Abstract
The overall DNA methylation level sharply decreases from the zygote to the blastocyst stage despite the presence of high levels of DNA methyltransferase (Dnmt1). Surprisingly, the enzyme is localized in the cytoplasm of early embryos despite the presence of several functional nuclear localization signals. We mapped a region in the NH2-terminal, regulatory domain of Dnmt1 that is necessary and sufficient for cytoplasmic retention during early development. Altogether, our results suggest that Dnmt1 is actively retained in the cytoplasm, which prevents binding to its DNA substrate in the nucleus and thereby contributes to the erasure of gamete-specific epigenetic information during early mammalian development.
Keywords: cytoplasmic retention, DNA methylation, DNA methyltransferase, genomic imprinting, preimplantation development
DNA methylation plays an important role in the regulation of gene expression and is essential for normal mammalian development. Until now very little has been known about the regulation of DNA methylation in mammalian cells. The predominant DNA methyltransferase, Dnmt11, is also involved in genomic imprinting and X-inactivation (Li et al. 1993; Panning and Jaenisch 1996), and Dnmt1-deficient mice die at mid-gestation (Li et al. 1992). Dnmt1 is composed of a NH2-terminal, regulatory domain and a COOH-terminal, catalytic domain that is closely related to the bacterial C5-methyltransferases (Bestor et al. 1988; Leonhardt and Bestor 1993). To date, two isoforms of Dnmt1 have been described, a longer, somatic form of ∼190 kD (Tucker et al. 1996; Yoder et al. 1996) and a shorter, oocyte-specific form of ∼170 kD that is generated by an alternative promoter and translation from a downstream start site (Gaudet et al. 1998; Mertineit et al. 1998).
Recently, three additional DNA methyltransferases, Dnmt2 (Okano et al. 1998b; Van den Wyngaert et al. 1998; Yoder and Bestor 1998) and Dnmt3 α and β (Okano et al. 1998a), have been identified but their role in vivo is still unknown.
The most dramatic changes in the DNA methylation pattern occur during gametogenesis and early development (Monk et al. 1987; Sanford et al. 1987). During oogenesis and spermatogenesis sex-specific methylation patterns are established that are essentially retained throughout development (Olek and Walter 1997; Tremblay et al. 1997) and control the expression of imprinted genes (reviewed in Constancia et al. 1998; Latham et al. 1995).
During preimplantation development the overall methylation level sharply decreases and reaches a low point at the blastocyst stage (Monk et al. 1987; Razin and Shemer 1995). The mechanism responsible for this demethylation is still unknown. DNA methylation activity (Howlett and Reik 1991; Monk et al. 1991) and Dnmt1 protein level are very high in oocytes and preimplantation embryos but immunofluorescence studies have shown that Dnmt1 is localized in the cytoplasm at these stages (Carlson et al. 1992; Mertineit et al. 1998).
We report here the mapping of a region in the regulatory domain of Dnmt1 that is necessary and sufficient for cytoplasmic localization during early development. Since the same region is also present in the nuclear, somatic form of Dnmt1, we postulate interacting factor(s) that are expressed during early development and sequester Dnmt1 in the cytoplasm. This separation of Dnmt1 from its nuclear DNA substrates may contribute to the regulation of DNA methylation during early development.
Materials and Methods
Translational Fusion Constructs
All translational fusions were generated by standard cloning protocols using the previously described plasmid pEMT that expresses the shorter, oocyte-specific Dnmt1 isoform (Czank et al. 1991). Dnmt1 sequences present in the fusions are indicated in the figures. In addition, amino acids 361–1061 of the Escherichia coli β-galactosidase were added in-frame at the COOH terminus. In two cases, a short nuclear localization signal (NLS) derived from SV-40 large T antigen (PKKKRKV) was added at the NH2 terminus of some deletion constructs. Plasmid DNA was purified using Qiagen columns according to the manufacturer's instructions.
Cell Culture and Transfection
Monkey COS1 cells and mouse fibroblast cells (C3H10T1/2 and NIH3T3) were grown in a humidified incubator at 37°C and 5% CO2 in DME supplemented with 10% fetal calf serum. Mouse C2C12 myoblast cells were grown as above except that the media was supplemented with 20% fetal calf serum.
COS1 cells were transfected with the DEAE-dextran pretreatment method and 2 d later, cells were scraped, extracted, and the fusion proteins analyzed by Western blotting as described in Leonhardt et al. 1992.
Mouse myoblast and fibroblast cells were transfected by the calcium phosphate-DNA coprecipitation method followed by glycerol shock treatment ∼8–12 h later. 36–48 h after DNA addition, cells were fixed and stained for the localization of the fusion protein.
Cultivation and Microinjection of Mouse Zygotes
Mouse embryos were obtained from FVB superovulated female mice mated with C57BL/6 males. Female mice (∼3–4 wk old) were injected intraperitoneally with 5 IU of pregnant mare's serum (PMS) followed by an intraperitoneal injection of 5 IU of human chorionic gonadotropin (HGG) 46–48 h later to induce superovulation, which is necessary to obtain an adequate amount of fertilized eggs upon mating. Matings were set up with fertile stud males after the HGG injection.
Zygotes were collected from the oviduct of 0.5 d post-coitum (p.c.) donors into flushing-holding medium (FHM) containing hyaluronidase (0.65 mg/ml), to remove the cumulus cells. After several washes in FHM, the eggs were transferred to kSOM medium in microdrop cultures and incubated in a humidified CO2 chamber at 37°C. Culture conditions and media compositions have been described in detail (Lawitts and Biggers 1993; Hogan et al. 1994).
