Abstract
Claudins, comprising a multigene family, constitute tight junction (TJ) strands. Clostridium perfringens enterotoxin (CPE), a single ∼35-kD polypeptide, was reported to specifically bind to claudin-3/RVP1 and claudin-4/CPE-R at its COOH-terminal half. We examined the effects of the COOH-terminal half fragment of CPE (C-CPE) on TJs in L transfectants expressing claudin-1 to -4 (C1L to C4L, respectively), and in MDCK I cells expressing claudin-1 and -4. C-CPE bound to claudin-3 and -4 with high affinity, but not to claudin-1 or -2. In the presence of C-CPE, reconstituted TJ strands in C3L cells gradually disintegrated and disappeared from their cell surface. In MDCK I cells incubated with C-CPE, claudin-4 was selectively removed from TJs with its concomitant degradation. At 4 h after incubation with C-CPE, TJ strands were disintegrated, and the number of TJ strands and the complexity of their network were markedly decreased. In good agreement with the time course of these morphological changes, the TJ barrier (TER and paracellular flux) of MDCK I cells was downregulated by C-CPE in a dose-dependent manner. These findings provided evidence for the direct involvement of claudins in the barrier functions of TJs.
Keywords: Clostridium perfringens enterotoxin, tight junction, claudin, epithelial barrier, freeze-fracture
In multicellular organisms, epithelial and endothelial cellular sheets function not only as diffusion barriers to establish compositionally distinct fluid compartments but are also involved in active transport of materials across the barrier to dynamically maintain the internal environment of each compartment. For cellular sheets to exert these physiological functions, there must be some seal to diffusion of solutes through the paracellular pathway. Tight junctions (TJs)1 have been shown to be responsible for this intercellular sealing (for reviews see Schneeberger and Lynch 1992; Gumbiner 1987, Gumbiner 1993; Anderson and van Itallie 1995; Goodenough 1999). When TJs are observed by ultrathin electron microscopy, the plasma membranes of adjoining cells can be seen to converge at multiple sites, where their outer leaflets appear to fuse to obliterate the intercellular space completely (Farquhar and Palade 1963). On freeze-fracture electron microscopy, TJs appear as a network of intramembranous particle strands (TJ strands) on the P-face (the outwardly facing cytoplasmic leaflet) with a complementary pattern of network of grooves on the E-face (the inwardly facing extracytoplasmic leaflet) (Staehelin 1973, Staehelin 1974).
To understand the molecular mechanism of the barrier function of TJs, clarification of the molecular architecture of TJ strands is necessary. Occludin, an ∼65-kD integral membrane protein bearing four transmembrane domains, was identified as the first component of TJ strands (Furuse et al. 1993; Ando-Akatsuka et al. 1996). Immunoreplica electron microscopy revealed that occludin is incorporated into TJ strands in situ (Fujimoto 1995; Furuse et al. 1996). Occludin was shown to be not only a structural but also a functional component of TJ strands; occludin has been shown to be directly involved in barrier functions (McCarthy et al. 1996; Balda et al. 1996; Chen et al. 1997; Wong and Gumbiner 1997). Recently, both alleles of the occludin gene were successfully disrupted in embryonic stem (ES) cells (Saitou et al. 1998). Surprisingly, when occludin-deficient ES cells were differentiated into epithelial cells, well-developed TJs were formed between adjacent cells and their barrier function did not appear to be affected. This finding not only led to reevaluation of the functional aspects of occludin reported previously, but also indicated that there are as yet unidentified TJ integral membrane protein(s) that can form strand structures without occludin.
Recently, two distinct types of integral membrane proteins were reported to be localized at TJs: JAM (Martin-Padura et al. 1998) and claudins (Furuse et al. 1998a). JAM (a junction-associated membrane protein) with a molecular mass of ∼40 kD has a single transmembrane domain and belongs to the immunoglobulin superfamily, but the relationship between JAM and TJ strands remains unclear. In contrast, recent evidence indicated that claudins are directly involved in the formation of TJ strands (Tsukita and Furuse 1999). Initially, two related ∼23-kD integral membrane proteins, claudin-1 and -2, were identified as proteins that were copartitioned with occludin during sonication and sucrose density gradient centrifugation of the junction-enriched fraction (Furuse et al. 1998a). Interestingly, when they were introduced singly into cultured L fibroblasts lacking TJs, they induced the formation of a well-developed network of TJ strands between stable L transfectants (Furuse et al. 1998b). Similarity searches in data bases identified many sequences similar to claudin-1 and -2, pointing to the existence of a new gene family, the claudin family (Morita et al. 1999a). To date, 15 members of this family (claudin-1 to -15) have been identified (Morita et al. 1999a,Morita et al. 1999b; Tsukita and Furuse 1999). These findings suggested that distinct species of claudin were copolymerized to form the backbone of TJ strands in situ.
To date, however, evidence is still lacking for the direct involvement of claudins in the barrier function of TJs. Reconstituted TJs in L transfectants did not surround individual cells continuously (Furuse et al. 1998b), making it difficult to measure their tightness as barriers. Coexpression of multiple species of claudins in epithelial cells complicated the evaluation of functions of claudins in this type of cells. In this study, to clarify the function of claudins, we used the bacterial toxin Clostridium perfringens enterotoxin (CPE). This enterotoxin, which consists of a single polypeptide chain with a molecular mass of ∼35 kD, is the causative agent of symptoms associated with Clostridium perfringens food poisoning in man (McClane et al. 1988). Katahira et al. 1997a cloned a cDNA encoding the receptor for CPE (CPE-R) from an expression library of enterotoxin-sensitive monkey Vero cells. They further found that previously reported RVP1 (rat ventral prostate-1; Briehl and Miesfeld 1991) showed marked similarity to CPE-R, and that RVP1 also functioned as a receptor for CPE (Katahira et al. 1997b), although the physiological functions of RVP1 and CPE-R remained unclear. Interestingly, identification of claudin-1 and -2 prompted us to notice the significant sequence similarity between claudin-1/-2 and RVP1/CPE-R, and to designate RVP-1 and CPE-R as claudin-3 and -4, respectively (Furuse et al. 1998a; Morita et al. 1999a). We speculated that CPE may also bind to other members of the claudin family, and that CPE could be used to modulate the function of claudins from the exterior of cells. There is accumulating evidence that the COOH-terminal half of this toxin (C-CPE) binds to CPE-R, and that its NH2-terminal half increases membrane permeability by forming small pores in the plasma membrane (Matsuda and Sugimoto 1979; McClane and McDonel 1981; Horiguchi et al. 1986, Horiguchi et al. 1987; Hanna et al. 1991, Hanna et al. 1992). Therefore, we used C-CPE as a peptide that specifically binds to at least claudin-3 and -4 to examine the function of claudins. We found that this peptide removed specific claudin species from TJ strands with concomitant downregulation of the TJ barrier function in cultured MDCK cells. This study provided the first evidence of the direct involvement of claudins in the barrier functions of TJs.
