Abstract
Glycine betaine is a potent osmoprotectant accumulated by Sinorhizobium meliloti to cope with osmotic stress. The biosynthesis of glycine betaine from choline is encoded by an operon of four genes, betICBA, as determined by sequence and mutant analysis. The betI and betC genes are separated by an intergenic region containing a 130-bp mosaic element that also is present between the betB and betA genes. In addition to the genes encoding a presumed regulatory protein (betI), the betaine aldehyde dehydrogenase (betB), and the choline dehydrogenase (betA) enzymes also found in Escherichia coli, a new gene (betC) was identified as encoding a choline sulfatase catalyzing the conversion of choline-O-sulfate and, at a lower rate, phosphorylcholine, into choline. Choline sulfatase activity was absent from betC but not from betB mutants and was shown to be induced indifferently by choline or choline-O-sulfate as were the other enzymes of the pathway. Unlike what has been shown in other bacteria and plants, choline-O-sulfate is not used as an osmoprotectant per se in S. meliloti, but is metabolized into glycine betaine. S. meliloti also can use this compound as the sole carbon, nitrogen, and sulfur source for growth and that depends on a functional bet locus. In conclusion, choline-O-sulfate and phosphorylcholine, which are found in higher plants and fungi, appear to be substrates for glycine betaine biosynthesis in S. meliloti.
When subjected to an environmental osmotic stress, the most common response mechanism developed by living organisms is to accumulate compatible solutes called osmolytes to maintain a positive cellular turgor essential for metabolic functions and structural integrity. Osmolytes identified until now are either ions (potassium) or small organic molecules such as amino acids (glutamate, proline), sugars (trehalose, glucosylglycerol), ectoine, dipeptides, and quartenary amines (stachydrine, glycine betaine) (1–5). Among these compatible solutes, glycine betaine (N,N,N-trimethylglycine) has been shown to be a very efficient osmolyte found in a wide range of bacterial and plant species, where it is accumulated at high cytoplasmic concentrations in response to osmotic stress. The potential importance of this molecule for stress resistance among agronomically important organisms has led to investigations of betaine biosynthesis and transport in Sinorhizobium meliloti. This soil bacterium has the ability to develop a symbiotic relationship with alfalfa within specific root nodules, where it can reduce atmospheric nitrogen into ammonia taken up by the plant host (6). However, variations of the osmotic environment within the rhizosphere may affect all steps of the plant–microbe interaction, from the root colonization to nodule development and function, and some osmoadaptative responses of the two partners have been characterized (7). Although it has been shown that S. meliloti has the ability to use different compatible solutes in presence of an osmotic stress, it appears that glycine betaine is the most potent osmoprotectant and strongly stimulates the growth rate of the bacteria in high-salt medium (8). Unlike enteric bacteria such as Escherichia coli, S. meliloti also catabolizes glycine betaine and uses it both as a carbon and a nitrogen source for growth (9, 10). The importance of this compound in response to osmotic stress is confirmed by the observation that the catabolic pathway is repressed in presence of high-salt concentration, favoring the accumulation of the compound over its metabolization (10). Glycine betaine either can be taken up directly from the environment by specific transport systems or synthesized from choline by a two-step pathway with betaine aldehyde as intermediate (10). This pathway appears to be conserved in bacteria and plants, but shows divergence in the enzymes involved. Gram-positive bacteria such as Arthrobacter pascens and A. globiformis and the fungus Cylindrocarpon didymun use a soluble choline oxidase to catalyze both steps (11, 12). Higher plants and Gram-negative bacteria both are using a conserved betaine aldehyde dehydrogenase to catalyze the betaine aldehyde to glycine betaine reaction. The choline-to-betaine aldehyde reaction, however, is catalyzed by a choline monooxygenase in plants and by a choline dehydrogenase in bacteria such as E. coli, Pseudomonas aeruginosa, and S. meliloti (9, 13–15). The glycine betaine biosynthesis pathway has been characterized at the molecular level in E. coli and Bacillus subtilis (16, 17), and the betBA genes of S. meliloti have been characterized previously (18).
Recently, choline-O-sulfate, synthesized from choline and 3′-phosphoadenosine 5′-phosphosulfate (PAPS), has been identified as a potent osmoprotectant in some plants (4, 19, 20) and fungi (21, 22) in which it seems to be accumulated essentially for sulfate storage. Choline-O-sulfate also is accumulated as an osmolyte in E. coli and B. subtilis that is subjected to an osmotic stress but, like glycine betaine in these organisms, cannot be metabolized further (ref. 19; E. Bremer, personal communication). We report here a genetic and molecular analysis of the S. meliloti bet region that revealed two additional genes encoding a BetI-like putative repressor and a choline sulfatase. We present evidence that the betC gene codes for a choline sulfatase activity that allows S. meliloti to convert choline-O-sulfate and, to a lesser extent, phosphorylcholine into choline.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Media.