For injection, DNA fragments containing all the regulatory sequences but lacking the prokaryotic vector part were used. DNA was purified with standard glassmilk absorption techniques (Qiagen), resuspended in TE buffer at a final concentration of 1–10 μg/ml. On average 1–2 pl were injected into one of the pronuclei.
Immunofluorescence and Microscopy
Tissue culture fibroblast or myoblast cells were washed once with PBS, fixed for 10 min in 3.7% formalin in PBS, and permeabilized with 0.25% Triton X-100 in PBS for 10 min. After blocking 30 min in 3% BSA or 0.2% gelatin in PBS, cells were incubated for 60 min with mouse anti–β-galactosidase antibody (Promega) diluted 1:1,000. After extensive washes with 0.1% NP-40 in PBS, cells were incubated for another 60 min in FITC-conjugated goat anti–mouse (Boehringer Mannheim) or in biotinylated goat anti–mouse IgG antibodies (Amersham), the latter followed by washing and 30 min incubation in streptavidin–Texas red (Amersham) as described before (Cardoso et al. 1993). DNA was counterstained with Hoechst 33258 and cells were mounted in mowiol with 2.5% DABCO (Cardoso and Leonhardt 1995). All dilutions were made in 3% BSA or 0.2% gelatin in PBS, and all incubations were carried out at room temperature.
Specimens were examined and photographed on Zeiss Axiophot and Axiovert microscopes equipped with phase-contrast and epifluorescence optics, using 63× and 100× plan-apochromat and plan-neofluor lenses. Pictures were taken with Kodak Ektar 100 film.
Mouse embryos were fixed and stained in microtiter plate wells and moved from one solution to the other with handmade capillaries under a stereo microscope. The staining procedure was as above using mouse anti–β-galactosidase antibody or rabbit antisera to Dnmt1 (Leonhardt et al. 1992), the latter followed by FITC-conjugated goat anti–rabbit IgG antibody (Boehringer Mannheim). Embryos were mounted in mowiol with 2.5% DABCO on 8-well multitest slides.
Stained embryos were analyzed on a Zeiss Axiophot microscope equipped with differential interference contrast (DIC), epifluorescence, and confocal laser scanning system using 40× and 63× plan-neofluor lenses. Epifluorescence and DIC images were recorded on film as described above. Optical sections were obtained using the Bio-Rad MRC-600 confocal imaging system and micrographs were taken on Kodak Tmax 400 film. Images were scanned, assembled, and annotated with Adobe Photoshop on a Power Macintosh computer and printed on a Phaser 440 dye sublimation printer (Tektronix).
Results and Discussion
Cytoplasmic Localization of Dnmt1 during Preimplantation Development
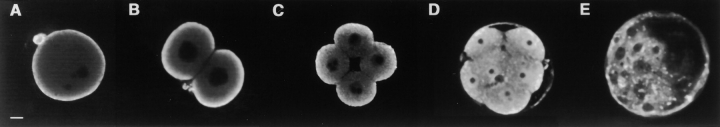
To study the mechanism responsible for the cytoplasmic localization of Dnmt1 during preimplantation development, we first tested whether this phenomenon can be reproduced in embryos that are cultured in vitro. Fertilized mouse eggs were collected at the one-cell stage and were cultured in vitro for up to 4 d. Under optimal conditions embryos develop until the blastocyst stage. Embryos were collected and fixed at different stages of preimplantation development and then stained for Dnmt1 with a specific polyclonal antiserum raised against the NH2-terminal domain of Dnmt1. The localization of Dnmt1 was then determined by confocal analyses (Fig. 1). Dnmt1 was localized in the cytoplasm of one-, two-, and four-cell embryos as well as in blastocysts. Only at the eight-cell stage was the enzyme found in the nucleus. These results are in good agreement with previous studies on naturally developed embryos (Carlson et al. 1992), and show that the developmentally controlled subcellular localization of Dnmt1 can be reproduced with in vitro cultured embryos.
Figure 1.
Localization of Dnmt1 in early mouse embryos. During preimplantation development, Dnmt1 localizes to the cytoplasm of one- (A), two- (B), and four- (C) cell embryos, it is partly imported into the nuclei with the remaining protein staying in the cytoplasm at the eight-cell stage (D), and it is again out of the nucleus at the blastocyst stage (E). Fertilized mouse embryos were collected and incubated in vitro. At different times, the embryos were fixed and stained with an anti-Dnmt1 polyclonal antibody and analyzed by confocal microscopy. The images show single confocal sections through embryos at different stages of development. Bar, 10 μm.
Dnmt1 Contains Several Functional NLS
In somatic cells, Dnmt1 is strictly localized in the nucleus and is targeted to replication foci during S-phase (Leonhardt et al. 1992). This raises the question how Dnmt1 enters the nucleus of somatic cells but remains in the cytoplasm during most of preimplantation development.
One possible explanation for this different localization could be the fact that different isoforms are expressed in these cells. Indeed, the Dnmt1 gene is transcribed from different promoters and a 118–amino acid shorter isoform of Dnmt1 was identified in oocytes and preimplantation embryos (Gaudet et al. 1998; Mertineit et al. 1998).
