Abstract
Neurotrophins play an essential role in the regulation of actin-dependent changes in growth cone shape and motility. We have studied whether neurotrophin signaling can promote the localization of β-actin mRNA and protein within growth cones. The regulated localization of specific mRNAs within neuronal processes and growth cones could provide a mechanism to modulate cytoskeletal composition and growth cone dynamics during neuronal development. We have previously shown that β-actin mRNA is localized in granules that were distributed throughout processes and growth cones of cultured neurons. In this study, we demonstrate that the localization of β-actin mRNA and protein to growth cones of forebrain neurons is stimulated by neurotrophin-3 (NT-3). A similar response was observed when neurons were exposed to forskolin or db-cAMP, suggesting an involvement of a cAMP signaling pathway. NT-3 treatment resulted in a rapid and transient stimulation of PKA activity that preceded the localization of β-actin mRNA. Localization of β-actin mRNA was blocked by prior treatment of cells with Rp-cAMP, an inhibitor of cAMP-dependent protein kinase A. Depolymerization of microtubules, but not microfilaments, inhibited the NT-3–induced localization of β-actin mRNA. These results suggest that NT-3 activates a cAMP-dependent signaling mechanism to promote the microtubule-dependent localization of β-actin mRNA within growth cones.
Keywords: mRNA localization, β-actin, neurotrophin, growth cone, neurite outgrowth
Neurotrophins have long been known to be chemoattractants (Levi-Montalcini 1976; Letourneau 1978; Gunderson and Barrett 1979). Local application of neurotrophin-3 (NT-3)1 and brain-derived neurotrophic factor (BDNF) from a micropipet can induce the turning and movement of the growth cone towards the neurotrophin source (Ming et al. 1997a,Ming et al. 1997b; Song et al. 1997). In addition, bath application of BDNF to these cultures rapidly induced the formation of lamellipodia, which may be an early event in the chemotactic response (Ming et al. 1997a,Ming et al. 1997b). Evidence indicates that neurotrophins may regulate actin polymerization and filopodial protrusion. Neurotrophin-dependent changes in F-actin content were observed in growth cones, as assayed by phalloidin labeling of fixed cells (Paves and Saarman, 1997). NGF-coated beads induced filopodial sprouting of axon collateral branches (Gallo and Letourneau 1998). Cytochalasin treatment abolished this sprouting suggesting involvement of actin polymerization in NGF stimulated protrusion. Disruption of actin filaments within filopodia abolished growth cone turning in response to chemotropic stimuli (Zheng et al. 1996). Axonal pathfinding decisions in vivo have also been shown to be adversely affected by cytochalasin-induced perturbation of actin filaments within growth cone filopodia (Bentley and Toroian-Raymond 1986; Chien et al. 1993).
Although the actin cytoskeleton has been shown to be required for neurotrophin stimulation of process outgrowth (Paves and Saarman, 1997; Gallo and Letourneau 1998), there have been no previous studies that identify the downstream events of neurotrophin signaling which could influence local actin polymerization. Our hypothesis was that neurotrophin stimulation promotes the localization of actin mRNA and protein within growth cones. The localization of mRNA has been shown to be an important mechanism to influence protein sorting and asymmetry within cells that can be regulated by growth factors (Bassell et al. 1999). A role for growth factor signaling in the localization of specific mRNAs within neurons has not been previously shown.
Many studies have addressed the question of how the neuron targets cytoskeletal precursors over considerable distances to reach the growth cone for their use in filopodial formation and control of process outgrowth. One established mechanism to provide axons and growth cones with specific cytoskeletal proteins is to actively transport them by a slow transport mechanism into processes and growth cones after their synthesis within the cell body (Okabe and Hirokawa 1990; Sabry et al. 1995; Takeda and Hirokawa 1995; Mills et al. 1996). In addition, evidence has accumulated that several mRNAs are localized into dendrites or axons, suggesting that local protein synthesis may provide an important mechanism to influence the distribution of cytoskeletal proteins (Garner et al. 1988; Kleiman et al. 1990; Litman et al. 1993; Hannan et al. 1995, Hannan et al. 1998; Olink-Coux and Hollenbeck 1996).
The localization of specific mRNAs and translational machinery within growth cones has also been demonstrated (Crino and Eberwine 1997; Bassell et al. 1998). We have shown that β-actin mRNA, ribosomal proteins and elongation factor 1α are present as granules within growth cones of developing dendritic and axonal processes (Bassell et al. 1998). These growth cones contained morphologically identifiable polyribosomes depicted by electron microscopy. Additionally, probes to β-actin mRNA colocalized with translational components in growth cones (Bassell et al. 1998). Dendritic growth cones have also been shown to contain translational machinery as demonstrated by their incorporation of radiolabeled amino acids into proteins after neurite transection (Davis et al. 1992). Ultrastructural analysis has revealed the presence of polyribosomes in growth cones of developing hippocampal neurons (Deitch and Banker 1993). Localized mRNAs could be translated directly within growth cones, providing immediate assembly of structural components where they are needed.
The localization of specific mRNAs to growth cones may be regulated by signaling pathways. We have previously shown that treatment of rat cortical neurons with db-cAMP resulted in the localization of β-actin mRNA, but not γ-actin mRNA, into processes and growth cones (Bassell et al. 1998). These results suggest that a cAMP intracellular signaling pathway may affect localization of distinct actin isoforms. We sought to elucidate the extracellular signals that activate a cAMP-dependent signal transduction pathway involved in β-actin mRNA localization to growth cones. We also investigated whether regulated localization of β-actin mRNAs could be correlated with changes in β-actin protein localization.
Materials and Methods
Cell Culture
Hippocampal or cortical cultures from embryonic rat brain have been previously used as a model system to study mRNA and protein localization as these cells differentiate distinct axonal and dendritic processes and have demonstrated similar molecular distributions to that observed in vivo (Goslin and Banker 1991). We have described the use of cerebrocortical cultures in studying the localization of β-actin mRNA to neuronal processes and growth cones (Bassell et al. 1998). In this study, we used a chick forebrain neuronal culture system, which also differentiates axon and dendrite-like neurites (Sensenbrenner et al. 1978; Chada et al. 1997), as an alternative to rat cortical neurons. We observed that neurons within these cultures frequently have larger growth cones that have a flattened, lamellar morphology that is characteristic of the type that localize β-actin mRNA (Bassell et al. 1998).
The general method of neuronal culture that we use has been described in detail (Goslin and Banker 1991) and modified for use with chick forebrain neurons (Chada et al. 1997). Cerebral hemispheres were dissected from 8-d-old chick embryos and trypsinized (0.15% in HBSS) at 37°C for 7 min, washed in HBSS, and placed in MEM with 10% FBS. Cells were mechanically dissociated by pipetting, washed three times in MEM, and plated at low density (6,000 cells/cm2) on poly-l-lysine (0.2 mg/ml, 16 h) and laminin (0.02 mg/ml, 12 min) coated coverslips or plates in MEM with 10% FBS for 2 h. Cells were inverted onto a monolayer of rat astrocytes in N2-conditioned medium with serum (2% FBS) and cultured for 4 d at 37°C in 5% CO2. N2-conditioned medium contained glutamate-free MEM supplemented with transferrin (100 μg/ml), insulin (5 μg/ml), progesterone (20 nM), putrescine (100 μM), selenium dioxide (30 nM), glucose (6 mg/ml), sodium pyruvate (1 mM), and ovalbumin (0.1%). The cells were then fixed in paraformaldehyde (4% in PBS) at room temperature for 15 min and washed in PBS containing 5 mM MgCl2 three times.
Drug and Growth Factor Treatments
Neurons were cultured in N2-conditioned medium with 2% FBS for 4 d and then starved in minimal essential medium (MEM) without N2 supplements or serum for a period of 6 h at 37°C. Before fixation, cells were treated at various time points (10 min through 2 h) with drugs or growth factors as follows: FBS (8%), forskolin (5 μM; Calbiochem-Novabiochem), neurotrophin-3 (NT-3) (25 ng/ml; Austral Biologicals), db-cAMP (N6, O2-dibutyryl-adenosine 3′,5′-cyclic monophosphate) (25 μM; Calbiochem-Novabiochem), brain derived neurotrophic factor (BDNF) (25 ng/ml; Sigma), nerve growth factor (25 ng/ml; GIBCO-BRL). In some experiments, the cells were treated with NT-3 in a series of time points, i.e., 1, 2, 5, and 10 min, after starvation in MEM to observe the changes of β-actin mRNA and protein localization in growth cones.
