Abstract
A stromal processing peptidase (SPP) cleaves a broad range of precursors targeted to the chloroplast, yielding proteins for numerous biosynthetic pathways in different compartments. SPP contains a signature zinc-binding motif, His-X-X-Glu-His, that places it in a metallopeptidase family which includes the mitochondrial processing peptidase. Here, we have investigated the mechanism of cleavage by SPP, a late, yet key event in the import pathway. Recombinant SPP removed the transit peptide from a variety of precursors in a single endoproteolytic step. Whereas the mature protein was immediately released, the transit peptide remained bound to SPP. SPP converted the transit peptide to a subfragment form that it no longer recognized. We conclude that SPP contains a specific binding site for the transit peptide and additional proteolysis by SPP triggers its release. A stable interaction between SPP and an intact transit peptide was directly demonstrated using a newly developed binding assay. Unlike recombinant SPP, a chloroplast extract rapidly degraded both the transit peptide and subfragment. A new degradative activity, distinguishable from SPP, was identified that is ATP- and metal-dependent. Our results indicate a regulated sequence of events as SPP functions during precursor import, and demonstrate a previously unrecognized ATP-requirement for transit peptide turnover.
Keywords: chloroplast, metallopeptidase, protein degradation, stromal processing peptidase, transit peptide
The chloroplast is the site of photosynthesis and also houses an amazing array of biosynthetic pathways needed for normal plant growth and development. Biogenesis of the chloroplast depends on the import of nuclear-encoded proteins from the cytoplasm. Typically, these proteins are synthesized with a transit peptide that facilitates multiple steps in a general import pathway, from receptor recognition of the precursor on the surface of the organelle to interactions with the translocation apparatus of the outer and inner membranes (Cline and Henry 1996; Fuks and Schnell 1997; Lübeck et al. 1997). Ultimately, transit peptides are removed as precursors enter the stroma. Processing is a key step in the import pathway, yet we understand very little about the specificity and regulation of precursor cleavage. Determinants for processing reside in the transit peptide itself and at the transit peptide-mature protein junction (Clark and Lamppa 1991; Archer and Keegstra 1993; Bassham et al. 1994; Pilon et al. 1995). However, given that transit peptides differ considerably in length (ranging from 31–142 amino acids) and primary sequence, and that a consensus sequence at the cleavage site is not well-defined (Gavel and von Heijne 1990), it has been difficult to formulate a good understanding of the molecular mechanism of precursor cleavage.
We initially identified a soluble metal-dependent chloroplast processing enzyme that cleaved the precursor for the light-harvesting chlorophyll a/b binding protein (preLHCP)1. Chloroplast extracts immunodepleted of this enzyme lost the ability to process preLHCP, as well as the precursors for the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (preRBCS) and acyl carrier protein (Oblong and Lamppa 1992). Using a recombinant form of the enzyme expressed in Escherichia coli, we recently demonstrated that it cleaves a large diversity of precursors imported into the chloroplast, and thus functions as a general stromal processing peptidase (SPP; Richter and Lamppa 1998). Furthermore, this analysis showed that SPP carries out the cleavage reaction in the absence of other chloroplast proteins. The role of SPP was also investigated by in vivo experiments. Downregulation of SPP via antisense gene expression disrupted chloroplast biogenesis at multiple levels, resulting in retarded shoot and root growth (Wan et al. 1998). Most notably, chloroplasts from the transgenic plants lost their ability to efficiently translocate precursors across the envelope during in vitro assays. These studies clearly establish a critical role for SPP as a component of the general import machinery.
SPP belongs to a relatively new family of metallopeptidases, the pitrilysins, that contain a signature zinc-binding motif His-X-X-Glu-His at the catalytic site (Rawlings and Barrett 1995). The family includes E. coli protease III, insulin-degrading enzyme, and significantly, the mitochondrial processing peptidase (MPP). Conservation extends beyond the His-X-X-Glu-His motif, where 25–30% identity is found in an NH2-terminal region of 150 residues of SPP, suggesting similar evolutionary origins of these metallopeptidases. Although they most probably share a common catalytic mechanism that depends on the zinc-binding domain (Becker and Roth 1992; Perlman et al. 1993; Kitada et al. 1995; Striebl et al. 1996), their substrate specificities are diverse. We do not understand how these metallopeptidases recognize their structurally distinct substrates.
The availability of SPP synthesized in E. coli allowed us to address some fundamental questions about the molecular mechanism of precursor processing. In particular, we tested the hypothesis that there are specific interactions between SPP and the precursor that depend on the transit peptide. We predicted this from our earlier experiments where preLHCP served as an affinity ligand to purify SPP under conditions that prevented precursor cleavage (Oblong and Lamppa 1992). We have found that SPP indeed contains a binding site for the intact transit peptide, and a specific protein–protein interaction is maintained after the mature protein is released.
During the course of our experiments, we discovered that SPP carries out a second reaction, converting the transit peptide to a subfragment form that SPP itself was unable to bind. The subfragments from three different precursors remained stable in the presence of recombinant SPP. Analysis of the fate of the transit peptide in a chloroplast extract revealed that both the transit peptide and a discrete subfragment form were produced upon precursor cleavage, yet both were rapidly degraded. In contrast, the mature protein remained stable providing strong evidence for selective protein degradation.
It has been estimated that an enormous amount of protein is imported by a single chloroplast, e.g., 2.5 × 104 molecules of the precursor for ferredoxin (preFD) are imported per minute during an in vitro assay (Pilon et al. 1992). In vivo, 2 × 107 molecules of preLHCP are predicted to be imported each 24 h (Pfisterer et al. 1982). With each precursor comes a transit peptide, yet transit peptides do not accumulate in the stroma in vivo or during in vitro assays, an indication that a proteolytic system exists to selectively eliminate them from the chloroplast. Therefore, we pursued the nature of the degradation activity we detected in the chloroplast extract. Our experiments have identified a soluble proteolytic activity that can be distinguished from SPP. Most importantly, degradation is ATP-dependent, suggesting that chloroplasts possess a special energy requirement for transit peptide turnover. A model integrating our findings is discussed in relationship to the general import pathway.
Materials and Methods
Preparation of Radioactive-labeled Substrates
Precursors labeled with [35S]methionine or [35S]cysteine were generated by translation in a rabbit reticulocyte lysate (Promega Corp.) using template RNA synthesized by either SP6 or T7 RNA polymerase (Lamppa and Abad 1987). During the course of the experiments, a nonspecific protease activity was found to be present in the reticulocyte lysate. In a typical control experiment, [35S]methionine-labeled preHSP21 was incubated with immobilized SPP (Richter and Lamppa 1998) for 30 min at 24°C. The supernatant containing the transit peptide subfragment was separated from the immobilized SPP fraction and incubated for 1 h at 24°C either without inhibitor or in the presence of 2 mM PMSF. Without inhibitor, the transit peptide subfragment was fully degraded. In contrast, in the presence of PMSF, ∼90% of the subfragment remained. Consequently, 2 mM PMSF was used in the experiments to minimize the nonspecific protease activity of the reticulocyte lysate.
Processing Assay
An extract of E. coli cells expressing recombinant SPP with a biotinylated peptide tag (Richter and Lamppa 1998), immobilized SPP, or a chloroplast extract (Abad et al. 1989) were used as sources of SPP. To ensure reproducible results, cultures of the E. coli strain carrying the expression construct for recombinant SPP were grown and prepared under identical conditions. Typically, an expression culture (120 ml) was started from an overnight culture (dilution 1:100), cultured at 30°C, and induced by 2 mM IPTG at OD560 nm 0.22–0.24 after 2 h and 15 min. The culture was continued for 3 h and 30 min (final OD560 nm 1.2–1.3) and cell extracts were prepared as described previously (Richter and Lamppa 1998). For processing reactions with either E. coli cell extract containing recombinant SPP or chloroplast extract, 10 μl extract was incubated in a total volume of 20 μl with 25 mM Hepes-KOH, pH 7.5, 2 mM PMSF, at 24°C. If immobilized SPP was used, SPP protein from 100 μl E. coli extract was bound to 50 μg streptavidin-coated magnetic beads and incubated in a total volume of 20 μl (Richter and Lamppa 1998). The amount of radioactive-labeled substrate per reaction was optimized for detection of small products, transit peptides and their subfragments, upon electrophoresis (preFD, 1 μl; preHSP21, 4 μl; preLHCP, 5 μl; preRBCA, 4 μl; preRBCS, 2 μl). Reactions were stopped by mixing with gel-loading buffer and boiling. However, using immobilized SPP, magnetic beads were removed by magnetic separation after boiling. If binding and release of processing products was studied, immobilized SPP was separated from supernatant, washed with 50 μl 25 mM Hepes-KOH, mixed with gel-loading buffer, and boiled. Tricine SDS-PAGE (at 4°C, 14 V/cm, 3 h; Schägger and von Jagow 1987) was employed for detection of transit peptides and their subfragments. Standard SDS-PAGE (at room temperature, 20 V/cm, 1 h; Laemmli 1970) was used to monitor the generation of mature FD and RBCA. All gels were analyzed by autoradiography.