Materials and Methods
Antibodies and Cells
Guinea pig anti–mouse claudin-1 pAb was raised against the GST fusion protein with the COOH-terminal cytoplasmic domain of claudin-1. Rabbit anti–mouse claudin-2 pAb, rabbit anti–mouse claudin-3 pAb, and rabbit anti–mouse claudin-4 pAb were raised and characterized, previously (Morita et al. 1999a; Furuse et al. 1999). These pAbs were affinity purified on nitrocellulose membranes with MBP (maltose-binding protein) fusion protein with the cytoplasmic domain of claudin-1, or GST fusion proteins with the cytoplasmic domains of claudin-2 to -4.
L transfectants expressing claudin-1 (C1L), claudin-2 (C2L), and claudin-3 (C3L) were established previously (Furuse et al. 1999), and those expressing claudin-4 (C4L) were obtained in this study. L transfectants were cultured on coverslips in DME supplemented with 10% FCS. MDCK I cells were cultured in Transwell™ chambers (polycarbonate membrane, filter pore size, 0.4 μm; Costar Corp.) in DME supplemented with 10% FCS.
Purification of CPE, Production of C-CPE, Cytotoxicity Assay, and Binding Assay
CPE was purified by the method of Sakaguchi et al. 1973. The COOH-terminal fragment (184-319 amino acids) of CPE with a 10-histidine tag was produced in E. coli and purified as described previously (Katahira et al. 1997a). The cytotoxic effect of CPE on L transfectants and MDCK I cells was determined by examining their morphological alterations 24 h after addition of CPE (500 ng /ml) to the culture medium. The binding of 125I-CPE to L transfectants expressing respective claudins was measured, and Scatchard analysis was performed as described previously (Katahira et al. 1997a).
Treatment of Cultured Cells with C-CPE
For treatment of C3L cells on coverslips, purified C-CPE was added into culture medium at a final concentration of 2.5 μg/ml. When MDCK I cells were cultured on Transwell™ chamber, purified C-CPE was added into the basolateral compartment at a final concentration of 2.5 μg/ml. These C-CPE–treated cells were processed for immunofluorescence microscopy, immunoblotting, and freeze-fracture replica electron microscopy.
SDS-PAGE and Immunoblotting
SDS-PAGE was performed according to the method of Laemmli 1970, and proteins were electrophoretically transferred from gels onto nitrocellulose membranes. The membranes were soaked in 5% skimmed milk and incubated with the primary antibodies. After washing, the membranes were incubated with the second antibodies to rabbit (Amersham-Pharmacia Biotechnology), or guinea pig (Chemicon) IgG as appropriate followed by incubation with streptavidin-conjugated alkaline phosphatase (Amersham-Pharmacia Biotechnology). Nitroblue tetrazolium and bromochloroindolyl phosphate were used as substrates to visualize the enzyme reaction.
Immunofluorescence Microscopy
MDCK I cells cultured on filters and L transfectants on coverslips were fixed with 10% trichloroacetic acid for 30 min on ice (Hayashi et al. 1999) and 1% formaldehyde for 10 min at room temperature, respectively. Cells were then washed with PBS, treated with 0.2% Triton X-100 in PBS for 10 min, washed with PBS several times, and soaked in 1% BSA in PBS. Samples were incubated with primary antibodies for 30 min in a moist chamber. Cy3-conjugated goat anti–rabbit IgG (Amersham-Pharmacia Biotechnology), rhodamine-conjugated goat anti–guinea pig IgG (Chemicon) and Cy2-conjugated goat anti–rabbit IgG (Jackson ImmunoResearch Laboratories Inc.) were used as secondary antibodies. Cells were washed three times with PBS and then mounted in 90% glycerol-PBS containing para-phenylenediamine and 1% n-propylgalate. Specimens were observed using a fluorescence Zeiss Axiophot photomicroscope (Carl Zeiss, Inc.), and the images were recorded with a Semsys™ cooled CCD camera system (Photometrics).
Freeze-Fracture Electron Microscopy
Cells were fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.3) overnight, washed with 0.1 M sodium cacodylate buffer three times, immersed in 30% glycerol in 0.1 M sodium cacodylate buffer for 2 h, and then frozen in liquid nitrogen. Frozen samples were fractured at −100°C and platinum-shadowed unidirectionally at an angle of 45° in Balzers Freeze Etching System (BAF060; BAL-TEC). Samples were immersed in household bleach, and the replicas floating off the samples were picked up on formvar-filmed grids, and examined with a JEOL 1200 EX electron microscope at an acceleration voltage of 100 kV.
Morphometrical analysis was performed on freeze-fracture replica images of TJs, which were printed at a final magnification of 20,000. The mean TJ strand number was determined by taking numerous counts along a line drawn perpendicular to the junctional axis at 200-nm intervals (Stevenson et al. 1988). The complexity of TJ strand networks was defined as the number of branch points per unit length (1 μm) of TJ strands (Wolburg et al. 1994). The free end number of TJ strands was defined here as the number of free ends per unit length (1 μm) of TJ strands.
Measurement of Transepithelial Electric Resistance and Paracellular Tracer Flux Assay
Confluent monolayers of MDCK I cells grown in Transwell™ chamber were used. Transepithelial electric resistance (TER) was measured using a Millicell-ERS epithelial volt-ohmmeter (Millipore Corp.) and normalized by the area of the monolayer. The background TER of blank Transwell™ filters was subtracted from the TER of cell monolayers.
For paracellular tracer flux assay, FITC-dextran with a molecular mass of 4, 10, or 40 kD (Sigma Chemical Co.) was dissolved in P buffer (10 mM Hepes, pH 7.4, 1 mM sodium pyruvate, 10 mM glucose, 3 mM CaCl2, and 145 mM NaCl) at the concentration of 1 mg/ml. MDCK I cells on filters were pretreated with 2.5 μg/ml C-CPE from the basolateral compartment for 24 h. The media in apical and basal compartments were then replaced with 200 μl of P buffer containing one type of FITC-dextran and 600 μl of P buffer, respectively. To evaluate the permeability of monolayers, basal compartment media were collected after 3-h incubation with FITC-dextran, and the amount of FITC-dextran in the media was measured with a fluorometer (excitation, 485 nm; emission, 535 nm).
Statistical Analysis
All values are given as mean ± SD. Statistical analysis was performed using the nonparametric Mann-Whitney test.
Results
Claudins Expressed in MDCK I Cells
In this study, to examine whether claudins are involved in the barrier function of TJs, as described below, we analyzed the effects of the COOH-terminal fragment of CPE on cultured MDCK I cells, which showed very high transepithelial electric resistance (TER) (Stevenson et al. 1988). First, we examined what types of claudins are expressed in this cell line. Preliminary immunofluorescence microscopic observations using available antibodies revealed that at least claudin-1 and -4 were expressed in large amounts. Since CPE was reported to bind to claudin-3/RVP1 and claudin-4/CPE-R, we focused on claudin-1 to -4 in MDCK I cells in this study.