K252a (200 nM; BioMol), an inhibitor for a broad range of serine and threonine protein kinases including Trk receptor tyrosine kinases (Knusel and Hefti 1992) and Rp-cAMP (50 μM; Calbiochem-Novabiochem), a nonhydrolyzable analogue competitor of cAMP (Bothelo et al. 1988), were both used to inhibit the NT-3 induction of β-actin mRNA localization. After 5 h of starvation in MEM, K252a or Rp-cAMP was added for 1 h followed by treatment with NT-3 (in the continued presence of the inhibitor). The cells were fixed in 4% paraformaldehyde in PBS after treatment with drugs or growth factors.
To disrupt microfilaments, cells were treated with 5 μg/ml of cytochalasin-D (Sigma Chemical Co.) in culture media at 37°C for 30 min before NT-3 stimulation. To depolymerize microtubules, cells were treated with colchicine (10 μg/ml, Sigma Chemical Co.) in culture media for 30 min before NT-3 stimulation. In each case, the cells were first starved in MEM for 5.5 h before addition of cytochalasin-D or colchicine. Stock solutions of cytochalasin-D and colchicine were made up in DMSO and ethanol, respectively, and the concentration of these solvents was diluted below 0.1% in the culture media, so as not to be toxic to neurons. Neurons were fixed in paraformaldehyde (4% in PBS with 5 mM MgCl2) after drug treatment and NT-3 stimulation.
In Situ Hybridization and Immunofluorescence Analysis
Six amino group modified oligonucleotides (50 bases each) complementary to 3′ untranslated sequences (3′-UTR) of chick β-actin mRNA were synthesized on a DNA synthesizer (Latham et al. 1994). Each oligonucleotide was modified at five positions within the sequence and chemically labeled using digoxigenin succinamide ester (Boehringer Mannheim). To ensure isoform specificity, probes were selected from unique regions within the 3′-UTR and were of identical length, guanine/cytosine (GC) content and hapten incorporation. Oligonucleotide probes complementary to β-galactosidase mRNA or Mu phage DNA were used as controls. In situ hybridization for β-actin mRNA was completed as previously described (Bassell et al. 1998). Cells were equilibrated in 1× SSC with 40% formamide for 5 min. Each coverslip was incubated at 37°C overnight in hybridization reactions containing 20 ng of oligonucleotide probe, 1× SSC, 40% formamide, 10% dextran sulfate, 0.4% BSA, 20 mM ribonucleotide vanadyl complex, salmon testes DNA (10 mg/ml), E. coli tRNA (10 mg/ml), and 10 mM sodium phosphate. Cells were washed twice with 4× SSC/40% formamide and then twice with 2× SSC/40% formamide, both at 37°C, and then with 2× SSC three times at room temperature.
The hybridized probes labeled with digoxigenin were detected using Cy3-conjugated monoclonal antibody (mAb) to digoxigenin and anti–mouse mAb-Cy3 (from Jackson ImmunoResearch Labs.). After blocking in TBS with BSA (2%) and FBS (2%) at 37°C for 1 h, the coverslips were incubated with Cy3-mAb to digoxigenin in TBS (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Triton X-100) with 1% BSA at 37°C for 1 h. After washes in TBS with 1% BSA, cells were mounted with n-propyl gallate (anti-fading agent). β-actin protein was detected with a mouse monoclonal antibody (Sigma) and secondary antibodies were conjugated with Cy3 (Jackson ImmunoResearch Labs.).
Microscopy and Digital Imaging
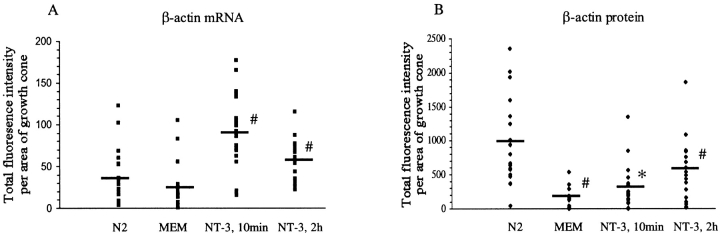
Immunofluorescence signal was viewed using an Olympus-IX70 microscope equipped with a 60× Plan-Neofluar objective and Nomarski (DIC) optics. Cells were viewed using a 100 watt mercury arc lamp and light was filtered using HiQ bandpass filters (ChromaTech). The images were captured with a cooled CCD camera (Photometrics) using a 35-mm shutter and processed using IP Lab Spectrum (Scanalytics) running on a Macintosh G3. After identification of growth cones using DIC optics, a fluorescence image was immediately acquired. All exposure times with the CCD camera were kept constant (1 s for β-actin mRNA, 0.5 s for β-actin protein) and below grey scale saturation to permit a linear response to light intensity and quantitative analysis of differences in fluorescence intensities. The perimeter of each growth cone was traced using the DIC image and IP Lab software to identify a region of interest (ROI) and measure total fluorescence intensity. For quantitative image analysis of β-actin mRNA and protein localization using this method (see Fig. 3 and Fig. 4), 20 cells were imaged for each cell culture condition.
Figure 3.
NT-3 stimulated localization of β-actin mRNA and protein analyzed using quantitative digital imaging microscopy. Neurons were fixed for in situ hybridization to β-actin mRNA (A) and immunofluorescence detection of β-actin protein (B). DIC and fluorescence images were captured using a cooled CCD camera. 20 growth cones were imaged for each condition with identical exposure times. Data expressed as fluorescence density (total intensity/growth cone area). NT-3 was observed to increase the density of fluorescence signal for both β-actin mRNA and protein within growth cones. #, P < 0.01 when MEM was compared with N2, or MEM was compared with NT-3, 10 min or NT-3, 2 h. *, P < 0.05 when MEM was compared to NT-3 at 10 min. N2, normal culture medium. MEM, starvation in minimum essential medium.
Figure 4.
Visualization of NT-3–stimulated β-actin mRNA localization in cells treated with cytoskeletal disrupting drugs. (A) β-actin mRNA localization in cytochalasin-D–treated cell. Hybridization signal was prominent in the cell body (arrow) and localized in granules within growth cones (arrowhead). (B) Disruption of F-actin in growth cones by cytochalasin-D. Note the absence of filamentous staining in growth cone (arrowhead). (C) In colchicine-treated cells, β-actin mRNA was not accumulated within growth cones in response to NT-3. (D) Lack of filamentous staining for tubulin was also observed in growth cones (arrowhead). There was no evidence that growth cones were collapsed by either drug. Quantitative analysis of growth once area failed to show any statistically significant reduction in size after 30-min exposures.
For quantitative analysis using a visual scoring method, 100 cells per coverslip were analyzed for each cell culture condition. Experiments were done with duplicate coverslips for each variable and each experiment was repeated at least three times. The scoring method involved visualization of the presence or absence of β-actin mRNA granules in the axon-like growth cone from each cell. Cells were scored as localized if several granules were observed, and scored as nonlocalized if the signal was not distinguishable from background levels (hybridization with control probe). Localized cells would be expected to have a higher amount of fluorescent signal in growth cones compared with nonlocalized cells. Examples of localized and nonlocalized cells are shown in Fig. 1. This scoring method was used to show that NT-3 promotes an increase in localization (see Fig. 2) and that the results were comparable to the quantitation of fluorescence intensity using the CCD camera (see Fig. 3). The value for each bar within the histogram reports the mean and standard deviation between independent samples (see Fig. 2). The Student's t test was used to compare the percentage of localized cells under a number of experimental conditions. This scoring method produced similar results to that of quantitation of fluorescence intensity within each growth cone using digital imaging analysis (see above). The advantage of the scoring method is that one can rapidly score hundreds of cells within a population and evaluate multiple variables.