Preparation of FD Transit Peptide and its Subfragment
Recombinant SPP protein from 1 ml E. coli extract was immobilized onto 500 μg of streptavidin-coated magnetic beads. [35S]methionine-labeled preFD (50 μl) was diluted in a total of 150 μl buffer (25 mM Hepes-KOH, pH 7.5, 2 mM PMSF), mixed with the immobilized SPP and incubated at 24°C. Separation of the supernatant from immobilized SPP was performed either after 3 min to obtain the FD transit peptide or after 120 min to obtain the subfragment. The supernatants were frozen and stored overnight at −70°C. During thawing of the supernatants, a protein precipitate formed, which was removed by centrifugation (5 min, 10,000 g). The supernatants were subsequently dialyzed (at 4°C for 5 h in 1 liter of 25 mM Hepes-KOH, pH 7.5) using a cellulose ester dialysis membrane with a cut off at mol wt 500 (Spectrum Medical Industries). Around 120 μl of sample was usually recovered from one preparation. To test for nonspecific protease activity, 10 μl of sample was incubated for 1 h at 24°C and compared with a nonincubated control sample upon tricine SDS-PAGE. No degradation was observed for either substrate.
Binding Assay
Immobilized SPP was incubated with 10 μl of substrate (preparations of FD transit peptide or its subfragment) in a 20-μl vol for 5 min using 25 mM Hepes-KOH, pH 7.5, at 24°C. The immobilized SPP fraction was separated from the supernatant, washed twice with 50 μl 25 mM Hepes-KOH, and once with 20 μl of solution of NaCl at different concentrations to elute bound substrate. The immobilized SPP fraction was finally resuspended in gel-loading buffer and liberated from magnetic beads by boiling before tricine SDS-PAGE. Gels were analyzed by autoradiography and scanned by a PhosphorImager (Molecular Dynamics) for quantification using ImageQuant software (Molecular Dynamics).
Degradation Assay
Typically, 10 μl chloroplast extract was incubated with 10 μl substrate (preparations of FD transit peptide or its subfragment) using 25 mM Hepes-KOH, pH 7.5, at 24°C. Reactions were stopped by mixing with gel-loading buffer and boiling before analysis by tricine SDS-PAGE for autoradiography and quantification.
Results
SPP Acts as an Endopeptidase
Our previous studies showed that processing of preFD by SPP generates two products: mature FD and its intact transit peptide (Richter and Lamppa 1998). The nature of each processing product was established using different amino acids for radioactive labeling of the precursor and by size estimates upon tricine SDS-PAGE. PreFD has four methionines that are found only in the transit peptide, and hence, only the transit peptide with an estimated size of ∼5 kD was detectable upon processing of [35S]methionine-labeled preFD. The five cysteines of preFD are present only in the mature protein. Consequently, using [35S]cysteine-labeled substrate in a processing reaction with SPP, only mature FD was detected.
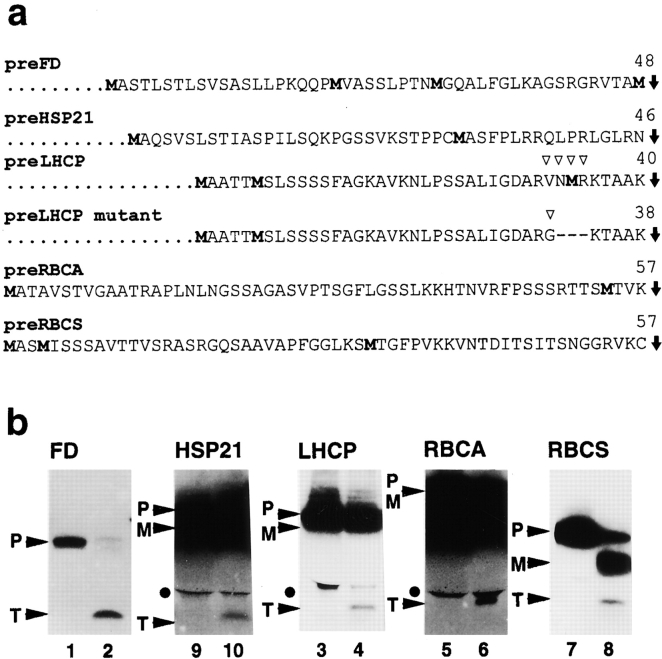
To determine if an intact transit peptide is usually an initial cleavage product, other chloroplast protein precursors were tested in processing reactions with recombinant SPP. In addition to preFD, the precursors of heat shock protein 21 (HSP21), LHCP, ribulose-1,5-bisphosphate carboxylase/oxygenase activase (RBCA), and RBCS, which contain two or more methionines in the transit peptide (Fig. 1 a), were synthesized by in vitro translation as [35S]methionine-labeled polypeptides. Each substrate was incubated with recombinant SPP for five minutes. A fragment that was similar in mobility to the FD transit peptide was observed for each reaction upon tricine SDS-PAGE (Fig. 1 b). Thus, in all cases, SPP initially generated what appeared to be an intact transit peptide by a single endoproteolytic step.
Figure 1.
SPP generates intact transit peptides. a, Amino acid sequences of transit peptides: preFD (Smeekens et al. 1985), preHSP21 (Vierling et al. 1988), preLHCP (Lamppa et al. 1985) and mutant of preLHCP with a modification indicated by open arrow heads (Clark et al. 1989), preRBCA (Werneke et al. 1988), and preRBCS (Coruzzi et al. 1984). Radiolabeled methionines incorporated into transit peptides during in vitro translation are indicated in bold. SPP processing sites are shown by filled arrows, and lengths of transit peptides are noted. b, Processing of [35S]methionine-labeled precursors by recombinant SPP. The long exposure times needed to detect the transit peptides (T) on the autoradiograms usually resulted in a loss of resolution of the precursor (P) and the mature protein (M). Closed circles mark an unidentified [35S]-labeled product detected in most of the substrate preparations. Name of each mature protein is given above each panel.
SPP Converts Transit Peptides to Subfragments
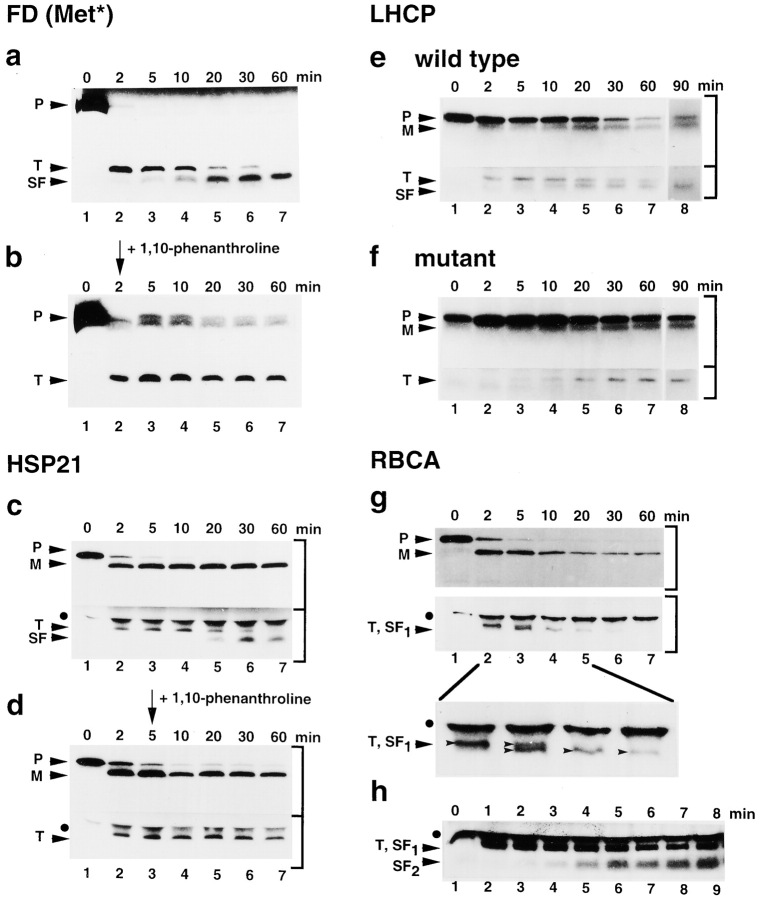
Despite the large mass of diverse transit peptides that enter the organelle, nothing is known about their turnover. To investigate if SPP is involved, time courses of processing reactions were carried out using the precursors shown in Fig. 1 a and recombinant SPP, which was immobilized onto magnetic beads (see Materials and Methods). After 2 min, the mature protein and the transit peptide appeared simultaneously in each reaction, although the processing of each precursor progressed at a different rate (Fig. 2, a, c, e, and g; not shown for RBCS). Interestingly, using the precursors for FD, HSP21 and LHCP, we found that the initial generation of intact transit peptide was followed by its conversion to one detectable subfragment. The transit peptide of RBCA gave rise to two subfragments (RBCA: Fig. 2g and Fig. h). Conversion of the RBCS transit peptide could not be directly monitored. Instead, it nearly disappeared after one hour, probably because the conversion products containing the labeled methionines were too small to be seen upon tricine SDS-PAGE. Progression of this trimming reaction differed considerably between the transit peptides examined. At one extreme, the conversion of the RBCA transit peptide to subfragment 1 was completed in <10 min (Fig. 2 g), whereas an incubation time of 90 min was needed for full conversion of the LHCP transit peptide to a smaller form (Fig. 2 e). Nevertheless, based on our results with four substrates, trimming of transit peptides as another function of SPP is apparently a common step upon precursor processing.