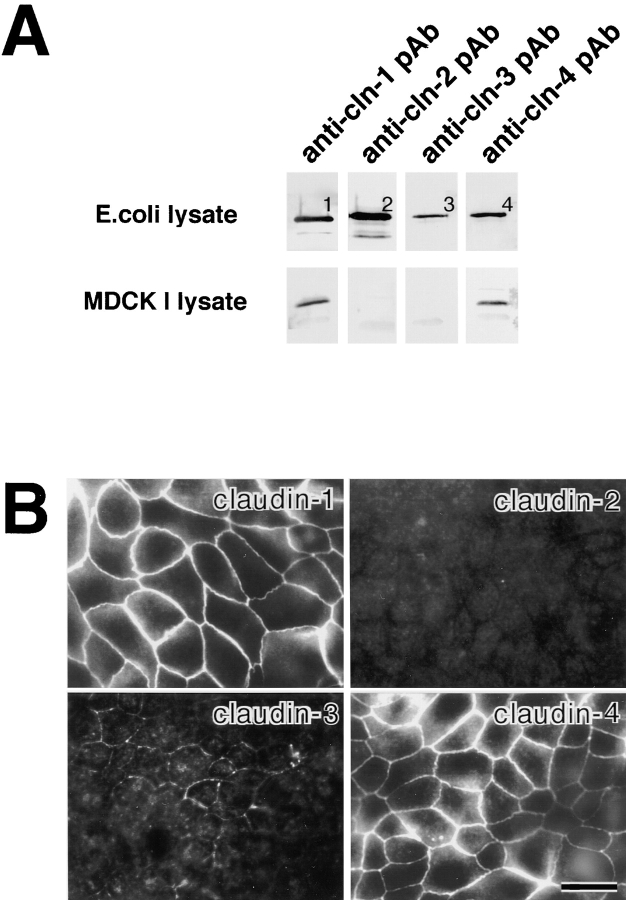
As shown in Fig. 1 A, in the total cell lysate of MDCK I cells, claudin-1 and -4 were reproducibly detected by immunoblotting as bands with expected molecular masses. When confluent cultures of MDCK I cells were immunofluorescently stained with antibodies for claudin-1 to -4, intense signals for claudin-1 and -4 and very weak signals for claudin-3 were detected at cell–cell borders, but claudin-2 was undetectable (Fig. 1 B). Confocal microscopy confirmed that claudin-1 and -4 were concentrated at the most apical part of lateral membranes of MDCK I cells (data not shown).
Figure 1.
Claudins expressed in MDCK I cells. (A) Immunoblotting. The total lysate of E. coli (E. coli lysate) expressing MBP fusion protein with the cytoplasmic domains of claudin-1 (1) and GST fusion proteins with the cytoplasmic domains of claudin-2 (2), -3 (3), and -4 (4), and the total lysate of MDCK I cells (MDCK I lysate) were separated by SDS-PAGE, followed by immunoblotting with anti–claudin-1 pAb (anti-cln-1 pAb), anti–claudin-2 pAb (anti-cln-2 pAb), anti–claudin-3 pAb (anti-cln-3 pAb), and anti–claudin-4 pAb (anti-cln-4 pAb). Only claudin-1 and -4 were detected in MDCK I cells. (B) Immunofluorescence microscopy. Confluent cultures of MDCK I cells were stained with anti–claudin-1 pAb (claudin-1), anti–claudin-2 pAb (claudin-2), anti–claudin-3 pAb (claudin-3), or anti–claudin-4 pAb (claudin-4). Both claudin-1 and -4 were concentrated at cell–cell borders in large amounts, where only a trace amount of claudin-3 was also detected. Claudin-2 signal was undetectable. Bar, 20 μm.
Binding Affinity of Clostridium perfringens Enterotoxin to Claudin-1 to -4
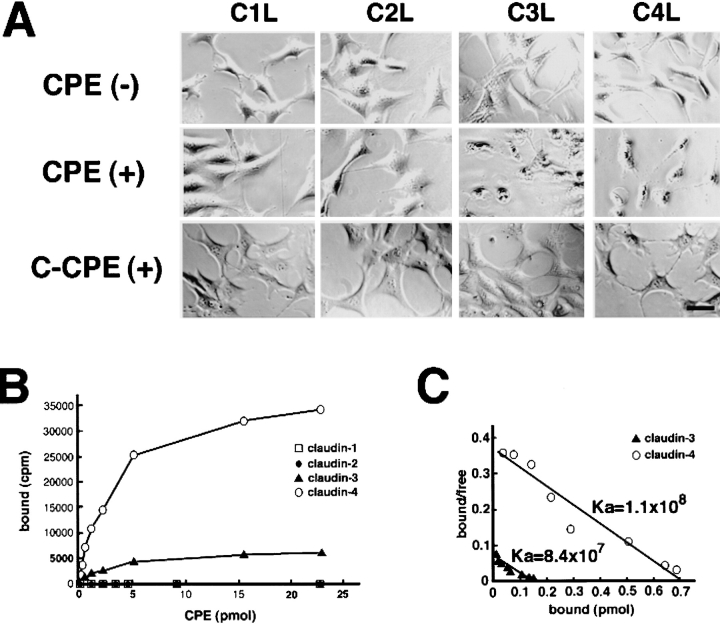
We next examined the sensitivity of claudin-1 to -4 to full-length CPE using L transfectants expressing respective claudins (C1L, C2L, C3L, and C4L cells). When purified CPE at 500 ng/ml was added into the culture medium followed by 1-h incubation, C3L and C4L cells formed characteristic bleb balloons and underwent cell death as expected from the results of previous studies (Katahira et al. 1997a,Katahira et al. 1997b), whereas either C1L or C2L cells did not exhibit any morphological changes up to 24-h incubation (Fig. 2 A), suggesting differences in the affinity to CPE between claudin-1/2 and claudin-3/4.
Figure 2.
Specific interaction of Clostridium perfringens enterotoxin (CPE) with claudin-3 and -4. (A) L transfectants expressing claudin-1 (C1L), claudin-2 (C2L), claudin-3 (C3L), or claudin-4 (C4L) were cultured in the absence (CPE(−)) or presence of 500 ng/ml full-length CPE (CPE(+)) or 2.5 μg/ml COOH-terminal half fragment of CPE (C-CPE(+)). Phase-contrast microscopic images taken at 1 h after incubation revealed that CPE, but not C-CPE, showed cytotoxicity only to C3L and C4L cells. Bar, 20 μm. (B) L transfectants (1 × 105 cells) expressing claudin-1, -2, -3, or -4 were incubated with various concentration of 125I-CPE, and binding was determined as reported previously (Katahira et al. 1997a). The binding of 125I-CPE to claudin-1 or -2 was negligible. Each symbol represents mean value (n = 6). (C) Scatchard plot of the specific binding of 125I-CPE to L transfectants expressing claudin-3 (closed triangles; Ka = 8.4 × 107 M−1) or claudin-4 (open circles; Ka = 1.1 × 108 M−1).