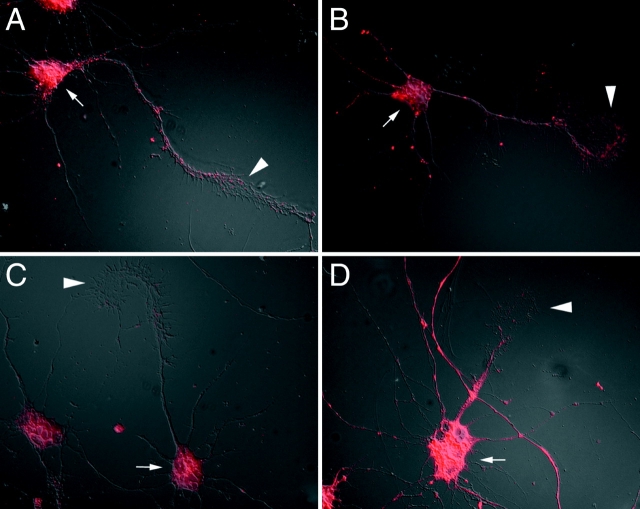
Figure 1.
β-actin mRNA and protein localization is influenced by neurotrophin-3 (NT-3). (A) β-actin mRNA was prominent in the cell body and localized within the axonal process and growth cone in the form of spatially distinct granules (arrow; example of localized cell). (B) Cells that were starved in minimal essential medium (MEM) for 6 h showed hybridization within the cell body but growth cones did not reveal mRNA granules (arrow; example of nonlocalized cell). (C) Cells that were starved for 6 h and then stimulated with NT-3 for 10 min were observed to relocalize β-actin mRNA granules within growth cones (arrow). (D) Relocalization of β-actin mRNA within growth cones after 2 h in NT-3–supplemented MEM (arrow). (E) Cells grown in normal defined medium showed intense staining for β-actin protein within the minor neurites and the peripheral margin of the axonal growth cone (arrow) (example of localized cell). (F) Starvation resulted in reduced staining for β-actin protein within growth cones (arrow) (example of nonlocalized cell). (G and H) NT-3 stimulation for 10 min and 2 h restored the localization of β-actin protein within the peripheral margin (arrows). Bar, 15 μm.
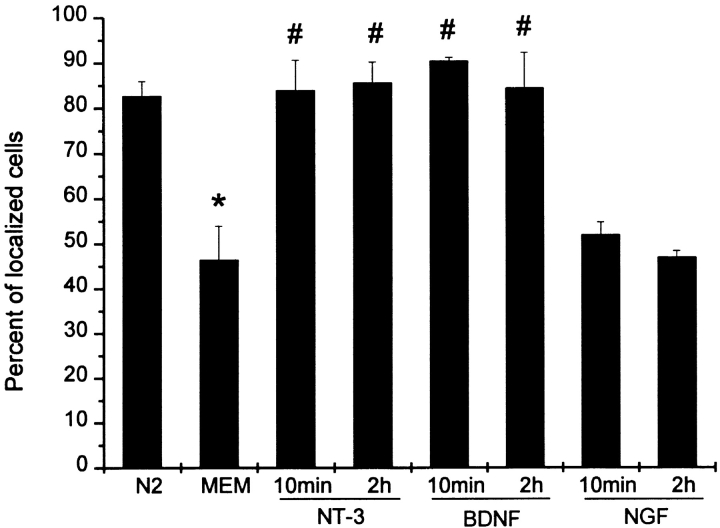
Figure 2.
Quantitative analysis of neurotrophin stimulated β-actin mRNA localization. Neurotrophin-3 (NT-3), brain-derived neurotrophic factor (BDNF), or NGF was added to the MEM (25 ng/ml) for the indicated time. NT-3 and BDNF, but not NGF, were observed to rapidly stimulate the localization of β-actin mRNA within growth cones. The axon-like neurite and growth cone from each cell was visualized for the presence of β-actin mRNA granules (see Materials and Methods). 100 cells were scored per coverslip. Histogram shows mean percentage and standard deviation between independent samples (n = 4). Examples of localized and nonlocalized cells are shown in Fig. 1. N2, normal control. MEM, starvation in minimum essential medium. *, P < 0.01 when MEM compared with N2. #, P < 0.01 when NT-3 or BDNF was compared with MEM.
PKA Activity
SignaTECT cAMP-Dependent Protein Kinase Assay (Promega) was used to monitor neurotrophin-3 stimulation of PKA activity in primary neuronal cultures (Cobb et al., 1992; Goueli et al. 1995; Walsh and Van Patten 1994). This assay uses a biotinylated Kemptide substrate, derived from pyruvate kinase (Piklis et al. 1980), which is highly specific for cAMP-dependent PKA (Km = 5–10 μM). The biotinylated peptide substrate is phosphorylated by cellular PKA using γ-[32P]ATP. The 32P-labeled biotin substrate is then recovered from the reaction mix and captured by a streptavidin-linked matrix (SAMTM Membrane). In brief, cultured neurons were treated with NT-3 as described, washed with HBSS, and then lysed in 0.3 ml cold extraction buffer (25 mM Tris-HCl, pH 7.4, 0.5 mM EDTA, 0.5 mM EGTA, 10 mM mercaptoethanol, 1 μg/ml leupeptin, 1 μg/ml aprotinin, and 0.5 mM PMSF). Extracts were centrifuged for 5 min at 4°C at 14,000 g and 5 μl of the supernatant was incubated with 20 μl of the reaction mixture containing biotinylated peptide substrate and γ-[32P]ATP for 5 min at 30°C. The reaction was terminated and 10 μl was spotted onto a biotin capture membrane. After washing and drying of the membrane, radioactivity was measured using a liquid scintillation counter. This assay detects 0.012 casein units (2.5 Kemptide units) or less of purified cAMP-dependent PKA. A linear regression analysis, using calibrated amounts of casein units, produced an R2 value >0.95. Triplicate samples were used for each time point and the PKA activities and standard deviation is shown in Fig. 8 (expressed relative to starved cells not exposed to NT-3).
Figure 8.
NT-3 signaling of β-actin mRNA localization through cAMP. NT-3 stimulation of β-actin mRNA localization was reduced by kinase inhibitors. K252a (200 nM) or Rp-cAMP (50 μM) was added to the MEM for 1 h after a 5-h starvation. Forskolin and db-cAMP stimulated β-actin mRNA localization in a similar fashion to NT-3. *, P < 0.01 when NT-3/K252a at 10min and NT-3/Rp-cAMP at 10min were compared with NT-3 at 10min. #, P < 0.01 when NT-3/K252a at 2h and NT-3/Rp-cAMP at 2h were compared with NT-3 at 2h.
Northern Blot Hybridization
Neurons (8 × 105 cells) were cultured in N2-conditioned medium supplemented with 2% fetal bovine serum on poly-l-lysine and laminin-coated plastic dishes for 4 d. Total cellular RNA was isolated using Tri Reagent following the product instructions (Molecular Research Center, Inc.). Cellular RNA was dissolved in DEPC-treated, distilled water. RNA (8 μg) was run in each lane in 0.8% formaldehyde-denatured agarose gel and transferred to Zeta-probe GT Genomic Tested Blotting Membrane (Bio-Rad Lab). The cDNA fragments (372 bp) complementary to the β-actin mRNA reading frame sequence were labeled with 32P-dCTP (Amersham Corp.) by using the Random Primers DNA Labeling System (GIBCO-BRL) and purified by Quick Spin Columns (Boehringer Mannheim). The RNA binding membrane was prehybridized in 5× SSPE supplemented with 5× Denhardt's, 0.2% SDS, 5% dextran sulfate, 300 μg/ml salmon testes DNA, and 50% formamide at 45°C for 4 h, and hybridized with 32P-labeled cDNA probes at 45°C overnight. After washes, the membrane was exposed to x-ray film (Kodak). Bands on the exposure film were scanned using a densitometer (Molecular Dynamics) and the optical densities were analyzed quantitatively using ImageQuant software.