Figure 2.
Transit peptides are converted to subfragments by SPP. Time courses of processing by immobilized SPP were carried out using [35S]methionine-labeled precursors as substrates. Separation of precursor (P) and products (mature protein, M; transit peptide, T; subfragment of transit peptide, SF) was done by tricine SDS-PAGE, if not otherwise noted. PreFD alone (a) and with 5 mM 1,10-phenanthroline added after 2 min (b). PreHSP21 without (c) and with 5 mM 1,10-phenanthroline added after 5 min (d). PreLHCP wild-type (e) and mutant (f). The 90-min incubation was done in a separate experiment (e and f, lane 8). PreRBCA (g), the top shows a standard SDS-PAGE used to monitor generation of mature RBCA, the middle shows generation of T and SF1, and the bottom shows an enlargement of lanes 2–5 of the middle panel. PreRBCA (h), to detect SF2, the time for tricine SDS-PAGE was reduced. Autoradiograms with different exposure times were used for presentation of the top and the bottom part of a gel, as indicated by brackets at right.
The chelator 1,10-phenanthroline inhibits precursor processing by SPP, which is a metallopeptidase (VanderVere et al. 1995; Richter and Lamppa 1998). We found that 1,10-phenanthroline also inhibited the conversion of the FD and HSP21 transit peptides to their respective subfragment forms (Fig. 2b and Fig. d). The transit peptides were stable for one hour in the presence of the chelator. This observation shows that both functions of SPP, precursor processing and transit peptide trimming, depend on metal ions.
To investigate the specificity of the proteolytic conversion of a transit peptide to a subfragment form, we took advantage of a preLHCP mutant that was previously used to study cleavage events upon in vitro import and in a chloroplast extract (Clark et al. 1989). The transit peptide of this precursor mutant has an altered COOH terminus, where a valine was changed to a glycine at position −9, and three amino acids were deleted from −8 to −6, relative to the site recognized by SPP in the chloroplast extract (Fig. 1 a). This alteration was sufficient to block the conversion of the preLHCP transit peptide to the subfragment, an indication that specific features of the transit peptide serve as determinants for the trimming reaction. When these features are disrupted, in this case by the COOH-terminal mutation, the transit peptide was not further processed by SPP.
Using immobilized SPP, two steps were observed during trimming of the RBCA transit peptide. The transit peptide was first converted into a slightly smaller subfragment 1 (Fig. 2 g) that was subsequently trimmed to subfragment 2, which was only detected under modified PAGE conditions (Fig. 2 h). However, neither fragment was detected after one hour incubation, which indicated that either inhibition of the intrinsic protease activities in the reticulocyte lysate by 2 mM PMSF (see Materials and Methods) was not sufficient to preserve these barely detectable amounts of peptide, or SPP converted them to smaller subfragments, which were not detectable upon tricine SDS-PAGE. Alternatively, SPP performed complete degradation of the transit peptide. The latter interpretation, however, is not supported by the results for the three other transit peptides, which suggested instead that SPP performed limited proteolysis of the transit peptide. Nevertheless, the fate of the RBCA transit peptide demonstrates that trimming of transit peptides by SPP can occur in more than one cleavage step that may depend on features specific to each transit peptide.
The results presented in Fig. 2 demonstrate that the mature protein and the transit peptide appear simultaneously in the course of a processing reaction, supporting the idea that transit peptide removal from a chloroplast precursor polypeptide initially occurs in one endoproteolytic step. Whereas the mature protein is generated by this single cleavage, the transit peptide is further trimmed by SPP.
SPP Has a High Affinity for Intact Transit Peptides
We investigated if SPP specifically binds the intact transit peptide and its subfragment using an in vitro binding assay (Fig. 3 a). A protocol was established for preparation of [35S]methionine-labeled FD transit peptide and its subfragment (see Materials and Methods). Aliquots of a FD transit peptide preparation were incubated with immobilized SPP for five minutes. Each immobilized SPP fraction was separated from the supernatant and individually washed with a solution of NaCl at different concentrations for elution of bound transit peptide. To determine the amount of transit peptide that remained bound to immobilized SPP at 1,000 mM NaCl, one SPP fraction was washed in 1,000 mM NaCl and the bound material was liberated for analysis by boiling in the presence of gel-loading buffer. A representative supernatant, all eluates, and the material released by boiling were subjected to tricine SDS-PAGE to analyze by both autoradiography (Fig. 3 b) and PhosphorImager scanning for quantification. The supernatant was separated from the immobilized SPP fraction and contained unbound transit peptide (Fig. 3 b, lane 2). Washing of the immobilized SPP fraction without NaCl did not elute detectable amounts of transit peptide (Fig. 2 b, lane 3). Washing with low NaCl (50 mM) released 3% of the substrate originally added to one reaction (Fig. 2 b, lane 4). Using a higher NaCl concentration (100 mM) for washing of another immobilized fraction, a substantially greater amount of transit peptide, 13%, was released (Fig. 3 b, lane 5). However, if parallel fractions were washed with even higher concentrations of NaCl, the amount of eluted transit peptide did not increase, not even at 1,000 mM NaCl (Fig. 3 b, lanes 6–8). Surprisingly, however, boiling of the SPP fraction after washing with 1,000 mM NaCl released an additional 7% of the original substrate added to the reaction (Fig. 3 b, lane 9). Together, the eluate at 1,000 mM NaCl and its corresponding SPP fraction contained in total 20%, i.e. 13% + 7%, of the original substrate, and thus was equivalent to the portion of transit peptide initially bound by immobilized SPP in one binding reaction. Two-thirds of the bound transit peptide was released from SPP at high NaCl concentration, but, importantly, one third remained bound. This observation suggested that SPP may have two states of transit peptide binding, and one of these has an especially high affinity for the intact transit peptide.
Figure 3.