We then compared the binding kinetics of CPE to claudin-1/2 with those of claudin-3/4 by incubating L transfectants with various concentrations of 125I-labeled CPE (Katahira et al. 1997a,Katahira et al. 1997b) (Fig. 2 B). Scatchard plot analyses indicated the binding of 125I-CPE to claudin-1 and -2 to be negligible, whereas claudin-3 and -4 gave Ka values of 8.4 × 107 M−1 and 1.1 × 108 M−1 for 125I-CPE binding, respectively (Fig. 2 C). Therefore, these findings suggested that MDCK I cells mainly expressed CPE-insensitive claudin-1 and CPE-sensitive claudin-4. In good agreement, MDCK I cells exhibited typical bleb formation in the presence of 500 ng/ml CPE (data not shown).
Effects of COOH-terminal Half Fragment of Clostridium perfringens Enterotoxin on the Subcellular Distribution of Claudin-1 to -4
In good agreement with previous observations (Horiguchi et al. 1986, Horiguchi et al. 1987; Hanna et al. 1991, Hanna et al. 1992), the COOH-terminal fragment (184-319 amino acids) of CPE (C-CPE) did not show any cytotoxicity to C3L or C4L cells (Fig. 2 A). The non-cytotoxicity of C-CPE allowed us to follow the subsequent behavior of claudin-3 and -4 after their specific binding to C-CPE within cells. Thus, in C1L, C2L, and C3L cells, we examined the effects of C-CPE on the subcellular distribution of claudin-1 to -3, respectively, by immunofluorescence microscopy, since for as yet unknown reasons in C4L cells exogenous claudin-4 was not concentrated efficiently into cell–cell borders. Similarly to claudin-1 and -2 (Furuse et al. 1998b), claudin-3 was also concentrated at cell–cell contact planes to reconstitute TJ strand networks when introduced into L fibroblasts. When these L transfectants were incubated with 2.5 μg/ml C-CPE for 4 h, claudin-3, but not claudin-1 and -2, which were concentrated at cell–cell contact planes, showed punctate distribution and gradually disappeared (Fig. 3, a and b). We then observed the cell–cell contact planes of C3L cells before and after 4-h incubation with C-CPE by conventional freeze-fracture replica electron microscopy. As shown in Fig. 3 d, in the presence of C-CPE, the well-developed network of claudin-3–based TJ strands disintegrated into thick belt-like aggregates of intramembranous particles of various lengths, and then disappeared.
Figure 3.
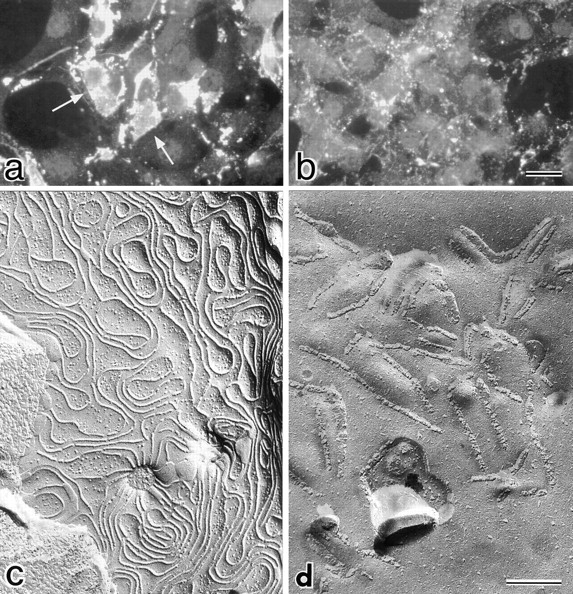
Effects of C-CPE on the subcellular distribution of claudin-3 and the morphology of reconstituted TJ strands in L transfectants expressing claudin-3 (C3L cells). (a and b) Immunofluorescence microscopy. When C3L cells were stained with anti–claudin-3 pAb (a), claudin-3 was shown to be concentrated at cell–cell borders as planes (arrows). At 4 h after incubation with 2.5 μg/ml C-CPE, these claudin-3-positive planes were fragmented into punctate structures and gradually disappeared (b). (c and d) Freeze-fracture replica electron microscopy. In the absence of C-CPE, a well-developed network of TJ strands was reconstituted between adjacent C3L cells (c). When cells were incubated with C-CPE for 4 h, TJ strands were disintegrated into fragmented belt-like aggregates of intramembranous particles (d). Bars: (a and b) 20 μm; (c and d) 200 nm.
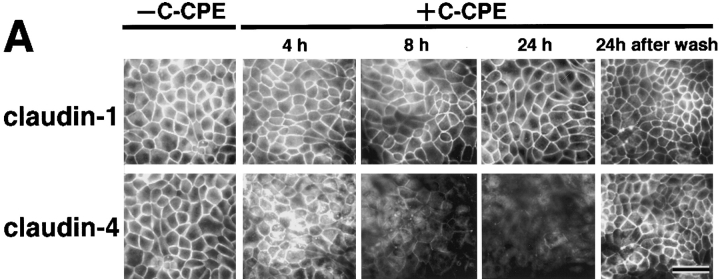
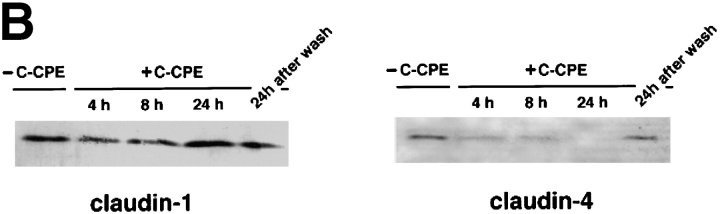
Next, in MDCK I cells plated at confluent density on filters, we examined the effects of C-CPE on the expression and distribution of claudin-1 and -4 (Fig. 4). Double-staining immunofluorescence microscopy with anti–claudin-1 and anti–claudin-4 pAbs revealed that C-CPE did not affect the subcellular distribution of claudin-1 even after 24-h incubation, but that within 4-h incubation claudin-4 began to be distributed in the cytoplasm with concomitant gradual disappearance from the junctional complex areas (Fig. 4 A). At 24 h after incubation, the claudin-4 signal became undetectable, but when C-CPE was washed out at this time point, the concentration of claudin-4 at the junctional region was completely recovered within 24 h. In good agreement, immunoblotting revealed that in the presence of C-CPE, claudin-4, but not claudin-1, decreased in amount with a similar time course to the disappearance of the immunofluorescence signal of claudin-4 (Fig. 4 B). These findings favored the notion that also in MDCK I cells claudin-1 and -4 were insensitive and sensitive for CPE, respectively. Furthermore, these findings were observed only when C-CPE was added to the basolateral compartment of MDCK I cells. C-CPE added to the apical compartment did not affect the subcellular distribution or the expression level of claudin-4 (data not shown).