Extraction with Rhodamine-labeled Actin
After 4 d of culturing in N2-conditioned medium with 2% FBS and 6 h of starvation in MEM, the cells were incubated in MEM with NT-3 (25 ng/ml) for 1, 2, 5, and 10 min. Cells were extracted in buffer (20 mM Hepes, pH 7.4, 138 mM KCl, 4 mM MgCl2, 3 mM EGTA, 0.1 mg/ml saponin, and 1 mM ATP) with 0.45 μM of rhodamine-labeled actin (rho-actin) at room temperature for 1 min and fixed for immunofluorescence microscopy (Chan et al. 1998). Rho-actin was rabbit G-actin labeled with rhodamine (provided by J. Condeelis, Albert Einstein College of Medicine). Rho-actin was thawed on ice and diluted in buffer (1 mM Hepes, pH 7.4, 0.2 mM MgCl2, and 0.2 mM ATP) to a final concentration of 12 μM. Rho-actin (75 μl) was added into 2 ml of extraction buffer just before application to the cells.
Results
β-Actin mRNA and Protein Are Localized by NT-3
To investigate the distribution of actin mRNA isoforms within neuronal processes and/or growth cones of chick forebrain neurons, chemically modified oligonucleotide probes were synthesized to specific β-actin mRNA sequences and conjugated for use with fluorescence in situ hybridization. With this approach it is possible to detect a signal that may not be apparent using conventional bright-field or epifluorescence microscopy. Also, the control of probe length, GC content, and hapten labeling efficiency using oligonucleotide probes has eliminated variability encountered previously with enzymatic labeling. Using these probes we can compare the signal obtained for specific mRNAs with that of control probe sequences such as lacZ which have identical probe complexity (not shown). We have previously used this method to show that specific mRNAs have distinct intracellular distributions in primary cultures and that β-actin mRNAs localize within axonal growth cones of cultured cortical neurons (Bassell et al. 1998). In chick forebrain neurons, hybridization signal to β-actin mRNA was observed in the cell body and extended into processes and growth cones in a highly punctate or granular pattern (Fig. 1 A).
To visualize β-actin within these forebrain neurons, an isoform-specific monoclonal antibody was used for immunofluorescence localization. In the somatodendritic compartment, β-actin labeling was highly enriched in the distal tips of minor neurites and no staining was observed within the cell body and proximal segments (Fig. 1 E). β-actin protein was highly enriched within the peripheral margin of the axon-like growth cone and filopodia (Fig. 1 E). In contrast to β-actin, γ-actin was distributed throughout the cell body, all processes and filopodia (not shown). The enrichment of the β-actin isoform within growth cones in these chick forebrain neurons was identical to that observed previously in rat cortical neurons (Bassell et al. 1998).
To investigate whether β-actin mRNA localization to growth cones is regulated by signal transduction mechanisms, we used an assay to study the effects of signaling molecules in the absence of the astrocyte coculture, which secretes neurotrophins, and other potential signaling molecules within the culture medium, e.g., insulin. This approach involved removing the neurons from their normal defined media with N2 supplements and placing them in MEM for 6 h, before challenging with specific growth factors. This brief deprivation of N2 supplements did not result in adverse effects on neuronal morphology or cytoskeletal integrity as judged by DIC microscopy or immunofluorescence staining with anti-tubulin antibody (not shown). The objective was to have the cells in a quiescent state but still sensitive to signaling mechanisms. It has been shown that starvation of nonneuronal cells in MEM causes them to enter a quiescent phase with reduced β-actin gene expression and mRNA levels (Greenberg and Ziff 1984; Latham et al. 1994). Serum stimulation promotes β-actin mRNA localization and enhances fibroblast motility (Latham et al. 1994; Kislauskis et al. 1997). This previous work provides a useful methodology to study the effects of neurotrophins on β-actin mRNA localization in neurons.
We investigated whether treatment of starved cells with neurotrophins could stimulate β-actin mRNA localization. Neurotrophins are a family of proteins that include NGF, BDNF, and the neurotrophins, NT3, NT4, and NT6. These proteins bind the tyrosine receptor kinases, TrkA, TrkB, and TrkC with selective affinity and activate several signaling pathways (Kaplan and Miller 1997). Both NT-3 and BDNF bind trk B receptors, whereas NT-3 also binds trk C (Kaplan and Miller 1997). NGF binds TrK A receptors and shows little affinity for Trk B/C receptors (Gallo et al. 1997).
We observed that addition of NT-3 to the medium of starved cells could promote the rapid localization of β-actin mRNA (compare Fig. 1B with C) within growth cones. Cells were scored as either localized or nonlocalized for β-actin mRNA (see Materials and Methods), as has been done previously, as a means to quantitate mRNA localization (Latham et al. 1994; Kislauskis et al. 1997). Cells were scored as localized if several granules were observed, and scored as nonlocalized if the signal was not distinguishable from background levels (hybridization with control probe). Localized cells would be expected to have a higher amount of fluorescent signal in growth cones compared with nonlocalized cells. After a 10-min stimulation with NT-3, >80% of the cells exhibited β-actin mRNA localization within growth cones, compared with only 45% of the starved cells (Fig. 2). BDNF, but not NGF, was also shown to promote localization of β-actin mRNA within >80% of the cells (Fig. 2). These results suggest involvement of specific neurotrophin-receptor interactions in the localization of β-actin mRNAs within growth cones.
The localization of β-actin protein within growth cones was also stimulated by NT-3. Starved cells showed little accumulation of β-actin protein within growth cones (Fig. 1 F). After 10 min in NT-3, a dramatic increase in the signal for β-actin protein within growth cone filopodia was observed (Fig. 1 G). This signal for β-actin protein colocalized with F-actin, as assayed using phalloidin, suggesting that there is the stimulation of new F-actin filaments (not shown).
To further quantitate the observations that NT-3 treatment results in an increase in the amount of detectable β-actin mRNA and protein within growth cones, images were acquired of both starved and NT-3–treated cells, using identical exposure times with a cooled CCD camera (see Materials and Methods). Fluorescence intensity values can be either expressed as a total amount or after dividing by growth cone area to get a measure of fluorescence density. Since growth cone area varies naturally from neuron to neuron, data expressed as fluorescence density was a more relevant parameter (total intensity/area). The perimeter of each growth cone was traced using DIC optics and the total fluorescence intensity was measured for each growth cone and divided by its area to obtain a measure of the density of mRNA and protein per pixel. NT-3 treatment resulted in a 3.3× increase in the mean pixel intensity for β-actin mRNA after 10 min in NT-3, and a 2.2× increase after 2 h in NT-3 (Fig. 3 A). The increase in the mean pixel intensity for β-actin protein was increased by 1.75× after 10 min in NT-3, and 3.25× after 2 h in NT-3 (Fig. 3 B). We also analyzed total fluorescence intensity, independent of growth cone area, and similarly observed an increase in β-actin mRNA and protein levels (not shown).
NT-3 Signaling of β-actin mRNA Localization Requires Microtubules and Not Actin Filaments
To determine whether the localization of β-actin mRNA particles to growth cones was dependent on the integrity of cytoskeletal filament systems, cells were pretreated with cytochalasin-D or colchicine to depolymerize microfilaments and microtubules, respectively. After treatment with cytochalasin-D, localization of β-actin mRNA by NT-3 was still observed within neuronal processes and growth cones (Fig. 4 A). Neurons treated with cytochalasin-D showed a disruption of filamentous actin in the peripheral margin of growth cones; labeling of F-actin was primarily localized to cytoplasmic aggregates (Fig. 4 B). The cytochalasin dose was tenfold above that used to delocalize β-actin mRNA from fibroblast lamellae (Sundell and Singer 1991). Interestingly, treatment with cytochalasin-D actually produced an increase in the density of β-actin mRNA in growth cones after NT-3, as measured by quantitative digital imaging microscopy (Fig. 5). Since cytochalasin-D can induce actin gene expression (Sympson and Geoghegan 1990), it was possible that new β-actin mRNA transcripts were also transported and localized in the presence of cytochalasin. An alternative explanation could be that cytochalasin-D may decrease growth cone area that would result in a higher fluorescence density even if mRNA levels were unaffected. However, we saw no evidence that growth cone area was affected by either cyotchalsin-D or colchicine treatment (Fig. 4B and Fig. D). Consistent with these images, quantitative analysis of growth cone area did not reveal any evidence for decreased growth cone area after 30 min in either cytochalasin or colchicine (not shown).
Figure 5.