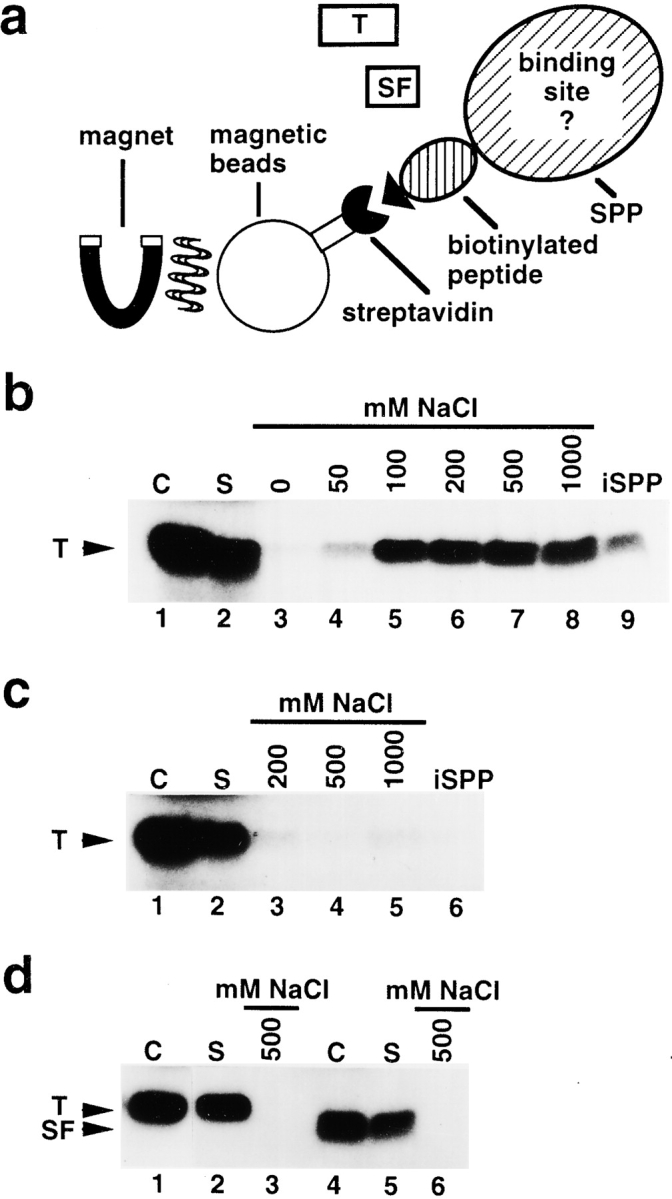
SPP contains a binding site for the intact transit peptide. a, Schematic representation of the binding assay (see Materials and Methods). Recombinant SPP was immobilized onto streptavidin-coated magnetic beads that bound to its biotin-containing peptide tag. [35S]methionine-labeled preparations of FD transit peptide (T) or its subfragment (SF) were added. Using a magnet, the supernatant containing the unbound substrate was separated from the immobilized SPP fraction with the bound substrate. b, Binding of FD transit peptide to immobilized SPP. Total amount of substrate used in one reaction, control lane 1; supernatant containing unbound substrate, lane 2; bound substrate independently eluted at NaCl concentrations between 0 and 1,000 mM, lanes 3–8; substrate still bound to immobilized SPP after washing at 1,000 mM NaCl, lane 9. c, Binding of FD transit peptide to immobilized SPP in the presence of 5 mM 1,10-phenanthroline. Substrate, control lane 1; supernatant, lane 2; bound substrate independently eluted at NaCl concentrations of 200, 500, and 1,000 mM, lanes 3–5; substrate bound to immobilized SPP after washing at 1,000 mM NaCl, lane 6. d, Binding of FD transit peptide to streptavidin-coated magnetic beads incubated with extract of E. coli cells. Substrate, control lane 1; supernatant, lane 2; bound substrate eluted from the magnetic beads fraction at 500 mM NaCl, lane 3. Binding of subfragment of FD transit peptide to immobilized SPP. Substrate, control lane 4; supernatant, lane 5; bound substrate eluted from immobilized SPP at 500 mM NaCl, lane 6.
SPP activity depends on metal ions. To investigate the importance of metal ions on the interactions between SPP and the transit peptide, binding assays were carried out in the presence of 1,10-phenanthroline. We found that neither eluates with different NaCl concentrations nor a SPP fraction after washing at 1,000 mM NaCl contained significant amounts of transit peptide (Fig. 3 c). Apparently, treatment by 1,10-phenanthroline causes conformational changes in SPP, preventing a stable interaction with the transit peptide.
In addition, it was tested whether SPP contains cysteines that are necessary for transit peptide binding by adding of the sulfhydryl group inhibitor N-ethylmaleimide (NEM) into a binding assay. Interactions of this inhibitor with the transit peptide could be excluded since the FD transit peptide does not have cysteines. Analysis of this experiment revealed that 10 mM NEM disrupted the interaction between SPP and FD transit peptide (not shown). Only unbound transit peptide, present in the supernatant, was detected. No transit peptide was found in eluates with different NaCl concentrations (200, 500, 1,000 mM) or in an immobilized SPP fraction after washing with 1,000 mM NaCl. Therefore, the sulfhydryl group of one or more cysteines is required for SPP's ability to bind a transit peptide.
To determine if the magnetic beads matrix used for immobilization of SPP nonspecifically binds the FD transit peptide that would interfere with our binding studies, magnetic beads were incubated with a control extract of E. coli cells not expressing SPP (Richter and Lamppa 1998). The binding assay was carried out as described above. Elution at 500 mM NaCl did not release detectable amounts of transit peptide from the matrix incubated with control extract (Fig. 3 d, lane 3). An aliquot of this matrix was boiled to release strongly bound transit peptide, but none was detected (not shown). Hence, the matrix alone did not bind the FD transit peptide establishing that the binding of the intact transit peptide to the matrix fraction was due to the presence of SPP.
The FD transit peptide is converted by SPP to a subfragment that remains relatively stable for at least 30 min (Fig. 2 a). We examined if SPP has an affinity for this subfragment. A subfragment preparation was incubated with immobilized SPP in a binding assay. Only unbound subfragment was found in the supernatant, whereas nothing was detected in a NaCl eluate (Fig. 3 d, lanes 4–6). We conclude that SPP did not recognize the subfragment. Apparently, conversion of the transit peptide into its subfragment causes the loss of features necessary for binding by SPP. The results presented in Fig. 3 demonstrate that there is a stable interaction between SPP and the intact transit peptide, which can be disturbed by metal ion chelator or sulfhydryl group inhibitor. In contrast, binding between SPP and the subfragment was not observed.
SPP Immediately Liberates the Mature Protein and Releases the Intact Transit Peptide Upon Trimming
The finding that SPP does not bind the FD transit peptide subfragment showed that trimming of the transit peptide alters SPP's affinity for the transit peptide, and suggested this may serve as a specific step to trigger its release from SPP. To understand the temporal relationship between transit peptide conversion to the subfragment form and release from SPP, time courses of precursor processing by immobilized SPP were carried out. Binding of the processing products to SPP was monitored by separate analysis of the supernatant and the immobilized SPP fraction. Using preFD and preHSP21 as substrates, during the first 10 min of processing, both mature proteins, FD and HSP21, and their respective transit peptides were produced simultaneously, but they were found in different fractions (Fig. 4). The mature proteins were immediately released into the supernatant (Fig. 4, a and d, lanes 2–4). In contrast, the intact transit peptides remained bound to immobilized SPP (Fig. 4c and Fig. e, lanes 2–4). Beginning at 10 min, proteolytic conversion of the transit peptide coincided with the release of its subfragment into the supernatant (Fig. 4b and Fig. d, lanes 5–7). This demonstrates that generation and release of the mature protein is accompanied by an initial accumulation of the intact transit peptide bound to SPP. Subsequently, limited proteolysis by SPP releases the subfragment, perhaps, to clear a binding site for new substrate.
Figure 4.
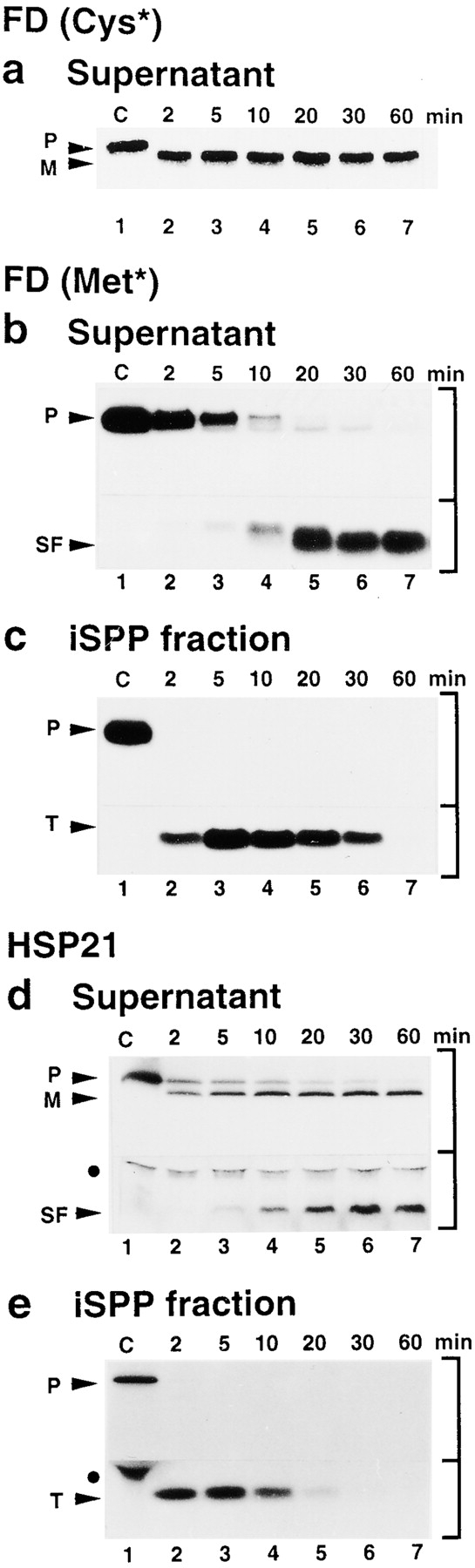
SPP has a different affinity for the products generated during processing. Aliquots of processing reactions with immobilized SPP using preFD and preHSP21 as substrates were collected at intervals over a 60-min period and immediately separated into the supernatant and the immobilized SPP fraction before analysis by standard SDS-PAGE (a) or tricine SDS-PAGE (b–e). [35S]cysteine-labeled preFD, used to monitor generation of mature FD, supernatant (a). [35S]methionine-labeled preFD, used to monitor generation of FD transit peptide and its subfragment, supernatant (b), and immobilized SPP fraction (c). [35S]methionine-labeled preHSP21 supernatant (d), and immobilized SPP fraction (e). Substrate control (C), lane 1.
