tetracycline-controlled transactivator
- DTA
diphtheria toxin A
- Cx43
connexin43
- MHC
myosin heavy chain
References
- 1.Hetts S W. J Am Med Assoc. 1998;279:300–307. doi: 10.1001/jama.279.4.300. [DOI] [PubMed] [Google Scholar]
- 2.Rinkenberger J L, Korsmeyer S J. Curr Opin Genet Dev. 1997;7:589–596. doi: 10.1016/s0959-437x(97)80004-4. [DOI] [PubMed] [Google Scholar]
- 3.MacLellan W R, Schneider M D. Circ Res. 1997;81:137–144. doi: 10.1161/01.res.81.2.137. [DOI] [PubMed] [Google Scholar]
- 4.Barbosa E R, Limongi J C, Cummings J L. Psychiatr Clin North Am. 1997;20:769–790. doi: 10.1016/s0193-953x(05)70344-0. [DOI] [PubMed] [Google Scholar]
- 5.Lev M. Cardiovasc Clin. 1972;4:159–186. [PubMed] [Google Scholar]
- 6.Ursell P C, Gardner P I, Albala A, Fenoglio J J, Jr, Wit A L. Circ Res. 1985;56:436–451. doi: 10.1161/01.res.56.3.436. [DOI] [PubMed] [Google Scholar]
- 7.Rosen M R. Am J Cardiol. 1988;61:2A–8A. doi: 10.1016/0002-9149(88)90735-7. [DOI] [PubMed] [Google Scholar]
- 8.Maxwell I H, Maxwell F, Glode L M. Cancer Res. 1986;46:4660–4664. [PubMed] [Google Scholar]
- 9.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu Z, Redfern C S, Fishman G I. Circ Res. 1996;79:691–697. doi: 10.1161/01.res.79.4.691. [DOI] [PubMed] [Google Scholar]
- 11.Passman R S, Fishman G I. J Clin Invest. 1994;94:2421–2425. doi: 10.1172/JCI117609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kramer K, Van Acker S A, Grimbergen J A, van den Berg D J, Van der Vijgh W J, Bast A. Gen Pharmacol. 1995;26:1403–1407. doi: 10.1016/0306-3623(94)00300-c. [DOI] [PubMed] [Google Scholar]
- 13.Mansier P, Medigue C, Charlotte N, Vermeiren C, Coraboeuf E, Deroubai E, Ratner E, Chevalier B, Clairambault J, Carre F, et al. Am J Physiol. 1996;271:H1465–H1472. doi: 10.1152/ajpheart.1996.271.4.H1465. [DOI] [PubMed] [Google Scholar]
- 14.Baxter W T, Davidenko J M, Loew L M, Wuskell J P, Jalife J. Ann Biomed Eng. 1997;25:713–725. doi: 10.1007/BF02684848. [DOI] [PubMed] [Google Scholar]
- 15.Gray R A, Jalife J, Panfilov A, Baxter W T, Cabo C, Davidenko J M, Pertsov A M. Circulation. 1995;91:2454–2469. doi: 10.1161/01.cir.91.9.2454. [DOI] [PubMed] [Google Scholar]
- 16.Wikswo J P, Jr, Lin S F, Abbas R A. Biophys J. 1995;69:2195–2210. doi: 10.1016/S0006-3495(95)80115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi S R, Key M E, Kalra K L. J Histochem Cytochem. 1991;39:741–748. doi: 10.1177/39.6.1709656. [DOI] [PubMed] [Google Scholar]
- 18.Fishman G I, Spray D C, Leinwand L A. J Cell Biol. 1990;111:589–598. doi: 10.1083/jcb.111.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmiter R D, Behringer R R, Quaife C J, Maxwell F, Maxwell I H, Brinster R L. Cell. 1987;50:435–443. doi: 10.1016/0092-8674(87)90497-1. [DOI] [PubMed] [Google Scholar]
- 20.Behringer R R, Mathews L S, Palmiter R D, Brinster R L. Genes Dev. 1988;2:453–461. doi: 10.1101/gad.2.4.453. [DOI] [PubMed] [Google Scholar]
- 21.Li Q, Karam S M, Gordon J I. J Biol Chem. 1996;271:3671–3676. [PubMed] [Google Scholar]
- 22.Harrington L, Klintworth G K, Secor T E, Breitman M L. Dev Biol. 1991;148:508–516. doi: 10.1016/0012-1606(91)90269-9. [DOI] [PubMed] [Google Scholar]
- 23.Breitman M L, Rombola H, Maxwell I H, Klintworth G K, Bernstein A. Mol Cell Biol. 1990;10:474–479. doi: 10.1128/mcb.10.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Assayag P, Carre F, Chevalier B, Delcayre C, Mansier P, Swynghedauw B. Cardiovasc Res. 1997;34:439–444. doi: 10.1016/s0008-6363(97)00073-4. [DOI] [PubMed] [Google Scholar]
- 25.Parker T G. Herz. 1993;18:245–255. [PubMed] [Google Scholar]
- 26.van Bilsen M, Chien K R. Cardiovasc Res. 1993;27:1140–1149. doi: 10.1093/cvr/27.7.1140. [DOI] [PubMed] [Google Scholar]