Differential effects of cytochalasin-D and colchicine on NT-3–stimulated β-actin mRNA localization. Cytochalasin-D (5 μg/ml) or colchicine (10 μg/ml) was added to the MEM for 30 min before NT-3 stimulation. Fluorescence intensity was measured using digital imaging microscopy and divided by growth cone area. Colchicine treatment, but not cytochalasin-D, inhibited the localization of β-actin mRNA by NT-3. *, P < 0.05 when NT-3 at 10 min was compared with MEM. #, P < 0.01 when cytochalasin-D/NT-3 at 10 min was compared with MEM.
The preservation of growth cone structure after treatment with cytoskeletal disrupting agents can largely be attributed to their brief exposure to the cells (only 30 min). These brief treatments were also insufficient to perturb the steady state distribution of β-actin mRNA in control cultures (not shown). We have previously shown that longer treatments with colchicine (1.5 h) can delocalize mRNA form processes, whereas cytochalasin-D has no effect (Bassell et al. 1994). These previous results are consistent with a hypothesis that mRNAs are anchored to microtubules. The current study focused instead on the dynamic aspects of mRNA localization in response to NT-3 stimulation. Here we show that perturbation of microtubules and not microfilaments can block the dynamic process of mRNA localization. These results suggest that β-actin mRNA transcripts can be localized into processes and growth cones by NT-3 signals despite the presence of disorganized actin filaments.
To confirm the role of microtubules in the localization of β-actin mRNA to processes, microtubules were depolymerized with colchicine before fixation and hybridization. A filamentous distribution for microtubules was not observed after colchicine treatment (Fig. 4 D). Also, minor alterations in neuronal morphology were evident, as most neurites were thinner with varicosities; however, the growth cones did not collapse (Fig. 4 D). Depolymerization of microtubules inhibited the NT-3 signaling of β-actin mRNA localization within processes and growth cones (Fig. 4 C and 5). These results indicate that the localization of β-actin mRNA within processes and the central domain of growth cones was dependent on drug-labile microtubules.
Neurotrophin-3 Rapidly Stimulates β-Actin Protein Localization and Actin Polymerization
The above results using cytochalasin suggest a specific mechanism to localize β-actin mRNA that is independent of any signals that NT-3 may directly impart on new actin synthesis and polymerization. We propose that the first neuronal response to NT-3 is to rapidly promote actin polymerization within the peripheral margin of the growth cone that is then followed by the microtubule-dependent targeting of β-actin mRNAs to the growth cone. We performed a time course experiment to determine whether β-actin protein localization may occur before and perhaps independent of mRNA localization. After starvation in MEM, 62% of the cells were scored as localized for β-actin protein (Fig. 6). NT-3 treatment for 2 min resulted in >85% of the cells being scored as localized for β-actin protein (Fig. 6). β-actin protein localization in response to NT-3 was initially concentrated in one or two foci within the peripheral margin, suggesting local accumulation and/or polymerization (Fig. 7 B). After several additional minutes, β-actin protein staining was observed throughout the entire peripheral margin (Fig. 7 C). An increase in β-actin mRNA localization was also observed after 2 min, although it took longer than β-actin protein to reach maximal levels, peaking at 10 min (Fig. 6). Therefore, the relocalization of β-actin mRNA was delayed relative to the protein localization. These results support the model that NT-3 signals may also promote the localization of β-actin protein by a mechanism independent of mRNA localization.
Figure 6.
Time course of β-actin mRNA and protein localization to growth cones after NT-3 treatment. Neurons were cultured for 4 d and starved in MEM for 6 h. NT-3 (25 ng/ml) was added to the medium for indicated time, and cells were fixed for in situ hybridization and immunofluorescence analysis. A rapid increase in β-actin protein localization (2 min) was observed before maximal localization of β-actin mRNA within growth cones (10 min).
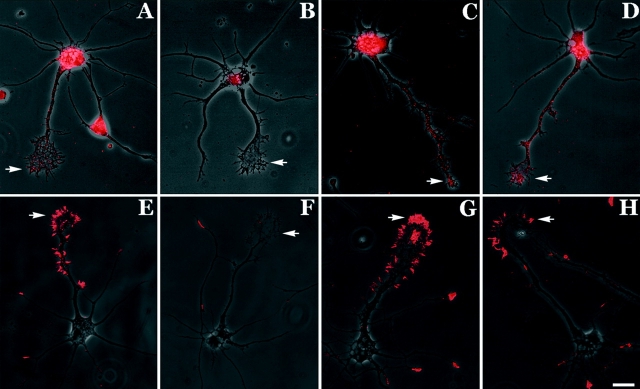
Figure 7.
Localization of β-actin protein and actin polymerization in response to NT-3. (A) Immunofluorescence localization of β-actin protein in starved cells. Weak staining was observed within growth cone (arrow) (example of nonlocalized cell). (B) Addition of NT-3 to the medium for 5 min resulted in the presence of focal staining within the peripheral margin of minor neurites and the axonal growth cone (arrow) (example of localized cell). (C) Addition of NT-3 to the medium for 10 min resulted in the localization of β-actin protein throughout the peripheral margin (arrow). (D) Starved cells were extracted with saponin (0.1 mg/ml) in buffer containing rhodamine-actin (0.45 mM) for 1 min, washed, and fixed in paraformaldehyde (4% in 1 PBS). There was no fluorescence staining in these untreated cells. Nucleus was stained with DAPI. (E) By contrast, stimulation of cells with NT-3 for 5 min resulted in visualization of fluorescence signal within the growth cone (arrow). (F) Stimulation with NT-3 for 10 min resulted in the incorporation of rhodamine-actin throughout the peripheral margin (arrow). Bar, 15 mm.
We hypothesized that NT-3 may signal the polymerization of monomeric actin within growth cones that may be diffuse within the cytoplasm. The de novo formation of F-actin within growth cones would create very high levels of actin within a given pixel consistent with our quantitative observations (Fig. 3 B). To address this issue, we used an actin polymerization assay developed to visualize actin polymerization at the leading edge of motile cells in response to EGF treatment (Chan et al. 1998). The assay involves extraction of live cells in the presence of rhodamine-labeled actin, which adds on to the growing barbed ends of actin filaments within the cell, allowing direct visualization of the location of NT-stimulated nucleation sites. Cells were treated with NT-3 and then extracted for 1 min with saponin in the presence of rhodamine-labeled G-actin. Cells were briefly washed, fixed, and examined by fluorescence microscopy. There was no incorporation of rhodamine-actin in control cells that were not treated with NT-3 (Fig. 7 D). After a 5-min treatment, incorporation of rhodamine-actin was observed within the cortex of the cell body as well within growth cones (Fig. 7 E). Staining levels increased progressively over the next several minutes. After a 10-min treatment with NT-3, rhodamine-actin incorporation was observed throughout the peripheral margin of the growth cone (Fig. 7 F). There was no appreciable level of rhodamine-actin incorporation within the neurite shaft. Peripheral cytoplasm was the preferred locus for continued actin polymerization. These results indicate that actin polymerization, β-actin protein localization, and β-actin mRNA localization are coordinately regulated by neurotrophin signals.
Neurotrophin Signaling of β-Actin mRNA Localization through a cAMP Signaling Pathway
Neurotrophin stimulation of growth cone motility (Song et al. 1997; Ming et al. 1997a,Ming et al. 1997b) may involve activation of a cAMP-dependent signaling pathway (Gunderson and Barrett 1980; Lohof et al. 1992; Song et al. 1997). We sought to determine whether NT-3 signaling of β-actin mRNA localization could be mimicked by activation of the cAMP signaling pathway, and whether inhibitors of intracellular kinases could block the NT-3 response. Treatment of starved forebrain cells with either db-cAMP, a membrane permeable analogue of cAMP, or forskolin, an activator of adenylate cyclase, was observed to rapidly stimulate a 40% increase in β-actin mRNA localization within growth cones after 10 min (Fig. 8). This stimulation in β-actin mRNA localization persisted throughout the various time points studied (up to 2 h). NT-3 signaling of β-actin mRNA localization could be blocked by prior incubation of the cells with K252a (Fig. 8), an inhibitor for a broad range of serine, and threonine protein kinases including Trk receptor tyrosine kinases (Knusel and Hefti 1992). A reduction in mRNA localization was also observed with Rp-cAMP (Fig. 8), a nonhydrolyzable analogue competitor of cAMP (Bothelo et al. 1988), which has been previously used to affect growth cone motility in response to neurotrophin treatment (Song et al. 1997). The reduction in mRNA localization observed with the specific PKA inhibitor, Rp-cAMP was less than that observed with K252a, suggesting the possibility that other signaling pathways may also be involved.