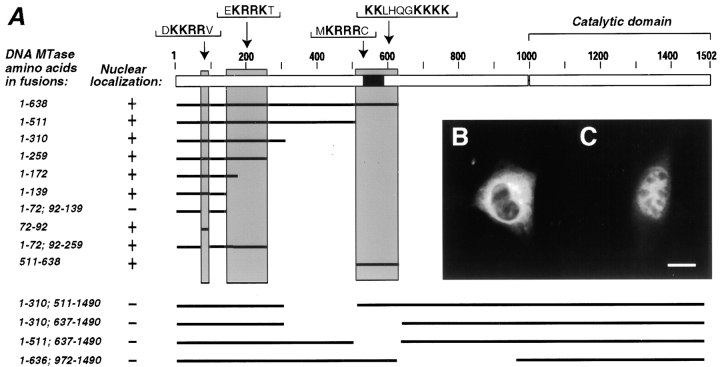
Therefore, we expressed this shorter isoform in different somatic cell lines including COS1, mouse fibroblasts, and myoblast cells. For visualization by immunofluorescence, the oocyte-specific isoform was fused with an unrelated β-galactosidase epitope. In all tested somatic cell lines the fusion protein showed a clear nuclear localization like the longer somatic isoform (data not shown). To further study the regulation of the nuclear uptake of Dnmt1, we generated a set of deletion mutants and determined their subcellular localization (Fig. 2). A series of deletions showed that the first 139 amino acids of the oocyte-specific form are sufficient for nuclear uptake of the β-galactosidase fusion in somatic cells. Deletion of one candidate NLS (KKRR from position 72–92) prevented nuclear uptake. This region clearly contains a functional NLS since this sequence alone allows nuclear uptake of the fusion protein (Fig. 2). The regulatory domain of Dnmt1, however, contains further stretches of basic amino acids that could serve as NLS. Further deletion constructs identified at least two independent additional NLS (positions 140–259 and 511–638). These results clearly show that the oocyte-specific Dnmt1 form enters the nucleus of somatic cells and contains at least three independent NLS. Though these three regions function as NLS, a set of internal deletions (from positions 310–637, 310–511, 511–637, and 636–972) that retain all three NLS or at least the two first NLS show cytoplasmic localization of the fusion protein in somatic cells (Fig. 2). Similar deletions constructs (not fused to β-galactosidase) expressed in mammalian cells are enzymatically inactive (Margot, J.B., A.M. Aguirre-Arteta, V. Di Giacco, S. Pradhan, R. Roberts, M.C. Cardoso, and H. Leonhardt, manuscript submitted for publication) suggesting an important role of this region in the proper folding of Dnmt1, rather than a specific effect on nuclear localization.
Figure 2.
Dnmt1 has at least three independent nuclear localization signals. (A) Diagram outlining the structure of the oocyte-specific murine Dnmt1 protein and indicating the different parts of the Dnmt1 present in fusions with the β-galactosidase epitope (for simplicity, the latter is not depicted). The different fusion constructs were expressed in murine myoblast or fibroblast cells, formalin fixed, stained with anti–β-galactosidase monoclonal antibody, and screened for their nuclear or cytoplasmic localization. B shows a representative example of a cytoplasmic fusion protein containing amino acids 1-72; 92-139 of Dnmt1 and C shows a nuclear fusion protein containing amino acids 1-72; 92-259 of Dnmt1, including only the second NLS. The analysis of all listed constructs lead to the identification of three regions (highlighted by grey shading) that contain independently functional NLS. Sequences representing possible NLS within each shaded area are specified below. The black box indicates the location of the cysteine-rich region that had been shown to bind zinc ions (Bestor 1992). Bar, 10 μm.
Mapping of a Cytoplasmic Retention Sequence in Dnmt1
The fact that Dnmt1 contains at least three independently functional NLS raises the question of how the oocyte-specific form is maintained in the cytoplasm during early development. This cytoplasmic localization of Dnmt1 could be caused by alternative splicing leading to the expression of an isoform without functional NLS. Alternatively, trans-acting factors could be present in the oocyte and early embryo that prevent nuclear localization of Dnmt1. To test these hypotheses, we established an experimental approach to analyze deletion constructs of Dnmt1 in early embryos (Fig. 3).
Figure 3.
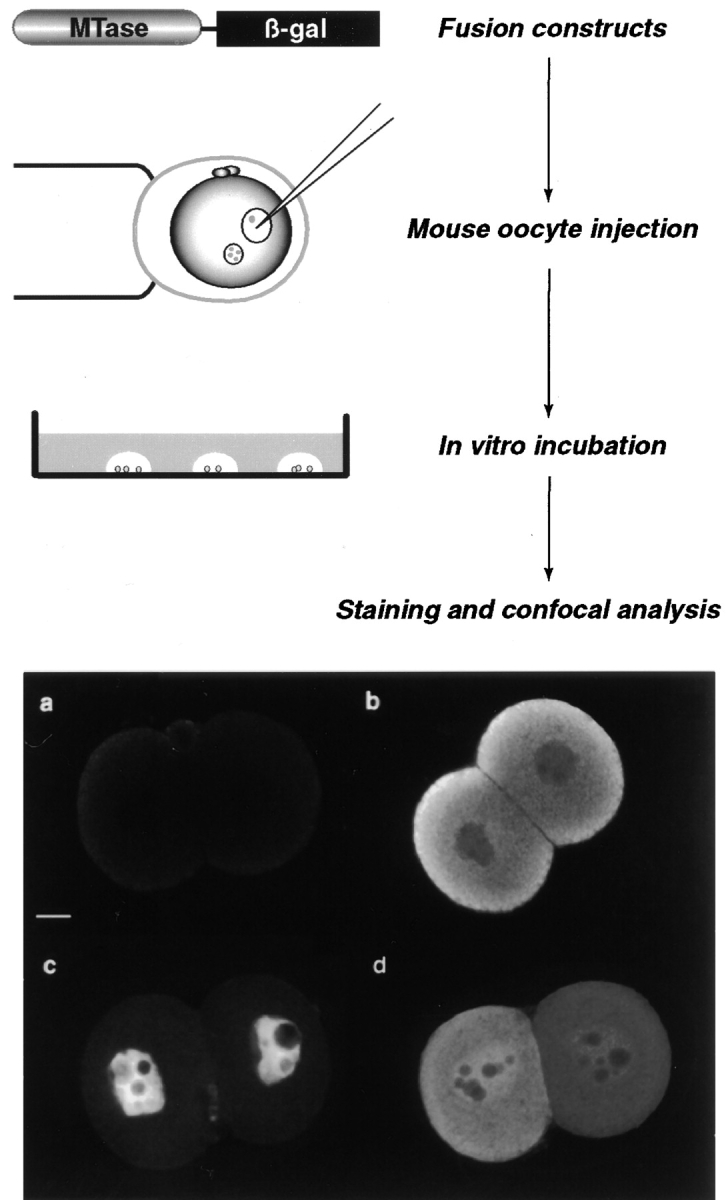
Experimental strategy to map domains of Dnmt1 responsible for cytoplasmic localization in preimplantation mouse embryos. A series of translational fusions containing different parts of Dnmt1 in frame with β-galactosidase was constructed in mammalian expression vectors. The corresponding plasmid DNA was then microinjected into one of the pronuclei of fertilized mouse eggs. The injected eggs were incubated for 1 or 2 d in vitro, after which they were formalin fixed and stained with anti–β-galactosidase mAb followed by FITC or rhodamine-conjugated secondary Ab. After mounting, the localization of the fusion protein was analyzed by confocal laser scanning microscopy. The background level was evaluated in each injection series by staining and analyzing TE buffer injected embryos in parallel with plasmid DNA (diluted in TE) injected ones. An optical section of one such TE negative control embryos is shown in a. In order for this assay to work, the oocyte-specific Dnmt1 fused to β-galactosidase should mimic the endogenous Dnmt1 protein localization. One representative confocal