- 27.Bayes-Genis A, Guindo J, Vinolas X, Tomas L, Elosua R, Duran I, Bayes de Luna A. Am J Cardiol. 1995;76:54D–59D. doi: 10.1016/s0002-9149(99)80493-7. [DOI] [PubMed] [Google Scholar]
- 28.Gidh-Jain M, Huang B, Jain P, el-Sherif N. Circ Res. 1996;79:669–675. doi: 10.1161/01.res.79.4.669. [DOI] [PubMed] [Google Scholar]
- 29.McLenachan J M, Dargie H J. Am J Cardiol. 1990;65:42G–44G. doi: 10.1016/0002-9149(90)90958-4. [DOI] [PubMed] [Google Scholar]
- 30.Swynghedauw B, Chevalier B, Charlemagne D, Mansier P, Carre F. Cardiovasc Res. 1997;35:6–12. doi: 10.1016/s0008-6363(97)00076-x. [DOI] [PubMed] [Google Scholar]
- 31.Takimoto K, Li D, Hershman K M, Li P, Jackson E K, Levitan E S. Circ Res. 1997;81:533–539. doi: 10.1161/01.res.81.4.533. [DOI] [PubMed] [Google Scholar]
- 32.Spach M S, Heidlage J F. Circ Res. 1995;76:366–380. doi: 10.1161/01.res.76.3.366. [DOI] [PubMed] [Google Scholar]
- 33.Spach M S, Boineau J P. Pacing Clin Electrophysiol. 1997;20:397–413. doi: 10.1111/j.1540-8159.1997.tb06199.x. [DOI] [PubMed] [Google Scholar]
- 34.Peters N S. Clin Sci (London) 1996;90:447–452. doi: 10.1042/cs0900447. [DOI] [PubMed] [Google Scholar]
- 35.Luke R A, Saffitz J E. J Clin Invest. 1991;87:1594–1602. doi: 10.1172/JCI115173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Severs N J. J Cardiovasc Electrophysiol. 1994;5:462–475. doi: 10.1111/j.1540-8167.1994.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 37.Peters N S, Coromilas J, Severs N J, Wit A L. Circulation. 1997;95:988–996. doi: 10.1161/01.cir.95.4.988. [DOI] [PubMed] [Google Scholar]
- 38.de Carvalho A C, Tanowitz H B, Wittner M, Dermietzel R, Roy C, Hertzberg E L, Spray D C. Circ Res. 1992;70:733–742. doi: 10.1161/01.res.70.4.733. [DOI] [PubMed] [Google Scholar]
- 39.Peters N S, Wit A L. Circulation. 1998;97:1746–1754. doi: 10.1161/01.cir.97.17.1746. [DOI] [PubMed] [Google Scholar]
- 40.Reaume A G, de Sousa P A, Kulkarni S, Langille B L, Zhu D, Davies T C, Juneja S C, Kidder G M, Rossant J. Science. 1995;267:1831–1834. doi: 10.1126/science.7892609. [DOI] [PubMed] [Google Scholar]
- 41.Guerrero P A, Schuessler R B, Davis L M, Beyer E C, Johnson C M, Yamada K A, Saffitz J E. J Clin Invest. 1997;99:1991–1998. doi: 10.1172/JCI119367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo C S, Reddy C P, Munakata K, Surawicz B. Eur Heart J. 1985;6:63–70. doi: 10.1093/eurheartj/6.suppl_d.63. [DOI] [PubMed] [Google Scholar]
- 43.Lazzara R, Scherlag B J. Am J Cardiol. 1988;61:20A–26A. doi: 10.1016/0002-9149(88)90737-0. [DOI] [PubMed] [Google Scholar]
- 44.Allessie M A, Schalij M J, Kirchhof C J, Boersma L, Huybers M, Hollen J. Eur Heart J. 1989;10:2–8. doi: 10.1093/eurheartj/10.suppl_e.2. [DOI] [PubMed] [Google Scholar]
- 45.Weissenburger J, Davy J M, Chezalviel F. Fundam Clin Pharmacol. 1993;7:29–38. doi: 10.1111/j.1472-8206.1993.tb00215.x. [DOI] [PubMed] [Google Scholar]
- 46.Moise N S, Gilmour R F, Jr, Riccio M L. J Cardiovasc Electrophysiol. 1997;8:98–103. doi: 10.1111/j.1540-8167.1997.tb00614.x. [DOI] [PubMed] [Google Scholar]
- 47.Geisterfer-Lowrance A A, Christe M, Conner D A, Ingwall J S, Schoen F J, Seidman C E, Seidman J G. Science. 1996;272:731–734. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- 48.James J F, Hewett T E, Robbins J. Circ Res. 1998;82:407–415. doi: 10.1161/01.res.82.4.407. [DOI] [PubMed] [Google Scholar]
- 49.Berul C I, Aronovitz M J, Wang P J, Mendelsohn M E. Circulation. 1996;94:2641–2648. doi: 10.1161/01.cir.94.10.2641. [DOI] [PubMed] [Google Scholar]
- 50.Berul C I, Christe M E, Aronovitz M J, Seidman C E, Seidman J G, Mendelsohn M E. J Clin Invest. 1997;99:570–576. doi: 10.1172/JCI119197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paulus W, Baur I, Oberer D M, Breakefield X O, Reeves S A. J Neurosurg. 1997;87:89–95. doi: 10.3171/jns.1997.87.1.0089. [DOI] [PubMed] [Google Scholar]