To determine whether NT-3 signaling can specifically activate a cAMP signaling pathway, we measured protein kinase A (PKA) activity in cell lysates after cultures were treated with NT-3 at various time points. This assay used a biotinylated Kemptide substrate that is highly specific for cAMP-dependent PKA (see Materials and Methods). PKA activity was observed to peak at 2 min (345% increase over control levels) and then declined to baseline levels (Fig. 9). We conclude from this assay that there is a rapid (within 2 min) increase in PKA activity that precedes our observations of β-actin mRNA localization.
Figure 9.
Activation of cAMP-dependent PKA in neurons treated with NT-3. Cells were cultured for 4 d and then incubated in MEM for 3 h. NT-3 (25 ng/ml) was added to the medium for the indicated time. Cells were extracted and the PKA activities were estimated by 32P-ATP incorporation assay (Signa TECT cAMP-dependent protein kinase assay system; Promega).
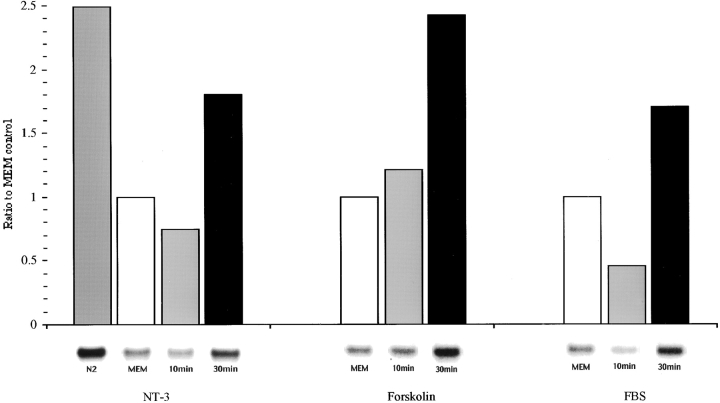
The localization of β-actin mRNA to growth cones in response to neurotrophin signaling is suggestive of a mechanism that promotes a redistribution in the preexisting mRNA population. In addition, neurotrophins may also enhance β-actin gene transcription that would result in increased levels of mRNA in the cytoplasm. We evaluated β-actin mRNA levels by Northern blot analysis in normal, starved, and stimulated cultures (Fig. 10). Starvation of cells for 6 h resulted in a 52% decrease in β-actin mRNA levels. Treatment of starved cells with NT-3, forskolin, or fetal bovine serum (also promotes mRNA localization, data not shown) did not result in an increase in β-actin mRNA levels within the 10-min time period during which mRNA localization occurred. These results suggest that mRNA localization can be due to a rapid redistribution of preexisting mRNAs. A 30-min treatment with NT-3 did result in an increase in β-actin mRNA levels (Fig. 9) which then declined at 2 h (not shown). These results indicate that neurotrophin signals can also upregulate β-actin mRNA levels, and that this response may be one of the last events in a signal transduction cascade that collectively affects many aspects of actin gene expression and localization which include actin polymerization, localization of β-actin mRNA and protein, and perhaps transcriptional activation.
Figure 10.
Stimulation of β-actin mRNA levels in neurons treated with NT-3, forskolin, or serum. Northern blot hybridization with 32P-labeled cDNA probes to β-actin mRNA (see Materials and Methods). An increase in β-actin mRNA levels occurred at 30 min which was after the relocalization of β-actin mRNA to growth cones.
Discussion
Regulation of β-Actin mRNA Localization and Localized Actin Polymerization
The elucidation of regulatory mechanisms for mRNA localization within neuronal growth cones is important as it would provide support for the hypothesis that external cues encountered by the growth cone during pathfinding could locally control protein synthetic activities. The regulated synthesis of cytoskeletal proteins could influence growth cone structure and affect the rate and/or direction of process outgrowth. In nonneuronal cells, the active transport of β-actin mRNA to the cell's leading edge was induced by serum and PDGF. This induction was, in fact, required to obtain maximal rates of cell motility (Latham et al. 1994; Kislauskis et al. 1997). The localization of β-actin mRNA within neurons could similarly influence local actin polymerization and process outgrowth.
In this study, we identify a new function for the neurotrophins: the localization of β-actin mRNA granules within growth cones. We suggest that β-actin mRNA localization into neuronal processes provides a local mechanism to control G-actin concentration and facilitates de novo nucleation of actin polymerization. Our results suggest that Trk receptor activation leads to the anterograde transport of β-actin mRNA granules from the cell body into growth cones. We speculate that this mechanism might involve Trk receptors within the cell body, which would signal the microtubule-dependent transport of mRNA through activation of a cAMP-dependent pathway. In this study, we have also observed that NT-3 stimulated the rapid polymerization of actin within growth cones (within 2 min) which is consistent with previous observations (Gallo and Letourneau 1998). This could be due to the binding of NT-3 or BDNF to tyrosine kinase receptors (Trk C and/or B) which are localized within growth cones. Receptor activation could stimulate the rapid and local polymerization of G-actin into F-actin within the growth cone and filopodia. We suggest that this initial stimulus for polymerization would be independent of mRNA localization and local protein synthesis. We propose that after the initial signal to polymerize actin, active transport of β-actin mRNA into the growth cone occurs, which further supports actin polymerization and long-term process outgrowth. An alternative hypothesis is that the anchoring of β-actin mRNA within the growth cone may be secondary to the polymerization of actin which is induced by NT-3. However, NT-3 treatment was observed to induce the localization of mRNA in cells that cannot polymerize new actin filaments. These data imply the presence of a specific signaling mechanism for mRNA localization.
Evidence in support of regulation of mRNA localization to growth cones was also obtained from a previous study using a micropipet to remove cytoplasm from growth cones and amplify a heterogeneous population of mRNAs, which included MAP2 and intermediate filament proteins (Crino and Eberwine 1997). The amount of mRNA within growth cones was dependent on the stage of neuronal development, and was varied for each mRNA species (Crino and Eberwine 1997). Translation of these mRNAs within growth cones was demonstrated by transfection of mRNA encoding an epitope tag and immunofluorescence localization. Collapse of growth cones with the calcium ionophore A23187 promoted transport of mRNAs encoding intermediate filaments into growth cones, further suggesting that local synthesis may be regulated (Crino and Eberwine 1997).
Our observations, along with the results of these recent studies, suggest a novel type of developmentally regulated sorting mechanism. Further work should provide new insight into the local control of cytoskeletal organization and growth dynamics.
Localization of β-Actin Isoform to Growth Cones
It is of fundamental interest to identify how the cytoskeletal composition of the growth cone differs from that of the perikarya and to identify mechanisms involved in cytoskeletal sorting and assembly. Not only must the neuron target cytoskeletal proteins to growth cones, it must also target a distinct set of proteins whose identity, stoichiometry, and structural organization differs from that of the perikarya. There is an increasing consensus that actin isoforms in several cell types are sorted to different intracellular compartments, and that β-actin has a specific role in regions of motile cytoplasm (Herman and D'Amore 1985; Otey et al. 1986; Shuster and Herman 1995; Von Arx et al. 1995; Yao and Forte 1995). β-actin may be the predominate isoform at submembranous sites where it is involved in local de novo nucleation of actin polymerization in response to extracellular signals (Shuster and Herman 1995). Our evidence here is that β-actin is highly concentrated within growth cones and filopodia. We propose that β-actin protein is the preferred isoform for de novo nucleation within growth cones and filopodia. The paucity of β-actin protein within the cell body and neurite shaft is consistent with a model in which β-actin mRNA is first transported to the growth cone and then translated locally to stimulate or bias the process of actin nucleation and polymerization near the peripheral margin.