section through an embryo injected with a construct containing amino acids 1–1,490 is shown in b and exhibits the same cytoplasmic localization as the endogenous protein (Fig. 1 and Carlson et al. 1992). As a further step in testing this experimental strategy, some deletion mutants of Dnmt1 should localize in the nucleus and therefore be scored as negative for cytoplasmic retention. In c is depicted one confocal section through an embryo injected with an expression plasmid containing only the first 259 amino acids of the oocyte-specific Dnmt1 isoform fused to β-galactosidase. This fusion protein is clearly not retained in the cytoplasm as the endogenous Dnmt1 at this stage of the preimplantation development. Instead, it shows exclusive nuclear localization with the exception of the nucleolar compartment as does the Dnmt1 present in somatic cells (Leonhardt et al. 1992). Finally, an intermediate phenotype with less efficient cytoplasmic retention is observed. In d is shown a confocal section through an embryo injected with an expression construct containing amino acids 1–638 of the oocyte-specific Dnmt1 isoform illustrating this latter phenotype. Bar, 10 μm.
To identify sequences controlling subcellular localization in early embryos and to visualize truncated proteins by immunofluorescence, we fused the shorter, oocyte-specific open reading frame of Dnmt1 with the unrelated β-galactosidase gene. These fusion constructs were cloned into a mammalian expression vector and injected into fertilized mouse eggs. These injected eggs were cultured in microdrops and fixed and stained at different time points. Fusion proteins were detected with β-galactosidase-specific antibodies and their localization analyzed by confocal microscopy. Fig. 3 b shows a two-cell embryo expressing an almost full-length Dnmt1 (amino acids 1–1,490) fused to β-galactosidase. This fusion protein is clearly localized in the cytoplasm just like the endogenous Dnmt1 protein at this stage. The same fusion protein has also been tested in somatic cells and was found in the nucleus ruling out possible artifacts caused by the fusion (data not shown). In other words, this experimental system reproduces the subcellular localization of Dnmt1: the fusion protein is localized in the nucleus of somatic cells and in the cytoplasm of early embryos. Since embryos are known to have a high level of autofluorescence control embryos were injected with TE buffer alone and analyzed in parallel (Fig. 3 a). The comparison with expressing embryos shows that the obtained signals are clearly distinguishable from the autofluorescence. Moreover, a truncated fusion protein comprising amino acids 1–259 including the first two NLS of Dnmt1 was clearly localized in the nucleus of two-cell embryos (Fig. 3 c) ruling out unspecific retention of the fusion protein caused by the β-galactosidase part. Finally, a fusion protein containing amino acids 1–638 exhibited an intermediate phenotype with less efficient cytoplasmic retention (Fig. 3 d). These results clearly show that this experimental system is suitable for the identification of sequences that control subcellular localization during early development.
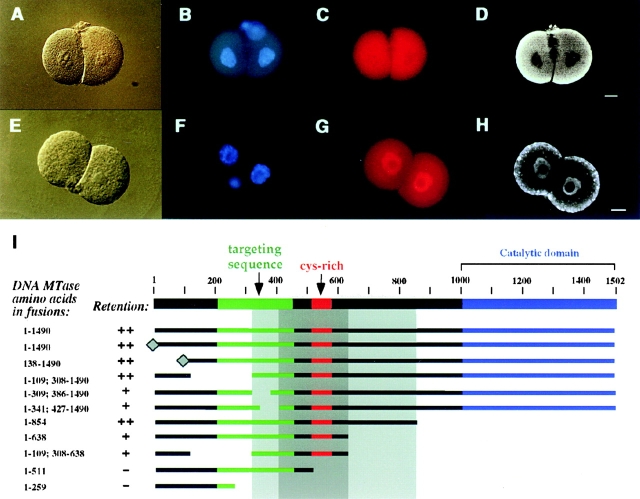
The comparison of the full-length and the truncated fusion protein in Fig. 3 suggests that Dnmt1 contains oocyte-specific retention sequences, since both fusions contain functional NLS. Therefore, we generated a set of deletions to map that putative retention sequence (Fig. 4). All constructs shown in this figure were first tested in somatic cells and showed a clear nuclear localization (data not shown). A deletion series coming from the COOH-terminal end indicated that the first 638 amino acids of the regulatory domain of Dnmt1 are sufficient for retention in the cytoplasm and that the catalytic domain is not required (Fig. 4 D, I). A similar deletion series starting from the NH2 terminus is not possible since the major NLS is located between amino acids 72 and 92. Therefore, internal deletions were done to further narrow down the region involved in cytoplasmic retention. These deletions showed that fusions containing the region from amino acid 308–854 are efficiently retained in the cytoplasm and constructs containing the region from amino acid 427–638 were still retained but less efficiently so that some signal could also be detected in the nucleus (see also Fig. 3 d). We propose that the binding interface mediating the cytoplasmic retention is complex and may involve several stretches of amino acids from different parts of the primary sequence. Deletion of some of these interacting parts may reduce but not totally abolish the affinity for the target(s). Similarly, deletions may affect the three-dimensional structure of the interface and thereby reduce the binding affinity and thus cause some leakage into the nucleus.