Figure 4.
Selective loss of claudin-4 from TJs in MDCK I cells. (A) Immunofluorescence microscopy. At 0 (−C-CPE), 4, 8, or 24 h after incubation with 2.5 μg/ml C-CPE in the basolateral compartment or at 24 h after C-CPE was removed, MDCK I cells plated at confluent density on filters were double stained with anti–claudin-1 pAb (claudin-1) and anti–claudin-4 pAb (claudin-4). C-CPE did not affect the subcellular distribution of claudin-1. At 4 h after incubation with C-CPE, claudin-4 was distributed in the cytoplasm with concomitant gradual disappearance from cell–cell borders. At 24 h after incubation, the claudin-4 signal was undetectable, but when C-CPE was washed out at this time point, the concentration of claudin-4 at the junctional region was recovered completely. Bar, 40 μm. (B) Immunoblotting. Total cell lysates of MDCK I cells were separated by SDS-PAGE, followed by immunoblotting with anti–claudin-1 pAb (claudin-1) or anti–claudin-4 pAb (claudin-4). In the presence of C-CPE, claudin-4, but not claudin-1, levels decreased with a similar time course to the disappearance of the immunofluorescence signal of claudin-4 (see A).
Effects of COOH-terminal Half Fragment of Clostridium perfringens Enterotoxin on the Structure and Functions of Tight Junctions in MDCK I Cells
The question naturally arose as to what types of morphological changes of TJ strands are associated with the C-CPE–induced disappearance of claudin-4 in MDCK I cells. MDCK I cells were incubated with C-CPE in their basolateral compartment for 4, 8, and 24 h, fixed with glutaraldehyde, and then examined by conventional freeze-fracture replica electron microscopy. TJs in nontreated MDCK I cells were characterized by well-developed anastomosing networks of TJ strands (Fig. 5 a). The mean strand number, which was determined by taking numerous counts along a line drawn perpendicular to the junctional axis (Stevenson et al. 1988), was 4.0 ± 1.3, and the complexity of TJs, defined as the number of branch points per unit length (1 μm) of TJ strands (Wolburg et al. 1994), was 11.4 (Table ). At 4 h after incubation with C-CPE, TJ strands facing toward the basolateral membrane domains began to disintegrate to thick belt-like aggregates of intramembranous particles (Fig. 5 b). Around 8 h after incubation, these belt-like particle aggregates mostly disappeared, leaving a fairly simple TJ strand network (Fig. 5 c). The mean strand number and the complexity of TJs at this time point were 2.6 ± 0.9 and 5.9, respectively (Table ). The number of free ends per unit length (1 μm) of TJ strands also increased with incubation period (Table ). This type of simple network of TJs was maintained until 24 h after addition of C-CPE (Table ).
Figure 5.
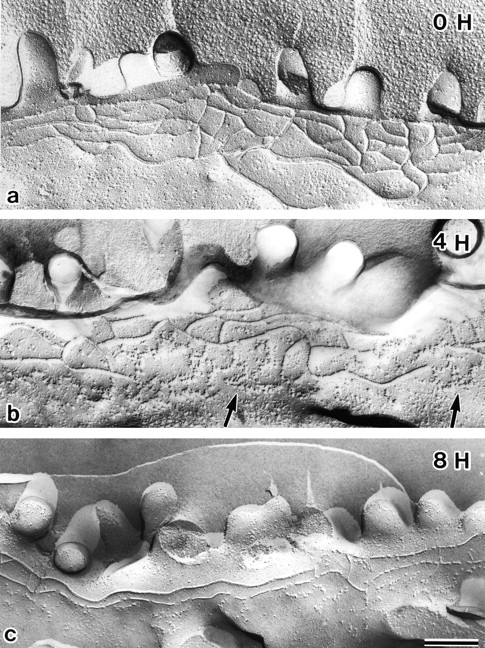
Freeze-fracture replica images of TJs in MDCK I cells incubated with C-CPE. At 0, 4, or 8 h after incubation with 2.5 μg/ml C-CPE in the basolateral compartment, MDCK I cells plated at confluent density on filters were fixed with glutaraldehyde, and processed for freeze-fracture replica electron microscopy. In the absence of C-CPE (0 H), MDCK I cells were characterized by well-developed anastomosing networks of TJ strands. At 4 h after incubation with C-CPE (4 H), TJ strands facing toward the basolateral membrane domains began to disintegrate to thick belt-like aggregates of intramembranous particles (arrows). Around 8 h after incubation (8 H), these belt-like particle aggregates had mostly disappeared, leaving a fairly simple TJ strand network. Bar, 20 μm.
Table 1.
Freeze-Fracture Quantification of C-CPE–induced Morphological Changes of TJ Network
| C-CPE incubation | 0 h | 4 h | 8 h | 24 h |
|---|---|---|---|---|
| Mean strand number | 4.0 ± 1.3 (258) | 2.8 ± 1.1 (228) | 2.6 ± 0.9 (205) | 2.2 ± 0.7 (166) |
| Complexity | 11.4 (49.8 μm) | 5.9 (43.6 μm) | 5.9 (39.0 μm) | 5.1 (30.2 μm) |
| Free end number | 0.2 (49.8 μm) | 1.7 (43.6 μm) | 1.5 (39.0 μm) | 1.9 (30.2 μm) |
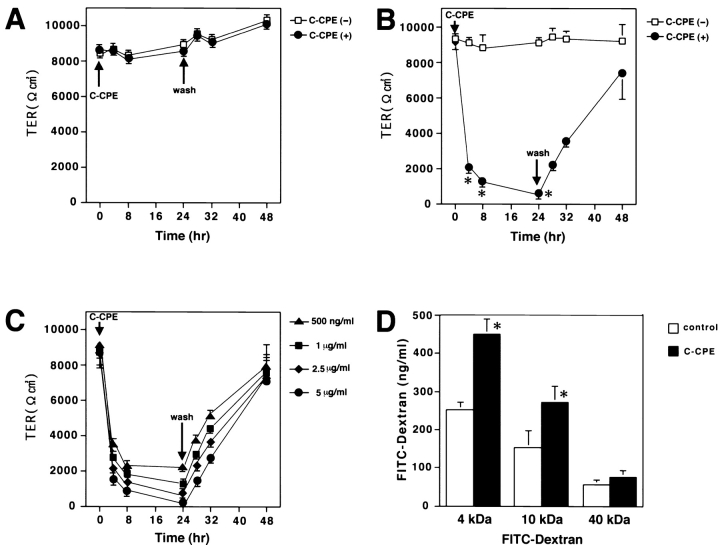
Next, we examined the effects of C-CPE on TER of MDCK I cells plated at confluent density on filters. When 2.5 μg/ml C-CPE was added in the apical compartment, the TER was not affected but remained at a level of 8,000–10,000 Ωcm2 (Fig. 6 A). In contrast, addition of C-CPE to the basolateral compartment resulted in an ∼4.5-fold reduction in TER from ∼9,000 Ωcm2 to ∼2,000 Ωcm2 within 4 h, and removal of C-CPE from the compartment induced gradual recovery of TER to the level of nontreated controls within one day (Fig. 6 B). Furthermore, this C-CPE–induced reduction of TER was dose dependent (Fig. 6 C). Finally, to exclude the possibility that this reduction of TER was caused by an increase in transcellular plasma membrane permeability to ions, we assessed the flux of membrane-impermeable paracellular tracers (FITC-dextran 4K, 10K, and 40K) across MDCK I cell monolayers. As shown in Fig. 6 D, C-CPE in the basolateral compartment caused an approximately twofold increase in the flux of FITC-dextran 4K and 10K, but not 40K. These findings indicated that C-CPE in the basolateral compartment downregulated the TJ barrier itself of MDCK I cells significantly with a similar time course to the disappearance of claudin-4 (see Fig. 4 A).