Localization of RNA Granules
Evidence indicates that RNA granules or particles are found in many different cell types and may represent a transported form for mRNAs and translational machinery (Bassell et al. 1999). These RNA particles may be translocated along cytoskeletal filaments via interactions of mRNA localization elements, motor molecules, and accessory proteins (Bassell et al. 1999). The microtubule-dependent transport of RNA granules into neuronal processes may provide a rapid mechanism for localized protein synthesis in distal compartments. Previous studies have demonstrated the presence of RNA granules within neuronal (Knowles et al. 1996; Bassell et al. 1998) and oligodendrocyte processes (Barbarese et al. 1995; Ainger et al. 1993, Ainger et al. 1997). These RNA granules were shown to contain specific mRNAs and translational components (Barbarese et al. 1995; Knowles et al. 1996; Bassell et al. 1998).
Granules containing myelin basic protein mRNA (MBP) moved into oligodendrocyte processes at 0.2 μm/s (Ainger et al. 1997), and microtubule depolymerizing drugs and antisense oligonucleotides to kinesin inhibited granule translocation into processes (Carson et al. 1997). RNA granules labeled with the nucleic acid binding dye, SYTO14, have been shown to move into minor neurites at an anterograde rate of 0.1 μm/s (Knowles et al. 1996). Translocation of SYTO labeled granules was blocked by prior depolymerization of microtubules with colchicine (Knowles et al. 1996). Our present work indicates that colchicine treatment inhibited the NT-3 stimulation of β-actin mRNA localization within growth cones, suggesting that microtubules were required for the active transport of RNA from the soma to the growth cone. Further work is needed to define the transport rates for granules containing β-actin mRNA which localize to neuronal processes and growth cones.
Previous work in live cells using the fluorescent nucleic acid binding dye, SYTO14, has shown that the anterograde translocation of RNA granules was induced by NT-3 (Knowles and Kosik 1997). It was proposed that Trk receptor signaling through receptors within the cell body could stimulate the movement of preexisting mRNA granules from the cell body into the process. It was not known previously whether the RNA granules induced by NT-3 contained specific mRNAs or whether they localized to growth cones. Our work here has shown that granules localized to growth cones in response to NT-3 contain β-actin mRNA. These new findings suggest an important function for neurotrophin-mediated mRNA localization: local synthesis of cytoskeletal precursors that are essential for actin polymerization and filopodial-directed outgrowth. The continued study of the dynamic regulation of mRNA localization should provide new insight into active mRNA transport mechanisms and the role of local protein synthesis in growth cone dynamics.
Acknowledgments
We thank John Condeelis for helpful discussions and for providing rhodamine-actin for these experiments. We thank Ken Kosik for helpful discussions. We thank Steve Braut for preparation of probes. We thank Shailesh Shenoy and Maritza Martinez for assistance with digital imaging and figure preparation.
This work was supported by the March of Dimes Foundation, National Science Foundation grant IBN9811384 and National Institutes of Health (NIH) grants RO1 GM55599 to G.J. Bassell and GM54887 and AR41480 to R.H. Singer. H.L. Zhang is a Research Associate supported by a NIH training grant to M. Bennett (IT32 NS-07439-01).
Footnotes
1.used in this paper: BDNF, brain-derived neurotrophic factor; GC, growth cone; NT-3, neurotrophin-3; ROI, region of interest
References
- Ainger K., Avossa D., Morgan F., Hill S.J., Barry C., Barbarese E., Carson J.H. Transport and localization of exogenous MBP mRNA microinjected into oligodendrocytes. J. Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainger K.A., Avossa D., Diana A.S., Barry C., Barbarese E., Carson J.H. Transport and localization elements in MBP mRNA. J. Cell Biol. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbarese E., Koppel D.E., Deutscher M.P., Smith C.L., Ainger K., Morgan F., Carson J.H. Protein translation components are colocalized in granules in oligodendrocytes. J. Cell Sci. 1995;108:2781–2790. doi: 10.1242/jcs.108.8.2781. [DOI] [PubMed] [Google Scholar]
- Bassell G.J., Oleynikov Y., Singer R.H. The travels of mRNAs through all cells large and small. FASEB J. 1999;13:447–454. doi: 10.1096/fasebj.13.3.447. [DOI] [PubMed] [Google Scholar]
- Bassell G.J., Singer R.H. mRNA and cytoskeletal filaments. Curr. Opin. Cell Biol. 1997;9:109–115. doi: 10.1016/s0955-0674(97)80159-7. [DOI] [PubMed] [Google Scholar]
- Bassell G.J., Singer R.H., Kosik K.S. Association of poly(A) mRNA with microtubules in cultured neurons. Neuron. 1994;12:571–582. doi: 10.1016/0896-6273(94)90213-5. [DOI] [PubMed] [Google Scholar]
- Bassell G.J., Zhang H.L., Byrd A.L., Femino A.M., Singer R.H., Taneja K.L., Lifshitz L.M., Herman I.M., Koisk K.S. Sorting of beta actin mRNA and protein to neurites and growth cones in culture. J. Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D., Toroian-Raymond A. Disoriented pathfinding by pioneer neurone growth cones deprived of filopodia. Nature. 1986;323:712–715. doi: 10.1038/323712a0. [DOI] [PubMed] [Google Scholar]
- Bothelo L.H., Rothermel J.D., Coombs R.V., Jastorff B. cAMP antagonists of cAMP action. Methods. Enzymol. 1988;159:159–172. doi: 10.1016/0076-6879(88)59017-1. [DOI] [PubMed] [Google Scholar]
- Carson J.H., Worboys K., Ainger K., Barbarese E. Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil. Cytol. 1997;38:318–328. doi: 10.1002/(SICI)1097-0169(1997)38:4<318::AID-CM2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Chada G., Lamoureux P., Buxbaum R.E., Heidemann S.R. Cytomechanics of neurite outgrowth from chick brain neurons. J. Cell Sci. 1997;110:1179–1186. doi: 10.1242/jcs.110.10.1179. [DOI] [PubMed] [Google Scholar]
- Chan A., Bailly M., Wyckoff J.B., Segall J.E., Condeelis J.S. EGF stimulates an increase in actin nucleation at the leading edge of the lamellipod in adenocarcinoma cells. J. Cell Sci. 1998;111:1–13. doi: 10.1242/jcs.111.2.199. [DOI] [PubMed] [Google Scholar]
- Chien C.B., Rosenthal D.E., Harris W.A., Holt C.E. Navigational errors made by growth cones without filopodia in the embryonic Xenopus brain. Neuron. 1993;11:237–251. doi: 10.1016/0896-6273(93)90181-p. [DOI] [PubMed] [Google Scholar]
- Crino P.B., Eberwine J. Molecular characterization of the dendritic growth cone. Neuron. 1997;17:1173–1187. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- Davis L., Ping D., DeWitt M., Kater S.B. Protein synthesis within neuronal growth cones. J. Neurosci. 1992;12:4867–4877. doi: 10.1523/JNEUROSCI.12-12-04867.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitch J.S., Banker G.A. An electron microscopic analysis of hippocampal neurons developing in cultureEarly stages in the emergence of polarity. J. Neurosci. 1993;13:4301–4315. doi: 10.1523/JNEUROSCI.13-10-04301.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo G., Lefcort F.B., Letourneau P.C. The trkA receptor mediates growth cone turning toward a localized source of nerve growth factor. J. Neurosci. 1997;17:5445–5454. doi: 10.1523/JNEUROSCI.17-14-05445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo G., Letourneau P. Localized sources of neurotrophins initiate axon collateral sprouting. J. Neurosci. 1998;18:5403–5414. doi: 10.1523/JNEUROSCI.18-14-05403.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner C.C., Tucker R.P., Matus A. Selective localization of mRNA for cytoskeletal protein MAP2 in dendrites. Nature. 1988;336:674–679. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- Goslin K., Banker G. Culturing nerve cells. MIT Press; London: 1991. [Google Scholar]