Figure 4.
Dnmt1 is actively retained in the cytoplasm of early mouse embryos via a sequence in the NH2-terminal, regulatory domain of the enzyme. A–D show the localization of a β-galactosidase fusion protein containing amino acids 1–638 of the oocyte-specific Dnmt1 isoform which is retained in the cytoplasm of two-cell mouse embryos. A shows the DIC image; B shows the DNA stained with Hoechst 33258; C shows the rhodamine detection of the β-galactosidase fusion protein; and D shows the corresponding confocal section clearly showing its cytoplasmic localization. E–H illustrate a cytoplasmic retention deficient β-galactosidase fusion protein containing amino acids 1–259 of the oocyte-specific Dnmt1 isoform, as DIC image (E), DNA staining (F), epifluorescence image of the fusion protein (G), and respective confocal section (H). The dotty cytoplasmic signal visible in the confocal section in H was not reproducible. Fertilized eggs were microinjected with the different fusion constructs schematically shown in I (β-galactosidase was fused at the COOH terminus and it is not shown), processed as described in Fig. 3, and assayed for their ability to be retained in the cytoplasm of two-cell mouse embryos. The previously identified targeting sequence that directs the protein to subnuclear sites of DNA replication (Leonhardt et al. 1992) is indicated in green. The diamond at the NH2 terminus of two fusions signifies the addition of an SV-40 large T antigen NLS. All the constructs listed show a clear nuclear localization in somatic cells (data not shown). The deletion endpoints of the different fusion proteins shown in I define a broad region between amino acids 308 and 854 of the oocyte-specific Dnmt1 isoform (light grey shading) which is necessary and sufficient for cytoplasmic retention (scored as ++) in preimplantation mouse embryos. Within this larger region the most crucial sequence (dark grey shading) lies between amino acids 427 and 638, since deletions in this region are clearly not retained in the cytoplasm (scored as −). Absence of amino acids 308–427 or 638–854 flanking this sequence caused a less efficient cytoplasmic retention (scored as +). Bars, 10 μm.
In the center of this retention sequence lies the cysteine-rich region that had been shown to bind zinc ions (Bestor 1992). To test whether this region is involved in the cytoplasmic retention we deleted this region in the full-length fusion construct. However, all deletions in this region as well as in neighboring regions caused an aberrant cytoplasmic localization even in somatic cells despite the fact that they all contained several functional NLS (Fig. 2 and data not shown).
One possible explanation could be that the remaining functional NLS were sequestered by an aberrant globular folding of the deletion construct. In fact, similar results were obtained in deletion studies to map the catalytic center of Dnmt1 (Margot, J.B., A.M. Aguirre-Arteta, V. Di Giacco, S. Pradhan, R. Roberts, M.C. Cardoso, and H. Leonhardt, manuscript submitted for publication; Zimmermann et al. 1997). Several deletions in the NH2-terminal domain affected the activity of the COOH-terminal, catalytic domain. These results suggest that the NH2-terminal domain has a complex folding that is required for efficient cytoplasmic retention as well as for protein functions located in other parts of the protein.
To test whether the retention sequence is dominant over heterologous NLS that are derived from an unrelated protein we added the SV-40 NLS at the NH2 terminus of the full-length fusion construct. As summarized in Fig. 4, the addition of the SV-40 NLS could not affect nuclear uptake suggesting that the regulatory domain of Dnmt1 in fact contains a dominant cytoplasmic retention sequence.
Dnmt1 Is Actively Sequestered in the Cytoplasm of Early Embryos
The fact that this retention sequence is dominant over a heterologous NLS suggests that it does not act through a specific modification or masking of the endogenous Dnmt1 NLS but through active retention by a retention factor and binding to some immobile cytoplasmic structures. Confocal images of endogenous Dnmt1 staining had shown stronger signal in the peripheral cytoplasm and were taken as indication that Dnmt1 binds to some structures at the cell membrane (Carlson et al. 1992). Similar results were also obtained in this study (Fig. 1) and were also reproduced with the fusion proteins (Fig. 3 and Fig. 4). However, very different results were obtained using regular fluorescence microscopy showing an uniform distribution throughout the cytoplasm (Fig. 4 C, G). Interestingly, the same embryos viewed by confocal microscopy showed again the peripheral signals (Fig. 4 D, H). These results suggest that the extreme peripheral staining signals seen in the confocal are most likely an optical artifact. Due to light absorption by the sample, the intensity of the excitation light decreases as it penetrates the specimen and the inner fluorophores are not excited to the same extent as the outer ones (Taylor and Salmon 1989). This inner filter effect is particularly evident in thick specimens like mouse embryos and results in stronger peripheral signal. However, in fluorescence microscopy, out of focus light from layers above and below is also detected compensating this absorption effect. In other words, a detailed localization of Dnmt1 is not possible with either method and further ultrastructural analyses by, e.g., electron microscopy, are needed to clarify this issue and to provide clues as to what structures Dnmt1 binds to in the cytoplasm of oocytes and early embryos.