Figure 6.
Effects of C-CPE on the TJ barrier function of MDCK I cells. (A–C) TER measurements. MDCK I cells were plated at confluent density on 12-mm filters. When 2.5 μg/ml C-CPE was added to the apical compartment at t = 0 (C-CPE), the TER was not affected but remained at the level of 8,000–10,000 Ωcm2 (A; n = 10 for each condition). Addition of C-CPE (2.5 μg/ml) in the basolateral compartment (at t = 0; C-CPE) resulted in an ∼4.5-fold reduction in TER from ∼9,000 Ωcm2 to ∼2,000 Ωcm2 within 4 h, and removal of C-CPE from the compartment (at t = 24 h; wash) induced gradual recovery of TER to the level of nontreated cells within one day (B; n = 10 for each condition). In B, asterisks denote significant difference from nonincubated cells at the corresponding time point (P < 0.01). The C-CPE–induced reduction of TER was dose-dependent (C; n = 4 for each condition). (D) Paracellular tracer flux assay (n = 4 for each condition). C-CPE (2.5 μg/ml) in the basolateral compartment caused an approximately twofold increase in the flux of FITC-dextran 4K and 10K (*P < 0.01), but not 40K. All error bars represent standard deviations.
Discussion
In previous studies, we demonstrated that claudin-1, -2, and -11 reconstituted strands/grooves within plasma membranes, which were morphologically indistinguishable from in situ TJ strands/grooves when introduced into L fibroblasts (Furuse et al. 1998b; Morita et al. 1999b). Furthermore, we recently confirmed that other claudin species also have the ability to reconstitute TJ stands in L fibroblasts (data not shown). In contrast, occludin, another TJ-specific integral membrane protein, did not reconstitute a well-developed network of TJ strands in L fibroblasts, although a small number of fragmented TJ strand-like structures were induced. Interestingly, when occludin was cotransfected into L fibroblasts together with claudin-1, occludin was incorporated into well-developed claudin-1–based strands (Furuse et al. 1998b). Therefore, at least from the structural point of view, we concluded that claudins, but not occludin, are major integral membrane proteins constituting the backbone of TJ strands (Tsukita and Furuse 1999).
To date, however, there have been no reports concerning the functional aspects of claudins. In this study, to examine the functions of claudins we used Clostridium perfringens enterotoxin (CPE), which has been reported to bind specifically to claudin-3/RVP1 and claudin-4/CPE-R (see introduction). The COOH-terminal half fragment of CPE (C-CPE), which did not show any cytotoxicity, did not bind to claudin-1 or -2, and caused disintegration reconstituted claudin-3–based TJ strands in L fibroblasts, but not those based on claudin-1 or claudin-2. Interestingly, TJ strands of MDCK I cells, which were composed of at least claudin-1 and -4, were also partly disintegrated in the presence of C-CPE with concomitant disappearance of claudin-4 from TJs. This C-CPE–induced disintegration of TJ strands and disappearance of claudin-4 were associated with a rapid increase in epithelial permeability, and removal of C-CPE from the medium resulted in reconcentration of claudin-4 at TJs as well as recovery of the epithelial barrier. Taking the specificity and high affinity of the binding between C-CPE and claudin-3/4 into consideration, we concluded that claudins are not only structural but also functional components of TJ strands, which are involved in the TJ barrier.
Occludin has been also shown to be involved in the barrier functions of TJs (McCarthy et al. 1996; Balda et al. 1996; Chen et al. 1997; Wong and Gumbiner 1997). When occludin lacking the COOH-terminal cytoplasmic domain was introduced into MDCK cells, endogenous occludin as well as introduced occludin mutant were aggregated in a punctate manner at cell–cell borders, but the morphology and continuity of TJ strands were not affected (Balda et al. 1996). Since claudins are now thought to be the major structural components of TJ strands, in these MDCK transfectants it is likely that the introduced occludin mutant removed endogenous occludin from the claudin-based TJ strands to aggregate in a punctate manner. Interestingly, in these transfectants, TER was not significantly affected, although the permeability for dextran was increased, suggesting that occludin itself is not directly involved in development and maintenance of TER. In contrast, as shown in this study, when claudin-4 was selectively removed from TJ strands in MDCK cells, their TER dropped rapidly. These findings indicated that claudins, but not occludin, play a central role in TJ barrier function. In good agreement, occludin-deficient visceral endoderm appeared to bear functional TJs (Saitou et al. 1998), and the very tight TJs in Sertoli cells in human testis lacked occludin (Moroi et al. 1998).
The molecular mechanism behind the C-CPE–induced disintegration of TJ strands in C3L and MDCK cells remains unclear. There are two alternative possible mechanisms. First, the direct binding of C-CPE to claudin-3 or -4 within TJ strands may induce depolymerization of the strands themselves, which may be homopolymers of claudin-3 in C3L cells and heteropolymers of at least claudin-1 and -4 in MDCK I cells (Tsukita and Furuse 1999). In L transfectants expressing exogenous claudins, strands were not observed on the free surface of the plasma membranes as a single strand, but instead were restricted to cell–cell adhesion sites to form strands paired laterally with those in apposing membranes (Furuse et al. 1998b). Therefore, if C-CPE causes dissociation of these paired strands into single strands by binding to the extracellular loops of claudin-3 or -4, it would result in the further depolymerization of single strands. The characteristic morphological changes of TJ strands of C3L and MDCK cells during the course of C-CPE–induced disintegration (see Fig. 3 d and 5 b) favored this depolymerization mechanism.