Transit Peptide Trimming and Degradation in Chloroplasts
The intact transit peptide was identified as an initial product of precursor processing by SPP. However, the fate of the transit peptides in vivo is unknown. They are not stable upon in vitro import into the chloroplast using preFD as a substrate (van't Hof and de Kruijff 1995; also, Richter, S., and G. Lamppa, unpublished results). SPP has the capability to trim transit peptides, but to complete their turnover in the chloroplast, one predicts that other degradative activities are needed. Precursor processing by a chloroplast extract was analyzed to explore this prediction. Using preFD and preHSP21 as substrates, within five minutes both the mature protein and the transit peptide appeared at the same time (Fig. 5, a–c, lanes 2 and 3). After ten minutes, conversion of the transit peptide to a subfragment was observed (Fig. 5b and Fig. c, lanes 4 and 5). The initial appearance and trimming of the transit peptide within the first 20 min of the reaction using the chloroplast extract resembled the pattern found for processing using immobilized SPP (Fig. 2, a and c). In sharp contrast to the results using immobilized SPP in a processing reaction, however, the subfragment was then quickly degraded by the chloroplast extract (Fig. 5b and Fig. c, lane 6). These results indicate that transit peptides are most likely trimmed in the chloroplast by SPP in a discrete step before their final degradation.
Figure 5.
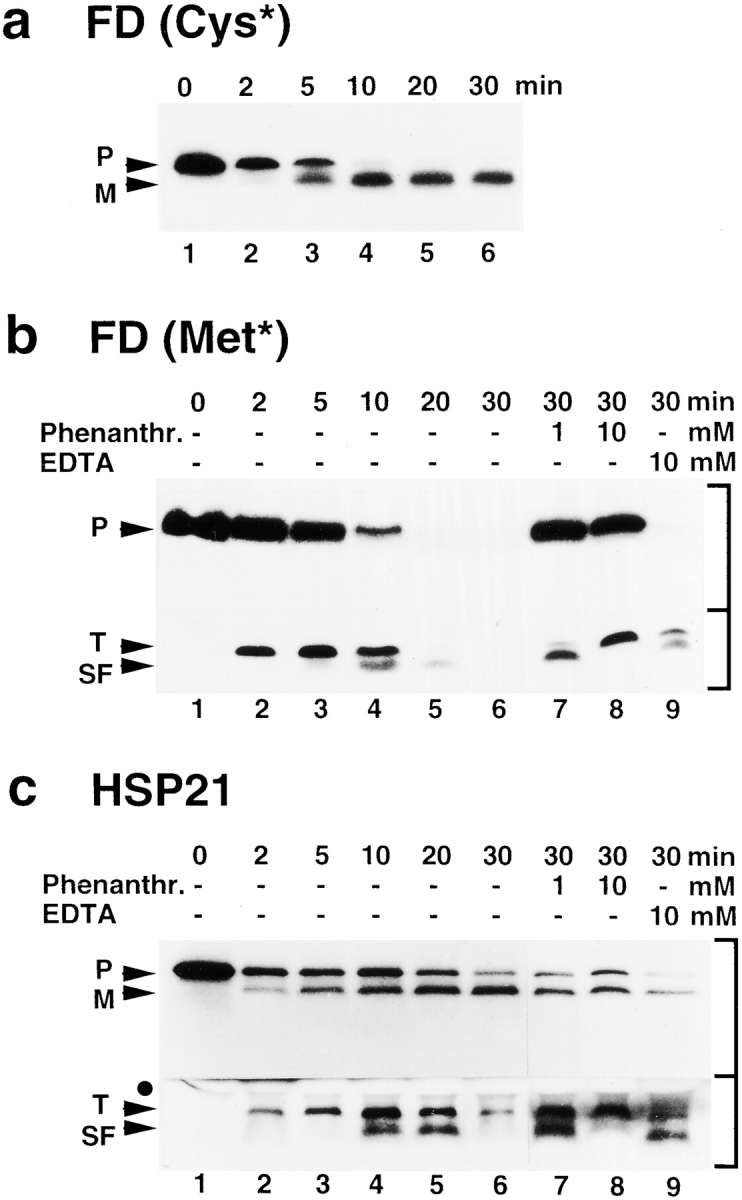
Transit peptide turnover by a chloroplast extract yields a discrete subfragment before degradation. Using preFD and preHSP21 as substrates, time courses of processing reactions with chloroplast extract were carried out and analyzed by standard SDS-PAGE (a) or tricine SDS-PAGE (b and c). If inhibition by 1,10-phenanthroline or EDTA was tested, the reactions were interrupted by adding the inhibitor and then continued for up to 30 min. a, [35S]cysteine-labeled preFD used to monitor generation of mature FD. b, [35S]methionine-labeled preFD used to monitor generation of FD transit peptide and its subfragment, lanes 1–6; inhibitors were added after 2 min, lanes 7–9. c, [35S]methionine-labeled preHSP21, lanes 1–6; inhibitors were added after 5 min, lanes 7–9.
Furthermore, degradation of both the FD and HSP21 transit peptides and their respective subfragments was sensitive to metal ion chelators. If 1,10-phenanthroline or EDTA was added during the processing reaction, they were stable in the chloroplast extract (Fig. 5b and Fig. c, lanes 7–9). However, interestingly, the two reactions, i.e. proteolytic conversion versus subfragment degradation, showed differential sensitivity to 1,10-phenanthroline. Conversion of the transit peptide to the subfragment needed as much as 10 mM 1,10-phenanthroline to be fully inhibited (Fig. 5b and Fig. c, lane 8). Subsequent degradation, in contrast, was efficiently inhibited at only 1 mM 1,10-phenanthroline (Fig. 5b and Fig. c, lane 7). These results support the idea that two distinct activities are involved in transit peptide turnover after precursor cleavage, the first dependent on SPP.
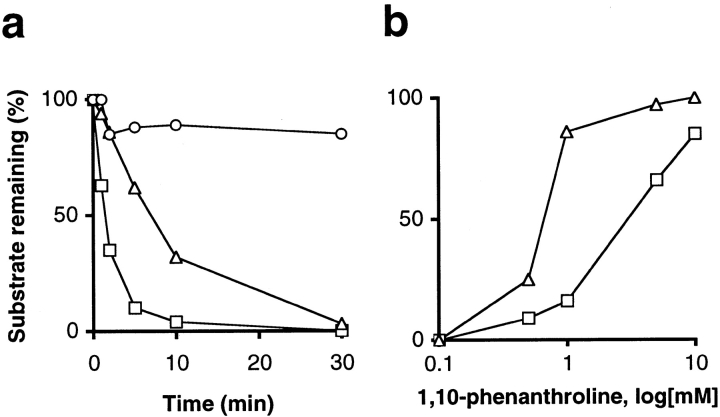
We analyzed the fate of the FD transit peptide and its subfragment separately. [35S]methionine-labeled FD transit peptide and its subfragment were prepared (see Materials and Methods) and used as substrates in separate reactions with chloroplast extract. We observed rapid conversion of the transit peptide and complete degradation of the subfragment within 30 min (Fig. 6 a). Mature FD was prepared by incubation (30 min) of [35S]cysteine-labeled preFD with immobilized SPP. The supernatant of this processing reaction was added directly to a chloroplast extract. Mature FD was relatively stable over 30 min (Fig. 6 a); we observed only a 15% loss of fragment within the first 2 min. The distinct stability of mature FD compared with its transit peptide in the chloroplast extract demonstrates that selective degradation of the latter occurred.