An interesting possible mechanism could involve an allosteric control through interacting factors. Since the folding of the NH2-terminal domain (in particular amino acids 300–1,000) seems to be complex and required for functions residing in other part of the protein, it seems possible that interacting proteins might affect the globular folding of Dnmt1 and thereby inactivate the NLS similar to the internal deletions in somatic cells (see Fig. 2).
One further prediction from these experiments is that if Dnmt1 is actively retained rather than modified in an enzymatic process, then retention should be saturable. In fact, injection of a four times higher amount of plasmid DNA resulted in cytoplasmic and nuclear localization of the fusion protein (data not shown). Also longer incubation of injected embryos lead to an accumulation of the fusion protein and gradual uptake into the nucleus (data not shown). Although these results were clear, one has to keep in mind that injecting these high amounts of plasmid DNA disturbs normal development of mouse embryos. With this cautionary note, these results suggest that the retention of Dnmt1 is indeed saturable.
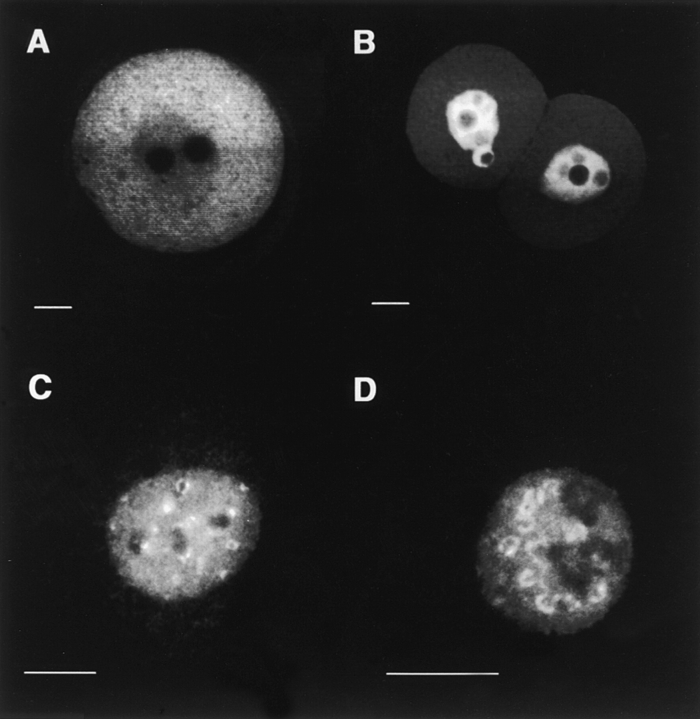
All tested Dnmt1 fusion constructs are relatively large, ranging in size from 100 kD to ∼250 kD. Although this size range does not make any difference for nuclear uptake in somatic cells, we generated a similar fusion construct of equal size using the DNA ligase I, which is involved in DNA replication and is like Dnmt1 targeted to nuclear replication foci (Cardoso et al. 1997). Fig. 5 shows the side-by-side comparison of both the Dnmt1 and the DNA ligase I construct. In somatic cells, both fusion constructs are targeted to nuclear replication foci, but only the DNA ligase fusion protein enters the nucleus of early mouse embryos. These results clearly show that the observed cytoplasmic retention of fusion proteins is specific for the Dnmt1 protein and is developmentally controlled. Moreover, the retention mechanism is active also in early preimplantation embryos and not only in oocytes, where the maternal stock of Dnmt1 is accumulated.
Figure 5.
Cytoplasmic retention of Dnmt1 is independent of protein size. Two β-galactosidase fusion constructs were generated, one containing amino acids 1–854 of Dnmt1 (depicted in the diagram, Fig. 4 I) and the other containing amino acids 1–796 of human DNA ligase I (Cardoso et al. 1997). The size of both fusion proteins is between 160–170 kD. Both plasmids were transfected into mouse fibroblasts and microinjected into fertilized mouse eggs. After a 1-d incubation, localization of the fusion proteins was assayed by immunofluorescence staining with anti–β-galactosidase monoclonal antibody. A shows a confocal section through the middle of a mouse embryo expressing the Dnmt1 fusion which is mostly cytoplasmic. B shows the DNA ligase I fusion expressed in mouse embryos, which clearly has an exclusive nuclear localization. C and D illustrate the localization of the same proteins as in A and B, respectively, in the nuclei of tissue culture cells, in these images undergoing S-phase as seen by the ring and dot-shaped pattern of subnuclear foci (Leonhardt et al. 1992; Cardoso et al. 1997). Bar 10 μm.
The data presented in this work suggest that Dnmt1 is actively retained in the cytoplasm, but cannot rule out the formal possibility that Dnmt1 is enriched in the cytoplasm by a nuclear export mechanism. However, the mapping of a cis-acting sequence allows now a specific search for interacting factors and the generation of dominant-negative mutants to further study the regulation and role of the cytoplasmic localization of Dnmt1 during early development.
The active retention of Dnmt1 in the cytoplasm from the oocyte to the blastocyst stage correlates well with the overall decrease in the genomic methylation level and thus is a likely mechanism to regulate DNA methylation by separating Dnmt1 from chromosomal DNA in the nucleus. Demethylation could occur by preventing maintenance methylation of newly synthesized DNA strands and/or by preventing remethylation of actively demethylated sites.
Recently, a cDNA coding for a protein with demethylase activity was isolated from HeLa cells and shown to be ubiquitously expressed at the mRNA level in somatic cells (Bhattacharya et al. 1999). It is still unknown whether it is active during early development and the fact that this demethylase is ubiquitous means that it cannot by itself cause the specific demethylation during preimplantation development.