Alternatively, C-CPE may bind to claudin-3 and -4 molecules on nonjunctional areas, which constitute the nonpolymerized pool, and this binding may suppress polymerization of these claudins into strands. If the network of TJ strands is dynamically maintained by the equilibrium between polymerization and depolymerization, this sequestering mechanism could explain the C-CPE–induced destruction of TJ strands. Interestingly, C-CPE downregulated the TJ barrier only when it was applied in the basolateral compartments of MDCK cells (see Fig. 6 A). The sequestering mechanism, but not the depolymerization mechanism, would readily explain this finding if claudins target the basolateral membranes as monomers or small oligomers and are then integrated into TJ strands similarly to occludin (Sakakibara et al. 1997; Matter and Balda 1998). These two depolymerization and sequestering mechanisms must be evaluated in future studies, but in either case C-CPE binding appeared to facilitate the degradation of claudin-4 (and probably also claudin-3) within cells as shown by immunoblotting in Fig. 4 B. In good agreement, the diffuse cytoplasmic distribution of claudin-4 in MDCK cells was observed within 4 h after addition of C-CPE to the culture medium (see Fig. 4 A). It should be clarified how these C-CPE–bound claudins were internalized and degraded in future studies.
The downregulation of TJs induced by binding of C-CPE to specific claudins does not appear to be the principal molecular mechanism responsible for the cytotoxicity of CPE itself. It is widely accepted that the NH2-terminal half of CPE increases membrane permeability by forming small pores in the plasma membrane (Matsuda and Sugimoto 1979; McClane and McDonel 1981; Horiguchi et al. 1986, Horiguchi et al. 1987; Hanna et al. 1991, Hanna et al. 1992). Therefore, binding of the COOH-terminal half of CPE to specific claudins may facilitate pore formation by its NH2-terminal half. Furthermore, the reason remained unclear why CPE, a luminal enterotoxin, binds only to the basolateral membranes of epithelial cells, but not to their apical membranes. It also remained elusive what types of claudins expressed in intestinal epithelial cells are responsible for the CPE-binding in situ. As shown in this study, however, C-CPE can be used as a powerful tool to modulate TJ barrier function. C-CPE bound with high affinity to claudin-3 and -4 but not to claudin-1 or -2, suggesting that claudins are subclassified into CPE-sensitive and nonsensitive types. Therefore, as CPE removes the CPE-sensitive claudins selectively from TJ strands, the barrier function of TJ strands containing larger amounts of CPE-sensitive claudins may be affected more strongly. Considering that the binding region for claudin-4/CPE-R can be narrowed down to the 30–amino acid sequence in the COOH-terminal half of CPE (Hanna et al. 1991), it may be worthwhile searching for ∼30–amino acid oligopeptides that specifically bind to individual claudin species. Selective removal of specific claudin species from TJ strands with the combination of these oligopeptides would provide a new way to modulate the TJ barrier function in situ and to improve bioavailability of drugs to targeted organs.
Acknowledgments
We thank all the members of our laboratory (Department of Cell Biology, Faculty of Medicine, Kyoto University) for helpful discussions. Our thanks are also due to Ms. K. Furuse for her excellent technical assistance in freeze-fracture replica electron microscopy. N. Sonoda expresses his gratitude to Reiko Sonoda for her continuous encouragement.
This study was supported in part by a Grant-in-Aid for Cancer Research and a Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Science and Culture of Japan to S. Tsukita.
Note Added in Proof. Positional cloning has identified a new member of the claudin family (paracellin-1), mutations in which cause hereditary renal hypomagnesemia in human (Simon, D.B., Y. Lu, K.A. Choate, H. Velazquez, E. Al-Sabban, M. Praga, G. Casari, A. Bettinelli, G. Colussi, J. Rodriguez-Soriano, D. McCredie, D. Milford, S. Sanjad, R.P. Lifton. 1999. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 285:103–106). This new claudin species is claudin-16.
Footnotes
1.used in this paper: C-CPE, COOH-terminal half fragment of Clostridium perfringens enterotoxin; CPE, Clostridium perfringens enterotoxin; CPE-R, Clostridium perfringens enterotoxin receptor; pAb, polyclonal antibody; TER, transepithelial electrical resistance; TJ, tight junction
References
- Anderson J.M., Van Itallie C.M. Tight junctions and the molecular basis for regulation of paracellular permeability. Am. J. Physiol. 1995;269:G467–G475. doi: 10.1152/ajpgi.1995.269.4.G467. [DOI] [PubMed] [Google Scholar]
- Ando-Akatsuka Y., Saitou M., Hirase T., Kishi M., Sakakibara A., Itoh M., Yonemura S., Furuse M., Tsukita Sh. Interspecies diversity of the occludin sequencecDNA cloning of human, mouse, dog, and rat-kangaroo homologues. J. Cell Biol. 1996;133:43–47. doi: 10.1083/jcb.133.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda M.S., Whitney J.A., Flores C., González S., Cereijido M., Matter K. Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical-basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J. Cell Biol. 1996;134:1031–1049. doi: 10.1083/jcb.134.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briehl M.M., Miesfeld R.L. Isolation and characterization of transcripts induced by androgen withdrawal and apoptotic cell death in the rat ventral prostate. Mol. Endocrinol. 1991;5:1381–1388. doi: 10.1210/mend-5-10-1381. [DOI] [PubMed] [Google Scholar]
- Chen Y.-H., Merzdorf C., Paul D.L., Goodenough D.A. COOH terminus of occludin is required for tight junction barrier function in early Xenopus embryos. J. Cell Biol. 1997;138:891–899. doi: 10.1083/jcb.138.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar M.G., Palade G.E. Junctional complexes in various epithelia. J. Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto K. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J. Cell Sci. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- Furuse M., Fujimoto K., Sato N., Hirase T., Tsukita Sa., Tsukita Sh. Overexpression of occludin, a tight junction-associated integral membrane protein, induces the formation of intracellular multilamellar bodies bearing tight junction-like structures. J. Cell Sci. 1996;109:429–435. doi: 10.1242/jcs.109.2.429. [DOI] [PubMed] [Google Scholar]
- Furuse M., Fujita K., Hiiragi T., Fujimoto K., Tsukita Sh. Claudin-1 and -2Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin J. Cell Biol. 141 1998. 1539 1550a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Hirase T., Itoh M., Nagafuchi A., Yonemura S., Tsukita Sa., Tsukita Sh. Occludina novel integral membrane protein localizing at tight junctions. J. Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Sasaki H., Fujimoto K., Tsukita Sh. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts J. Cell Biol. 143 1998. 391 401b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Sasaki H., Tsukita Sh. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J. Cell Biol. 1999;20:429–435. doi: 10.1083/jcb.147.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough D.A. Plugging the leaks. Proc. Natl. Acad. Sci. USA. 1999;96:319–321. doi: 10.1073/pnas.96.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B. Structure, biochemistry, and assembly of epithelial tight junctions. Am. J. Physiol. 1987;253:C749–C758. doi: 10.1152/ajpcell.1987.253.6.C749. [DOI] [PubMed] [Google Scholar]