Figure 6.
Selective degradation of the FD transit peptide and its subfragment in the chloroplast. [35S]methionine-labeled FD transit peptide (open square) and its subfragment (open triangle), as well as [35S]cysteine-labeled mature FD (open circle), were used as substrates in degradation assays with chloroplast extract. Reactions were analyzed by standard SDS-PAGE (mature FD) or tricine SDS-PAGE (transit peptide, subfragment) and quantified. a, Percent of substrate remaining upon incubation in chloroplast extract. b, Percent of substrate remaining upon 30 min incubation in chloroplast extract in the presence of 1,10-phenanthroline.
Transit peptide conversion to a subfragment and its subsequent degradation showed differential sensitivity to 1,10-phenanthroline in the course of a processing reaction (Fig. 5). To refine this analysis using separate reactions, preparations of FD transit peptide and its subfragment were incubated in a chloroplast extract at different 1,10-phenanthroline concentrations ranging from 0.1 mM to 10 mM. Transit peptide conversion was less sensitive to 1,10-phenanthroline than subfragment degradation (Fig. 6 b). At 1 mM 1,10-phenanthroline, only 16% of transit peptide was left after 30 min, whereas 86% of subfragment remained after the same incubation time. At 10 mM 1,10-phenanthroline, 80% of the transit peptide remained and no degradation was found for the subfragment. Hence, in the chloroplast, transit peptide turnover is metal-dependent and can be dissected into two different steps, initial conversion and subsequent degradation, each sensitive to a different concentration of the chelator 1,10-phenanthroline.
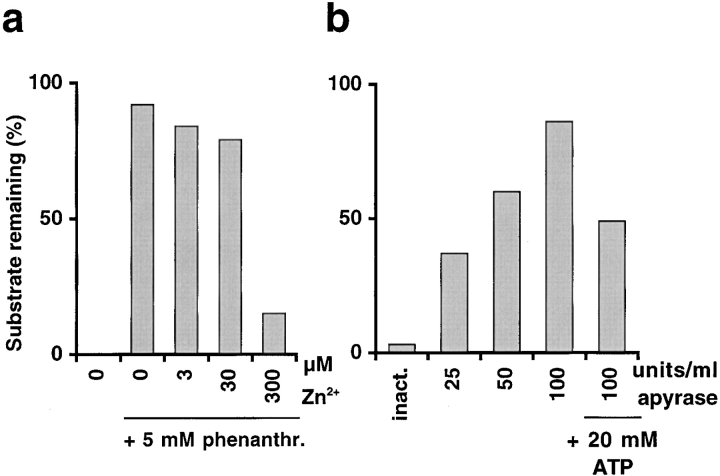
To characterize the nature of the activity that carries out the subsequent degradation step, the subfragment of FD transit peptide was used as a substrate to test group specific protease inhibitors for their ability to block degradation in the chloroplast extract. Divalent metal ion chelators other than 1,10-phenanthroline also inhibited the degradation activity, although to a lower extent. Using 10 mM EDTA, 73% of the originally added subfragment was detected in the sample after 30 min and using 10 mM EGTA, 46% of the original amount remained. The divalent metal ion requirement was directly tested. Degradation of the subfragment, which was inhibited at 5 mM 1,10-phenanthroline, was assayed in the presence of zinc ions (Fig. 7 a). At a concentration of 300 μM Zn2+, the degradation activity was restored.
Figure 7.
Degradation of FD transit peptide subfragment in the chloroplast depends on metal ions and ATP. Degradation assays with chloroplast extract were carried out using [35S]methionine-labeled subfragment as substrate and analyzed by tricine SDS-PAGE for quantification. a, Reactivation of degradation activity by Zn2+ in the presence of 1,10-phenanthroline. Percent of subfragment remaining upon 30 min incubation in chloroplast extract without inhibitor and with 5 mM 1,10-phenanthroline at different Zn2+ concentrations. b, Reactivation of degradation activity by ATP after apyrase preincubation causes a loss of subfragment. Percent subfragment remaining after 30 min incubation in pretreated chloroplast extract without ATP or at 20 mM ATP. Chloroplast extract was preincubated with apyrase inactivated by boiling (inact.) or with different concentrations of active apyrase for 1 h at 37°C.
Other group specific inhibitors were tested as well, including NEM (10 mM), PMSF (10 mM), which inhibits serine proteases, E64 (100 μM), specific for cysteine proteases, and pepstatin (100 μM), which inhibits some aspartic proteases. None of these inhibitors significantly decreased subfragment degradation within 30 min incubation (not shown). However, the degradation activity and SPP showed a different sensitivity to NEM. NEM did not significantly decrease degradation activity, but it inhibited SPP cleavage of precursors (not shown). The inhibitor profile found for the degradation of transit peptide subfragments suggested that this reaction is carried out by a metallopeptidase that is most probably distinct from SPP.
We tested whether degradation activity is ATP-dependent. A chloroplast extract was preincubated at different concentrations of apyrase to hydrolyze ATP. The subfragment of the FD transit peptide was subsequently added to assay degradation. We found a decrease of degradation with increasing concentration of apyrase (Fig. 7 b). At the highest apyrase concentration, 100 units/ml, 86% of the originally added substrate remained after 30 min incubation. If ATP (20 mM) was added back to this reaction, 49% of the original substrate was detected, demonstrating that ATP partially restored degradation activity. No significant inhibition of degradation was observed when the extract was preincubated with apyrase inactivated by boiling. These results indicate that an ATP-dependent protease is involved in transit peptide turnover.
Other properties of the degradation activity were established. Using a chloroplast extract clarified by ultracentrifugation (1 h at 139,000 g), all activity was found in the supernatant and no significant degradation was observed using the pellet containing the membrane vesicle fraction (not shown). Thus, the degradation activity is soluble and does not reside in the membrane. Furthermore, this activity showed a broad pH optimum (between pH 5.0 and 9.0, using 20 mM Hepes-KOH buffer). Complete substrate degradation occurred within 30 min at temperatures between 24°C and 42°C. It was slightly reduced at 16°C, whereas 43% of the added subfragment remained at 4°C. Taken together, these results show that SPP trims transit peptides to subfragments that are specifically degraded by an ATP-dependent, soluble metallopeptidase activity with broad pH and temperature optima.
Discussion
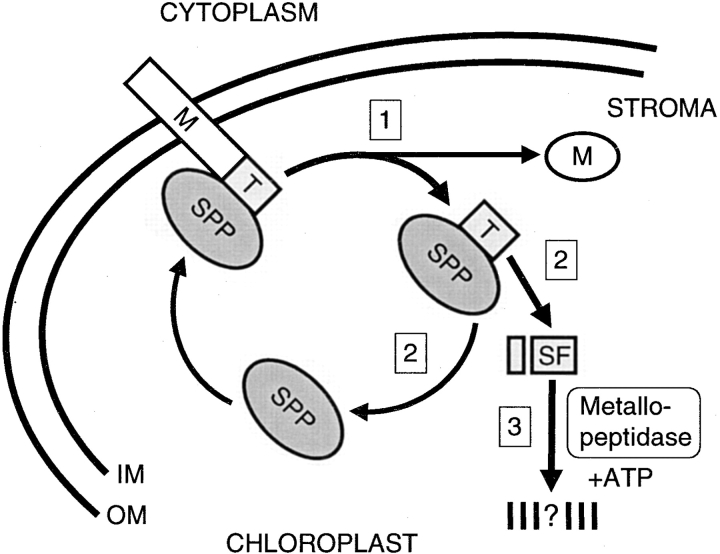
The experiments presented in this study provide evidence that transit peptide removal and turnover are regulated processes, and significantly, degradation of the transit peptide is an ATP-dependent step not previously recognized in the general import pathway. Our observations have been incorporated into a model (Fig. 8) that makes a number of important predictions about how SPP carries out its function during precursor import.
Figure 8.
Schematic representation of transit peptide removal and turnover in the chloroplast. Step 1, A precursor entering the stroma with its NH2-terminal transit peptide first is recognized by SPP. A single endoproteolytic cleavage by SPP releases the mature protein. The transit peptide remains bound by SPP. Step 2, SPP releases the transit peptide by its conversion to a subfragment. The regenerated SPP is free for a new enzyme–substrate interaction. Step 3, The liberated subfragment is degraded by an ATP-dependent, soluble metallopeptidase.