Cytoplasmic accumulation of maternal nuclear gene products has been extensively observed during oocyte maturation and early development in Xenopus (Dabauvalle and Franke 1984; Paine 1984; Dreyer 1987). Some transcription factors (e.g., Oct-1 and Xnf7; Li et al. 1994; Veenstra et al. 1999) are retained in the cytoplasm until the mid-blastula stage when transcriptional activity is restarted. Nuclear exclusion in this developmental system has been proposed as a mechanism to control the function of maternal proteins until the time during development when they are required. These results parallel the regulation of Dnmt1 nuclear uptake during mammalian preimplantation development. Until the blastocyst stage, Dnmt1 is retained in the cytoplasm with the exception of the eight-cell stage when it briefly enters the nucleus (Fig. 1 D; Carlson et al. 1992). Also, in growing oocytes, Dnmt1 enters the nucleus at a time when parasitic sequences are heavily methylated (Walsh et al. 1998). Analogously, it is conceivable that specific sequences are methylated at the eight-cell stage. In that regard, it is interesting to note that modifications occur in eight-cell stage embryos that make them unable to recapitulate the normal program of gene expression when transplanted into one-cell embryos (Latham et al. 1994). This different behavior of transplanted nuclei from two- and eight-cell embryos might in part be caused by methylation of specific genomic DNA sequences around the eight-cell stage. In fact, the Igf2r gene locus is methylated exactly at the eight-cell stage and could thus be one such candidate genes (Birger et al. 1999).
The high level of the enzyme in oocytes possibly represents a maternal stock to be used for maintenance and/or de novo methylation in later cell cycles. This large amount would last through subsequent cell divisions and might thus also allow development of Dnmt1-deficient mice till mid-gestation. In this context, it is noteworthy that the oocyte-specific Dnmt1 isoform is fully active, can restore DNA methylation in Dnmt1-null embryonic stem cells, and can thus rescue their capacity to differentiate in vivo (Gaudet et al. 1998).
In addition, one cannot exclude that Dnmt1 might also have a function in the cytoplasm during early development, in particular since this cytoplasmic store is enzymatically active. Interestingly, the bacterial restriction-modification system is also localized in the periplasmic space and serves as a first line of defense against foreign DNA. One possible function of the high concentration of Dnmt1 in the cytoplasm could be to protect the developing embryo and the future germ line by methylating intruding DNA and thus rendering it transcriptionally inactive. The identification and characterization of a cytoplasmic retention sequence makes it now possible to directly investigate functions of Dnmt1 during early development and to screen for interacting factors.
Acknowledgments
We are indebted to Jay Baltz, Joel Lawitts, and Sandra Schieferl for valuable help and advice concerning the work with mouse embryos. We would like to thank Bernardo Nadal-Ginard for his very special support and always enticing discussions.
This work was supported by grants from The Council for Tobacco Research and the Deutsche Forschungsgemeinschaft.
Footnotes
1.used in this paper: DIC, differential interference contrast; Dnmt, DNA methyltransferase; FHM, flushing-holding medium; HGG, human chorionic gonadotropin; NLS, nuclear localization signal; p.c., post-coitum; PMS, pregnant mare's serum
References
- Bestor T., Laudano A., Mattaliano R., Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Bestor T.H. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO (Eur. Mol. Biol. Organ.) J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S.K., Ramchandani S., Cervoni N., Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Birger Y., Shemer R., Perk J., Razin A. The imprinting box of the mouse Igf2r gene. Nature. 1999;397:84–88. doi: 10.1038/16291. [DOI] [PubMed] [Google Scholar]
- Cardoso M.C., Joseph C., Rahn H.P., Reusch R., Nadal-Ginard B., Leonhardt H. Mapping and use of a sequence that targets DNA ligase I to sites of DNA replication in vivo. J. Cell Biol. 1997;139:579–587. doi: 10.1083/jcb.139.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso M.C., Leonhardt H. Immunofluorescence techniques in cell cycle studies. In: Pagano M., editor. Cell CycleMaterials and Methods. Springer-Verlag; Heidelberg: 1995. pp. 15–28. [Google Scholar]
- Cardoso M.C., Leonhardt H., Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replicationcyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- Carlson L.L., Page A.W., Bestor T.H. Properties and localization of DNA methyltransferase in preimplantation mouse embryosimplications for genomic imprinting. Genes Dev. 1992;6:2536–2541. doi: 10.1101/gad.6.12b.2536. [DOI] [PubMed] [Google Scholar]
- Constancia M., Pickard B., Kelsey G., Reik W. Imprinting mechanisms. Genome Res. 1998;8:881–900. doi: 10.1101/gr.8.9.881. [DOI] [PubMed] [Google Scholar]
- Czank A., Hauselmann R., Page A.W., Leonhardt H., Bestor T.H., Schaffner W., Hergersberg M. Expression in mammalian cells of a cloned gene encoding murine DNA methyltransferase. Gene. 1991;109:259–263. doi: 10.1016/0378-1119(91)90618-l. [DOI] [PubMed] [Google Scholar]
- Dabauvalle M.C., Franke W.W. Karyophobic proteins. A category of abundant soluble proteins which accumulate in the cytoplasm. Exp. Cell Res. 1984;153:308–326. doi: 10.1016/0014-4827(84)90603-7. [DOI] [PubMed] [Google Scholar]