- Goueli B.S., Hsiao K., Teraba A., Goueli S.A. A novel and simple method to assay the activity of individual protein kinases in a crude tissue extract. Anal. Biochem. 1995;225:10–17. doi: 10.1006/abio.1995.1100. [DOI] [PubMed] [Google Scholar]
- Greenberg M.E., Ziff E.B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–435. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Gunderson R.W., Barrett J.N. Neuronal chemotaxischick dorsal root axons turn toward high concentrations of NGF. Science. 1979;206:1079–1080. doi: 10.1126/science.493992. [DOI] [PubMed] [Google Scholar]
- Gunderson R.W., Barrett J.N. Characterization of the turning response of dorsal root neurites toward NGF. J. Cell Biol. 1980;87:546–554. doi: 10.1083/jcb.87.3.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan A.J., Gunning P., Jeffrey P.L., Weinberger R.P. Structural compartments within neuronsdevelopmentally regulated organization of microfilament isoform mRNA and protein. Mol. Cell Neurosci. 1998;11:289–304. doi: 10.1006/mcne.1998.0693. [DOI] [PubMed] [Google Scholar]
- Hannan A.J., Schevzov G., Gunning P., Jeffrey P.L., Weinberger R.P. Intracellular localization of tropomyosin mRNA and protein is associated with development of neuronal polarity. Mol. Cell Neurosci. 1995;6:397–412. doi: 10.1006/mcne.1995.1030. [DOI] [PubMed] [Google Scholar]
- Herman I.M., D'Amore P.A. Microvascular periocytes contain muscle and nonmuscle actins. J. Cell Biol. 1985;101:43–52. doi: 10.1083/jcb.101.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.R., Miller F.D. Signal transduction by the neurotrophin receptors. Curr. Opin. Cell Biol. 1997;9:213–221. doi: 10.1016/s0955-0674(97)80065-8. [DOI] [PubMed] [Google Scholar]
- Kislauskis E., Zhu X., Singer R. β-Actin messenger RNA localization and protein synthesis augment cell motility. J. Cell Biol. 1997;136:1263–1270. doi: 10.1083/jcb.136.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman R., Banker G., Steward O. Differential subcellular localization of particular mRNAs in hippocampal neurons in culture. Neuron. 1990;5:821–830. doi: 10.1016/0896-6273(90)90341-c. [DOI] [PubMed] [Google Scholar]
- Knowles R.B., Kosik K.S. Neurotrophin-3 signals redistribute RNA in neurons. Proc. Natl. Acad. Sci. USA. 1997;94:14804–14808. doi: 10.1073/pnas.94.26.14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles R.B., Sabry J.H., Martone M.E., Deerinck T.J., Ellisman M.H., Bassell G.J., Kosik K.S. Translocation of RNA granules in living neurons. J. Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knusel B., Hefti F. K-252a compoundsmodulators of neurotrophin signal transduction. J. Neurochem. 1992;59:1987–1996. doi: 10.1111/j.1471-4159.1992.tb10085.x. [DOI] [PubMed] [Google Scholar]
- Latham V.M., Kislauskis E.H., Singer R.H., Ross A.F. Beta actin mRNA localization is regulated by signal transduction mechanisms. J. Cell Biol. 1994;126:1211–1219. doi: 10.1083/jcb.126.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneau P.C. Chemotactic responses of nerve fiber elongation to NGF. Dev. Biol. 1978;66:183–196. doi: 10.1016/0012-1606(78)90283-x. [DOI] [PubMed] [Google Scholar]
- Levi-Montalcini R. The nerve growth factorits role in growth, differentiation and function of the sympathetic adrenergic neuron. Prog. Brain Res. 1976;45:235–258. doi: 10.1016/S0079-6123(08)60993-0. [DOI] [PubMed] [Google Scholar]
- Litman P., Barg J., Rindzoonski L., Ginzburg I. Subcellular localization of tau mRNA in differentiating neuronal cell cultureimplications for neuronal polarity. Neuron. 1993;10:627–638. doi: 10.1016/0896-6273(93)90165-n. [DOI] [PubMed] [Google Scholar]
- Lohof A.M., Quillan M., Dan Y., Poo M.-m. Asymmetric modulation of cytosolic cAMP activity induces growth cone turning. J. Neurosci. 1992;12:1253–1261. doi: 10.1523/JNEUROSCI.12-04-01253.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.G., Minamide L.S., Yuan A., Bamburg J.R., Bray J.J. Slow axonal transport of soluble actin with actin depolymerizing factor, cofilin, and profilin suggests actin moves in an unassembled form. J. Neurochem. 1996;67:1225–1234. doi: 10.1046/j.1471-4159.1996.67031225.x. [DOI] [PubMed] [Google Scholar]
- Ming G., Lohof A.M., Zheng J.Q. Acute morphogenic and chemotropic effects or neurotrophins on cultured embryonic xenopus spinal neurons J. Neurosci. 17 1997. 7860 7871a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming G., Song H., Berninger B., Holt C.E., Tessier-Lavigne M., Poo M.-m. cAMP-development growth cone guidance by netrin-1 Neuron 19 1997. 1225 1235b [DOI] [PubMed] [Google Scholar]
- Okabe S., Hirokawa N. Turnover of fluorescently labeled tubulin and actin in the axon. Nature. 1990;343:479–482. doi: 10.1038/343479a0. [DOI] [PubMed] [Google Scholar]
- Olink-Coux M., Hollenbeck P.J. Localization and active transport of mRNA in axons of sympathetic neurons. J. Neurosci. 1996;16:1346–1358. doi: 10.1523/JNEUROSCI.16-04-01346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otey C.A., Lessard J.L., Bulinski J.C. Immunolocalization of the gamma isoform of nonmuscle actin. J. Cell Biol. 1986;102:1732–1737. doi: 10.1083/jcb.102.5.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paves H., Saarma M. Neurotrophins as in vitro guidance molecules for embryonic sensory neurons. Cell Tissue Res. 1997;290:285–297. doi: 10.1007/s004410050933. [DOI] [PubMed] [Google Scholar]
- Piklis S.J., Tager H.S., Heinrikson R.L. Phosphorylation of rat hepatic fructose-1,6-biphosphatase and pyruvate kinase. J. Biol. Chem. 1980;255:2770–2775. [PubMed] [Google Scholar]
- Sabry J., O'Connor T.P., Kirschner M.W. Axonal transport of tubulin in Ti1 pioneer neurons. Neuron. 1995;14:1247–1256. doi: 10.1016/0896-6273(95)90271-6. [DOI] [PubMed] [Google Scholar]
- Sensenbrenner M., Maderspach K., Latzkovits L., Jaros G.G. Neuronal cells from chick embryo cerebral hemispheres cultivated on polylysine-coated surfaces. Dev. Neurosci. 1978;1:90–101. doi: 10.1159/000112560. [DOI] [PubMed] [Google Scholar]
- Shuster C.B., Herman I.M. Indirect association of ezrin with F-actinisoform specificity and calcium sensitivity. J. Cell Biol. 1995;128:837–848. doi: 10.1083/jcb.128.5.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Ming G., Poo M.-m. cAMP-induced switching in turning direction of nerve growth cones. Nature. 1997;388:275–279. doi: 10.1038/40864. [DOI] [PubMed] [Google Scholar]
- Sundell C.L., Singer R.H. Requirement of microfilaments in sorting of actin mRNAs. Science. 1991;253:1275–1277. doi: 10.1126/science.1891715. [DOI] [PubMed] [Google Scholar]
- Sympson C.L., Geoghegan T.E. Actin gene expression in murine erythroleukemia cells treated with cytochalasin D. Exp. Cell Res. 1990;189:28–32. doi: 10.1016/0014-4827(90)90252-6. [DOI] [PubMed] [Google Scholar]
- Takeda S., Hirokawa N. Tubulin dynamics in neuronal axons. Neuron. 1995;14:1257–1264. doi: 10.1016/0896-6273(95)90272-4. [DOI] [PubMed] [Google Scholar]
- Von Arx P., Bantle S., Soldati T., Perriard J. Dominant negative effect of cytoplasmic actin isoproteins on cardiomyocyte cytoarchitecture and function. J. Cell Biol. 1995;131:1759–1773. doi: 10.1083/jcb.131.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D.A., Van Patten S.M. Multiple pathway signal transduction by cAMP dependent protein kinase. FASEB J. 1994;8:1227–1236. doi: 10.1096/fasebj.8.15.8001734. [DOI] [PubMed] [Google Scholar]
- Yao X., Forte J.G. Polarized distribution of actin isoforms in gastric parietal cells. Mol. Biol. Cell. 1995;6:541–557. doi: 10.1091/mbc.6.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J.Q., Wan J., Poo M.-m. Essential role of filopodia in chemotrophic turning of nerve growth cone induced by a glutamate gradient. J. Neurosci. 1996;16:1140–1149. doi: 10.1523/JNEUROSCI.16-03-01140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]