First, we propose that SPP initially recognizes, binds, and cleaves the transit peptide from the precursor as it is translocated across the inner membrane. This is based on our finding that SPP has a strong affinity for the transit peptide as a separate domain of the precursor, as demonstrated in both binding assays and time courses of precursor processing. It is also supported by studies showing that translocation intermediates can be cleaved before complete transport into the stroma, a stage when the precursor is likely to be in an unfolded conformation while still spanning the envelope membranes (Schnell and Blobel 1993; Schnell et al. 1994). Once the precursor is cleaved and the mature protein released, SPP continues to interact with the intact transit peptide, although the duration and equilibrium of this interaction remains to be fully explored.
Second, SPP then carries out a second processing reaction and converts the transit peptide to a subfragment form which it no longer recognizes. Specific, yet uncharacterized, features of the transit peptide are necessary to promote its conversion, which was shown by the failure of SPP to trim a mutated LHCP transit peptide with a three amino acid deletion near its COOH terminus. The dissociation of the subfragment also frees SPP, which becomes available for additional rounds of precursor processing at the inner membrane. Thus, transit peptide trimming may serve as a recycling step for SPP. In fact, immobilized SPP can be used after a processing reaction again in another reaction (Richter, S., and G. Lamppa, unpublished result).
Third, we predict that the subfragment, upon being released from SPP, becomes a target for rapid ATP-dependent degradation by a stromal metallopeptidase. That the reaction depends on ATP suggests an energy requirement. Either the subfragment is degraded to oligopeptides no longer detectable in our assays, or it is fully hydrolyzed to free amino acids. Overall, we propose a regulated sequence of events that is mediated by SPP and results in transit peptide turnover by an ATP-dependent proteolytic machinery.
Aspects of the model (Fig. 8) may apply to certain other members of the pitrilysin family that share the His-X-X-Glu-His zinc-binding motif (Rawlings and Barrett 1995). Representatives include the mitochondrial processing peptidase (MPP; Hawlitschek et al. 1988; Yang et al. 1988), the human insulin-degrading enzyme responsible for β-endorphin processing among other proposed functions (Safavi et al. 1996), and a yeast homologue of insulin-degrading enzyme needed for processing of pro-a-factor (Adames et al. 1995). In general, we propose that each metalloendopeptidase recognizes a precursor with a removable NH2- or COOH-terminal extension peptide because of the enzyme's high affinity for this domain. This interaction determines the substrate specificity of the peptidase. A single endoproteolytic cleavage occurs that immediately releases the mature product. However, to disrupt the interaction with the cleaved extension peptide, the enzyme carries out one or more internal cleavages of the extension, which may yield a subfragment for complete degradation.
The observation that SPP specifically binds the transit peptide, but does not interact with its subfragment, suggested that another proteolytic activity exists in the chloroplast for transit peptide turnover. Indeed, the degradation activity identified in our work can be distinguished from SPP. SPP activity is ATP-independent in vitro (Robinson and Ellis 1984; Oblong and Lamppa 1992) and is blocked by the sulfhydryl group inhibitor NEM. Furthermore, SPP was found to be about tenfold less sensitive to the metal ion chelator 1,10-phenanthroline than the degradation activity. Little is known about the pathways employed by the chloroplast for protein degradation in general. Two ATP-dependent proteolytic systems have been described, but their natural substrates in the organelle remain to be established. One system is comprised of ClpP and ClpC, which are homologues of E. coli serine protease ClpP and a chaperone-like ClpA activity (Moore and Keegstra 1993; Shanklin et al. 1995). The other system is the homologue of E. coli FtsH, a membrane anchored metallopeptidase (Ostersetzer and Adam 1997). However, neither proteolytic system fully resembles the activity responsible for transit peptide subfragment degradation. On the one hand, we found that the degradation activity is insensitive to the serine protease inhibitor PMSF. On the other, the degradation activity was found in the soluble fraction and not the thylakoid membranes where FtsH is localized in chloroplasts. At present, it appears that a novel activity separate from SPP is needed for transit peptide turnover.
SPP cleaves a large diversity of precursors targeted to the chloroplast, all with transit peptides of different length and primary sequence (see introduction). Despite this variability, our results indicate that the transit peptide alone contains sufficient information to mediate the interaction of the precursor with SPP. What does SPP recognize that determines a highly specific association with the intact transit peptide? There are some indications that critical determinants for processing are located near the COOH terminus of the transit peptide (Clark and Lamppa 1991; Archer and Keegstra 1993; Bassham et al. 1994; Pilon et al. 1995). As a corollary to this question, where in SPP is the binding site for the transit peptide? One can speculate where the binding domain of SPP will be found based on a comparison with the structure of MPP. Unlike SPP, which is a single polypeptide of ∼125 kD (based on its primary sequence), MPP is comprised of α and β subunits (Hawlitschek et al. 1988; Yang et al. 1988). Only subunit α is necessary for the binding step of the processing reaction (Luciano et al. 1997; Shimokata et al. 1998). Subunit β contains the catalytic His-X-X-Glu-His zinc-binding motif within a domain that shows sequence conservation with the NH2 terminus of SPP (Kitada et al. 1995; Striebl et al. 1996). Thus, it is reasonable to predict that the transit peptide binding site of SPP will be found in a downstream COOH-terminal region. The binding site of SPP, as well as that of MPP, remains to be mapped, and its structural features characterized in order to elucidate the molecular mechanism of precursor recognition and cleavage.
What is the nature of the proteolytic machinery that is responsible for transit peptide degradation? As discussed above, our current results suggest that a novel activity, separate from SPP, functions after transit peptide conversion to a subfragment form. However, an answer to this question depends on knowing more about the identity of the degradation activity. Another important goal is to establish which step during transit peptide turnover exhibits an ATP requirement. Given the enormous amount of transit peptide that must be digested as proteins are imported, the degradation process is likely to be essential for normal chloroplast biogenesis.
Acknowledgments
This work was supported in part by National Science Foundation grant MCB-9407739, U.S. Department of Agriculture grant 98-35304-6744, and The University of Chicago Biological Sciences Faculty Research Fund-97.