- Dreyer C. Differential accumulation of oocyte nuclear proteins by embryonic nuclei of Xenopus . Development. 1987;101:829–846. doi: 10.1242/dev.101.4.829. [DOI] [PubMed] [Google Scholar]
- Gaudet F., Talbot D., Leonhardt H., Jaenisch R. A short DNA methyltransferase isoform restores methylation in vivo. J. Biol. Chem. 1998;273:32725–32729. doi: 10.1074/jbc.273.49.32725. [DOI] [PubMed] [Google Scholar]
- Hogan B., Beddington R., Constantini F., Lacy E. Manipulating the Mouse EmbryoA Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1994. [Google Scholar]
- Howlett S.K., Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991;113:119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- Latham K.E., Garrels J.I., Solter D. Alterations in protein synthesis following transplantation of mouse 8-cell stage nuclei to enucleated 1-cell embryos. Dev. Biol. 1994;163:341–350. doi: 10.1006/dbio.1994.1153. [DOI] [PubMed] [Google Scholar]
- Latham K.E., McGrath J., Solter D. Mechanistic and developmental aspects of genetic imprinting in mammals. Int. Rev. Cytol. 1995;160:53–98. doi: 10.1016/s0074-7696(08)61553-3. [DOI] [PubMed] [Google Scholar]
- Lawitts J.A., Biggers J.D. Culture of preimplantation embryos. Methods Enzymol. 1993;225:153–164. doi: 10.1016/0076-6879(93)25012-q. [DOI] [PubMed] [Google Scholar]
- Leonhardt H., Bestor T.H. Structure, function and regulation of mammalian DNA methyltransferase. In: Jost J.P., Saluz H.P., editors. DNA MethylationMolecular Biology and Biological Significance. Vol. 64. Birkäuser Verlag; Basel, Switzerland: 1993. pp. 109–119. [DOI] [PubMed] [Google Scholar]
- Leonhardt H., Page A.W., Weier H.U., Bestor T.H. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- Li E., Beard C., Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T.H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li X., Shou W., Kloc M., Reddy B.A., Etkin L.D. Cytoplasmic retention of Xenopus nuclear factor 7 before the mid blastula transition uses a unique anchoring mechanism involving a retention domain and several phosphorylation sites. J. Cell Biol. 1994;124:7–17. doi: 10.1083/jcb.124.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertineit C., Yoder J.A., Taketo T., Laird D.W., Trasler J.M., Bestor T.H. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development. 1998;125:889–897. doi: 10.1242/dev.125.5.889. [DOI] [PubMed] [Google Scholar]
- Monk M., Adams R.L., Rinaldi A. Decrease in DNA methylase activity during preimplantation development in the mouse. Development. 1991;112:189–192. doi: 10.1242/dev.112.1.189. [DOI] [PubMed] [Google Scholar]
- Monk M., Boubelik M., Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Okano M., Xie S., Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases Nat. Genet. 19 1998. 219 220a [DOI] [PubMed] [Google Scholar]
- Okano M., Xie S., Li E. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells Nucleic Acids Res 26 1998. 2536 2540b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olek A., Walter J. The preimplantation ontogeny of the H19 methylation imprint. Nat. Genet. 1997;17:275–276. doi: 10.1038/ng1197-275. [DOI] [PubMed] [Google Scholar]
- Paine P.L. Diffusive and nondiffusive proteins in vivo. J. Cell Biol. 1984;99:188–195. doi: 10.1083/jcb.99.1.188s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panning B., Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 1996;10:1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- Razin A., Shemer R. DNA methylation in early development. Hum. Mol. Genet. 1995;4:1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]
- Sanford J.P., Clark H.J., Chapman V.M., Rossant J. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes Dev. 1987;1:1039–1046. doi: 10.1101/gad.1.10.1039. [DOI] [PubMed] [Google Scholar]
- Taylor D.L., Salmon E.D. Basic fluorescence microscopy. In: Wang Y.-L., Taylor D.L., editors. Fluorescence Microscopy of Living Cells in Culture. Vol. 29. Academic Press; San Diego, CA: 1989. pp. 207–237. [PubMed] [Google Scholar]
- Tremblay K.D., Duran K.L., Bartolomei M.S. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker K.L., Talbot D., Lee M.A., Leonhardt H., Jaenisch R. Complementation of methylation deficiency in embryonic stem cells by a DNA methyltransferase minigene. Proc. Natl. Acad. Sci. USA. 1996;93:12920–12925. doi: 10.1073/pnas.93.23.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Wyngaert I., Sprengel J., Kass S.U., Luyten W.H. Cloning and analysis of a novel human putative DNA methyltransferase. FEBS Lett. 1998;426:283–289. doi: 10.1016/s0014-5793(98)00362-7. [DOI] [PubMed] [Google Scholar]
- Veenstra G.J., Mathu M.T., Destree O.H. The Oct-1 POU domain directs developmentally regulated nuclear translocation in Xenopus embryos. Biol. Chem. 1999;380:253–257. doi: 10.1515/BC.1999.033. [DOI] [PubMed] [Google Scholar]
- Walsh C.P., Chaillet J.R., Bestor T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Yoder J.A., Bestor T.H. A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum. Mol. Genet. 1998;7:279–284. doi: 10.1093/hmg/7.2.279. [DOI] [PubMed] [Google Scholar]
- Yoder J.A., Yen R.W.C., Vertino P.M., Bestor T.H., Baylin S.B. New 5′ regions of the murine and human genes for DNA (cytosine-5)-methyltransferase. J. Biol. Chem. 1996;271:31092–31097. doi: 10.1074/jbc.271.49.31092. [DOI] [PubMed] [Google Scholar]
- Zimmermann C., Guhl E., Graessmann A. Mouse DNA methyltransferase (MTase) deletion mutants that retain the catalytic domain display neither de novo nor maintenance methylation activity in vivo. Biol. Chem. 1997;378:393–405. doi: 10.1515/bchm.1997.378.5.393. [DOI] [PubMed] [Google Scholar]