- Gumbiner B. Breaking through the tight junction barrier. J. Cell Biol. 1993;123:1631–1633. doi: 10.1083/jcb.123.6.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna P.C., Mietzner T.A., Schoolnik G.K., McClane B.A. Localization of the receptor-binding region of Clostridium perfringens enterotoxin utilizing cloned toxin fragments and synthetic peptides. J. Biol. Chem. 1991;17:11037–11043. [PubMed] [Google Scholar]
- Hanna P.C., Wieckowski E.H., Mietzner T.A., McClane B.A. Mapping of functional regions of Clostridium perfringens type A enterotoxin. Infect. Immun. 1992;60:2110–2114. doi: 10.1128/iai.60.5.2110-2114.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Yonemura S., Matsui T., Tsukita Sa., Tsukita Sh. Immunofluorescence detection of ezrin/radixin/moesin (ERM) proteins with their carboxyl-terminal threonine phosphorylated in cultured cells and tissuesApplication of a novel fixation protocol using trichloroacetic acids (TCA) as a fixative. J. Cell Sci. 1999;112:1149–1158. doi: 10.1242/jcs.112.8.1149. [DOI] [PubMed] [Google Scholar]
- Horiguchi Y., Akai T., Sakaguchi G. Isolation and function of a Clostridium perfringens enterotoxin fragment. Infect. Immun. 1987;55:2912–2915. doi: 10.1128/iai.55.12.2912-2915.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi Y., Uemura T., Kamata Y., Kozaki S., Sakaguchi G. Production and characterization of monoclonal antibodies to Clostridium perfringens enterotoxin. Infect. Immun. 1986;52:31–35. doi: 10.1128/iai.52.1.31-35.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Inoue N., Horiguchi Y., Matsuda M., Sugimoto N. Molecular cloning and functional characterization of the receptor for Clostridium perfringens enterotoxin J. Cell Biol. 136 1997. 1239 1247a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Sugiyama H., Inoue N., Horiguchi Y., Matsuda M., Sugimoto N. Clostridium perfringens enterotoxin utilizes two structurally related membrane proteins as functional receptors in vivo J. Biol. Chem. 272 1997. 26652 26658b [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Martin-Padura I., Lostaglio S., Schneemann M., Williams L., Romano M., Fruscella P., Panzeri C., Stoppacciaro A., Ruco L., Villa A. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 1998;142:117–127. doi: 10.1083/jcb.142.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M., Sugimoto N. Calcium-independent and dependent steps in action of Clostridium perfringens enterotoxin on hela and vero cells. Biochem. Biophys. Res. Commun. 1979;91:629–636. doi: 10.1016/0006-291x(79)91568-7. [DOI] [PubMed] [Google Scholar]
- Matter K., Balda M.S. Biogenesis of tight junctionsthe C-terminal domain of occludin mediates basolateral targeting. J. Cell Sci. 1998;111:511–519. doi: 10.1242/jcs.111.4.511. [DOI] [PubMed] [Google Scholar]
- McCarthy K.M., Skare I.B., Stankewich M.C., Furuse M., Tsukita Sh., Rogers R.A., Lynch R.D., Schneeberger E.E. Occludin is a functional component of the tight junction. J. Cell Sci. 1996;109:2287–2298. doi: 10.1242/jcs.109.9.2287. [DOI] [PubMed] [Google Scholar]
- McClane B.A., McDonel J.L. Protective effects of osmotic stabilizers on morphological and permeability alterations induced in Vero cells by Clostridium perfringens enterotoxin. Biochim. Biophys. Acta. 1981;641:401–409. doi: 10.1016/0005-2736(81)90496-x. [DOI] [PubMed] [Google Scholar]
- McClane B.A., Wnek A.P., Hulkower K.I., Hanna P.C. Divalent cation involvement in the action of Clostridium perfringens type A enterotoxin. J. Biol. Chem. 1988;263:2423–2435. [PubMed] [Google Scholar]
- Morita K., Furuse M., Fujimoto K., Tsukita Sh. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands Proc. Natl. Acad. Sci. USA. 96 1999. 511 516a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Sasaki H., Fujimoto K., Furuse M., Tsukita Sh. Claudin-11/OSP–based tight junctions in myelinated sheaths of oligodendrocytes and Sertoli cells in testis J. Cell Biol. 145 1999. 579 588b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroi S., Saitou M., Fujimoto K., Sakakibara A., Furuse M., Yoshida O., Tsukita Sh. Occludin is concentrated at tight junctions of mouse/rat but not human/guinea pig Sertoli cells in testes. Am. J. Physiol. 1998;274:C1708–C1717. doi: 10.1152/ajpcell.1998.274.6.C1708. [DOI] [PubMed] [Google Scholar]
- Saitou M., Fujimoto K., Doi Y., Itoh M., Fujimoto T., Furuse M., Takano H., Noda T., Tsukita Sh. Occludin-deficient embryonic stem cells can differentiate into polarized epithelial cells bearing tight junctions. J. Cell Biol. 1998;141:397–408. doi: 10.1083/jcb.141.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi G., Uemura T., Riemann H. Simplified method for purification of Clostridium perfringens type A enterotoxin. Appl. Microbiol. 1973;27:762–767. doi: 10.1128/am.26.5.762-767.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara A., Furuse M., Saitou M., Ando-Akatsuka Y., Tsukita Sh. Possible involvement of phosphorylation of occludin in tight junction formation. J. Cell Biol. 1997;137:1393–1401. doi: 10.1083/jcb.137.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger E.E., Lynch R.D. Structure, function, and regulation of cellular tight junctions. Am. J. Physiol. 1992;262:L647–L661. doi: 10.1152/ajplung.1992.262.6.L647. [DOI] [PubMed] [Google Scholar]
- Staehelin L.A. Further observations on the fine structure of freeze-cleaved tight junctions. J. Cell Sci. 1973;13:763–786. doi: 10.1242/jcs.13.3.763. [DOI] [PubMed] [Google Scholar]
- Staehelin L.A. Structure and function of intercellular junctions. Int. Rev. Cytol. 1974;39:191–283. doi: 10.1016/s0074-7696(08)60940-7. [DOI] [PubMed] [Google Scholar]
- Stevenson B.R., Anderson J.M., Goodenough D.A., Mooseker M.S. Tight junction structure and ZO-1 constant are identical in two strains of Madin-Darby Canine Kidney cells which differ in transepithelial resistance. J. Cell Biol. 1988;107:2401–2408. doi: 10.1083/jcb.107.6.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita Sh., Furuse M. Occludin and claudins in tight junction strandsleading or supporting players? Trend. Cell Biol. 1999;9:268–273. doi: 10.1016/s0962-8924(99)01578-0. [DOI] [PubMed] [Google Scholar]
- Wolburg H., Neuhaus J., Kniesel U., Krauss B., Schmid E.-M., Öcalan M., Farrell C., Risau W. Modulation of tight junction structure in blood-brain barrier endothelial cells. Effects of tissue culture, second messengers and cocultured astrocytes. J. Cell Sci. 1994;107:1347–1357. doi: 10.1242/jcs.107.5.1347. [DOI] [PubMed] [Google Scholar]
- Wong V., Gumbiner B.M. A synthetic peptide corresponding to the extracellular domain of occludin perturbs the tight junction permeability barrier. J. Cell Biol. 1997;136:399–409. doi: 10.1083/jcb.136.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]