Footnotes
1.used in this paper: FD, ferredoxin; HSP21, heat shock protein 21; LHCP, light-harvesting chlorophyll a/b binding protein; MPP, mitochondrial processing peptidase; NEM, N-ethylmaleimide; preFD, precursor ferredoxin; preHSP21, precursor heat shock protein 21; preLHCP, precursor light-harvesting chlorophyll a/b binding protein; preRBCA, precursor ribulose-1,5-bisphosphate carboxylase/oxygenase activase; preRBCS, precursor ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit; RBCA, ribulose-1,5-bisphosphate carboxylase/oxygenase activase; RBCS, ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit; SPP, stromal processing peptidase
References
- Abad M.S., Clark S.E., Lamppa G.K. Properties of a chloroplast enzyme that cleaves the chlorophyll a/b binding protein precursor. Plant Physiol. 1989;90:117–124. doi: 10.1104/pp.90.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adames N., Blundell K., Ashby M.N., Boone C. Role of yeast insulin-degrading enzyme homologs in propheromone processing and bud site selection. Science. 1995;270:464–467. doi: 10.1126/science.270.5235.464. [DOI] [PubMed] [Google Scholar]
- Archer E.K., Keegstra K. Analysis of chloroplast transit peptide function using mutations in the carboxyl-terminal region. Plant Mol. Biol. 1993;23:1105–1115. doi: 10.1007/BF00042345. [DOI] [PubMed] [Google Scholar]
- Bassham D.C., Creighton A.M., Karnauchov I., Herrmann R.G., Klösgen R.B., Robinson C. Mutations at the stromal processing peptidase cleavage site of a thylakoid lumen protein precursor affect the rate of processing but not the fidelity. J. Biol. Chem. 1994;269:16062–16066. [PubMed] [Google Scholar]
- Becker A.B., Roth R.A. An unusual active site identified in a family of zinc metalloendopeptidases. Proc. Natl. Acad. Sci. USA. 1992;89:3835–3839. doi: 10.1073/pnas.89.9.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Lamppa G.K. Determinants for cleavage of the chlorophyll a/b binding protein precursora requirement for a basic residue that is not universal for chloroplast imported proteins. J. Cell Biol. 1991;114:681–688. doi: 10.1083/jcb.114.4.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Abad M.S., Lamppa G.K. Mutations at the transit peptide–mature protein junction separate two cleavage events during chloroplast import of the chlorophyll a/b-binding protein. J. Biol. Chem. 1989;264:17544–17550. [PubMed] [Google Scholar]
- Cline K., Henry R. Import and routing of nucleus-encoded chloroplast proteins. Annu. Rev. Cell Dev. Biol. 1996;12:1–26. doi: 10.1146/annurev.cellbio.12.1.1. [DOI] [PubMed] [Google Scholar]
- Coruzzi G., Broglie R., Edwards C., Chua N.H. Tissue-specific and light-regulated expression of a pea nuclear gene encoding the small subunit of ribulose-1,5-bisphosphate carboxylase. EMBO (Eur. Mol. Biol. Organ.) J. 1984;3:1671–1679. doi: 10.1002/j.1460-2075.1984.tb02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks B., Schnell D.J. Mechanism of protein transport across the chloroplast envelope. Plant Physiol. 1997;114:405–410. doi: 10.1104/pp.114.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavel Y., von Heijne G. A conserved cleavage-site motif in chloroplast transit peptides. FEBS Lett. 1990;261:455–458. doi: 10.1016/0014-5793(90)80614-o. [DOI] [PubMed] [Google Scholar]
- Hawlitschek G., Schneider H., Schmidt B., Tropschug M., Hartl F.U., Neupert W. Mitochondrial protein importidentification of processing peptidase and of PEP, a processing enhancing protein. Cell. 1988;53:795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- Kitada S., Shimokata K., Niidome T., Ogishima T., Ito A. A putative metal-binding site in the beta subunit of rat mitochondrial processing peptidase is essential for its catalytic activity. J. Biochem. 1995;117:1148–1150. doi: 10.1093/oxfordjournals.jbchem.a124836. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamppa G.K., Abad M.S. Processing of a wheat light-harvesting chlorophyll a/b protein precursor by a soluble enzyme from higher plant chloroplasts. J. Cell Biol. 1987;105:2641–2648. doi: 10.1083/jcb.105.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamppa G.K., Morelli G., Chua N.H. Structure and developmental regulation of a wheat gene encoding the major chlorophyll a/b-binding polypeptide. Mol. Cell. Biol. 1985;5:1370–1378. doi: 10.1128/mcb.5.6.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübeck J., Heins L., Soll J. Protein import into chloroplasts. Physiol. Plant. 1997;100:53–64. [Google Scholar]
- Luciano P., Geoffroy S., Brandt A., Hernandez J.F., Geli V. Functional cooperation of the mitochondrial processing peptidase subunits. J. Mol. Biol. 1997;272:213–225. doi: 10.1006/jmbi.1997.1231. [DOI] [PubMed] [Google Scholar]
- Moore T., Keegstra K. Characterization of a cDNA clone encoding a chloroplast-targeted Clp homologue. Plant Mol. Biol. 1993;21:525–537. doi: 10.1007/BF00028809. [DOI] [PubMed] [Google Scholar]
- Oblong J.E., Lamppa G.K. Identification of two structurally related proteins involved in proteolytic processing of precursors targeted to the chloroplast. EMBO (Eur. Mol. Biol. Organ.) J. 1992;11:4401–4409. doi: 10.1002/j.1460-2075.1992.tb05540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostersetzer O., Adam Z. Light-stimulated degradation of an unassembled Rieske FeS protein by a thylakoid-bound proteasethe possible role of the FtsH protease. Plant Cell. 1997;9:957–965. doi: 10.1105/tpc.9.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman R.K., Gehm B.D., Kuo W.L., Rosner M.R. Functional analysis of conserved residues in the active site of insulin-degrading enzyme. J. Biol. Chem. 1993;268:21538–21544. [PubMed] [Google Scholar]
- Pfisterer J., Lachmann P., Kloppstech K. Transport of proteins into chloroplasts. Binding of nuclear-coded chloroplast proteins to the chloroplast envelope. Eur. J. Biochem. 1982;126:143–148. doi: 10.1111/j.1432-1033.1982.tb06758.x. [DOI] [PubMed] [Google Scholar]
- Pilon M., Weisbeek P.J., de Kruijff B. Kinetic analysis of translocation into isolated chloroplasts of the purified ferredoxin precursor. FEBS Lett. 1992;302:65–68. doi: 10.1016/0014-5793(92)80286-p. [DOI] [PubMed] [Google Scholar]
- Pilon M., Wienk H., Sips W., de Swaaf M., Talboom I., van't Hof R., de Korte-Kool G., Demel R., Weisbeek P., de Kruijff B. Functional domains of the ferredoxin transit sequence involved in chloroplast import. J. Biol. Chem. 1995;270:3882–3893. doi: 10.1074/jbc.270.8.3882. [DOI] [PubMed] [Google Scholar]
- Rawlings N.D., Barrett A.J. Evolutionary families of metallopeptidases. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- Richter S., Lamppa G.K. A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc. Natl. Acad. Sci. USA. 1998;95:7463–7468. doi: 10.1073/pnas.95.13.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C., Ellis R.J. Transport of proteins into chloroplasts. Partial purification of a chloroplast protease involved in the processing of important precursor polypeptides. Eur. J. Biochem. 1984;142:337–342. doi: 10.1111/j.1432-1033.1984.tb08291.x. [DOI] [PubMed] [Google Scholar]
- Safavi A., Miller B.C., Cottam L., Hersh L.B. Identification of gamma-endorphin-generating enzyme as insulin-degrading enzyme. Biochem. 1996;35:14318–14325. doi: 10.1021/bi960582q. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schnell D.J., Blobel G. Identification of intermediates in the pathway of protein import into chloroplasts and their localization to envelope contact sites. J. Cell Biol. 1993;120:103–115. doi: 10.1083/jcb.120.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell D.J., Kessler F., Blobel G. Isolation of components of the chloroplast protein import machinery. Science. 1994;266:1007–1012. doi: 10.1126/science.7973649. [DOI] [PubMed] [Google Scholar]
- Shanklin J., DeWitt N.D., Flanagan J.M. The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpAan archetypal two-component ATP-dependent protease. Plant Cell. 1995;7:1713–1722. doi: 10.1105/tpc.7.10.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokata K., Kitada S., Ogishima T., Ito A. Role of alpha-subunit of mitochondrial processing peptidase in substrate recognition. J. Biol. Chem. 1998;273:25158–25163. doi: 10.1074/jbc.273.39.25158. [DOI] [PubMed] [Google Scholar]
- Smeekens S., van Binsbergen J., Weisbeek P. The plant ferredoxin precursornucleotide sequence of a full length cDNA clone. Nucleic Acids Res. 1985;13:3179–3194. doi: 10.1093/nar/13.9.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebl H.-M., Rysavy P., Adamec J., Spizek J., Kalousek F. Mutational analysis of both subunits from rat mitochondrial processing peptidase. Arch. Biochem. Biophys. 1996;335:211–218. doi: 10.1006/abbi.1996.0500. [DOI] [PubMed] [Google Scholar]
- VanderVere P.S., Bennett T.M., Oblong J.E., Lamppa G.K. A chloroplast processing enzyme involved in precursor maturation shares a zinc-binding motif with a recently recognized family of metalloendopeptidases. Proc. Natl. Acad. Sci. USA. 1995;92:7177–7181. doi: 10.1073/pnas.92.16.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Hof R., de Kruijff B. Characterization of the import process of a transit peptide into chloroplasts. J. Biol. Chem. 1995;270:22368–22373. doi: 10.1074/jbc.270.38.22368. [DOI] [PubMed] [Google Scholar]
- Vierling E., Nagao R.T., DeRocher A.E., Harris L.M. A heat shock protein localized to chloroplasts is a member of a eukaryotic superfamily of heat shock proteins. EMBO (Eur. Mol. Biol. Organ.) J. 1988;7:575–581. doi: 10.1002/j.1460-2075.1988.tb02849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Bringloe D., Lamppa G.K. Disruption of chloroplast biogenesis and plant development upon down-regulation of a chloroplast processing enzyme involved in the import pathway. Plant J. 1998;15:459–468. [Google Scholar]
- Werneke J.M., Zielinski R.E., Ogren W.L. Structure and expression of spinach leaf cDNA encoding ribulosebisphosphate carboxylase/oxygenase activase. Proc. Natl. Acad. Sci. USA. 1988;85:787–791. doi: 10.1073/pnas.85.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Jensen R.E., Yaffe M.P., Oppliger W., Schatz G. Import of proteins into yeast mitochondriathe purified matrix processing protease contains two subunits which are encoded by the nuclear MAS1 and MAS2 genes. EMBO (Eur. Mol. Biol. Organ.) J. 1988;7:3857–3862. doi: 10.1002/j.1460-2075.1988.tb03271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]