All Sinorhizobium meliloti strains used in this study are derived from 102F34R, a rif derivative of wild-type 102F34 (18). The pCHO341 plasmid contains the S. meliloti bet genes cloned as a 8.6-kb BglII fragment into the TcR vector pRK415 (18). For growth, Luria broth (Luria-Bertani; LB) was used as complex medium for E. coli and supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2 (LBmc) for S. meliloti. Defined minimal medium used for S. meliloti was either carbon-free M9 (23) or carbon- and nitrogen-free S medium (10). LAS [S medium + 0.1% (vol/vol) lactic acid + 0.1% (wt/vol) aspartic acid] (9) was used for analysis of the osmotolerance phenotype. Cells were grown in MCAA [S medium + 0.1% (wt/vol) malic acid + 0.1% (wt/vol) casamino acids] (10) for preparation of cell extracts for enzyme assays. Sulfur-free M9 medium was prepared by replacing 1 mM MgSO4 with 1 mM MgCl2. Choline, glycine betaine, and choline-O-sulfate were prepared as 0.5 M solutions and sterilized by filtration before incorporation into defined media. Their final concentration was 1 mM for osmoprotection assays and 10 mM when used as the carbon, nitrogen, and sulfur source. When required, antibiotics were added at concentrations described previously (18, 24).
Chemicals.
Choline chloride and the choline esters were purchased from Sigma/Aldrich. Choline-O-sulfate was prepared from choline chloride and sulfuric acid according to Stevens and Vohra (25). After the two ethanol precipitations, the purity of the choline-O-sulfate formed was confirmed by a modified Dragendorff reagent assay (26), which would have detected traces of residual choline.
Radiolabeled choline ([1,2-14C], 259 MBq/mmol) and phosphorylcholine ([methyl-14C], 1.9 GBq/mmol) were purchased from New England Nuclear. Radiolabeled choline-O-sulfate ([1, 2-14C]-) was synthesized from radiolabeled choline by using the protocol of Stevens and Vohra (25) slightly modified as follows: 200 μl (740 kBq) [1,2-14C]choline was lyophilized in an Eppendorf tube and resuspended in 25 μl concentrated H2SO4. After 6 hr of incubation at 95°C, the tube was cooled slowly to room temperature and 500 μl 1 M Tris⋅HCl, pH8, was added. The [1,2-14C]choline-O-sulfate was purified by ion-exchange chromatography through a 0.5 × 4 cm column packed with Dowex-50×4–200 on H+ form (Sigma). The choline stayed bound to the column whereas the choline-O-sulfate was eluted directly with 2 ml H2O as it remains zwitterionic at very low pH values. The amount of remaining choline could be determined by elution with 2 ml 2 M HCl.
Genetic Techniques.
Triparental bacterial matings were performed by using E. coli MT616 as helper strain (24). Site-directed Tn5 mutagenesis of the pCHO341 plasmid was carried out in E. coli MT614, followed by mobilization into E. coli MT609 as described previously (27). The Ω interposon (Spr) (28) was inserted as an EcoRI restriction fragment in the EcoRI sites of pCHO341. The Tn5 and Ω insertions were recombined into the 102F34R genome by the plasmid-incompatibility technique (29) as described previously (30).
DNA Manipulations and Sequencing.
Standard methods were used for plasmid DNA isolation, restriction analysis, agarose gel electrophoresis, Southern blotting, DNA ligation, and E. coli transformation (31). S. meliloti genomic DNA was prepared by the method of Meade et al. (32). Hybridizations were performed at 65°C with either 32P-labeled [[α-32P]dCTP (Amersham) and Prime-a-Gene (Promega)] or digoxigenin-labeled (DIG DNA Labeling and Detection Kit, Boehringer Mannheim) probes.
The DNA sequence of a HindIII-ApaI 2,785-bp fragment from pCHO341 was determined after subcloning into pBluescript II SK(−) (Stratagene) or pUC118 (33). The sequencing reactions and readings were done by Genome Express (Grenoble, France) by using the fluorescent-terminator technique. Both DNA strands were sequenced. DNA and derived protein sequences were analyzed by using wisconsin genetics computer group (34), blast (35), and clustalv (36) software. The Tn5 insertions in pCHO341 were subcloned into pUC118 as SalI fragments, and the insertion sites were mapped by DNA sequence analysis using an IS50-specific primer (5′-TCACATGGAAGTCAGATCCT-3′).
Enzymology.
Bacterial cultures were grown in MCAA or MCAA + 0.3 M NaCl, supplemented, when desired, with 7 mM choline or choline-O-sulfate, to late log phase. The cells were washed once with the same volume of S medium and resuspended in 0.1 M K2HPO4, pH 7.6 (for BADH and MDH) or 0.1 M Tris⋅Cl, pH 7.6 (for CHS) at 1/100th of the original volume. Choline dehydrogenase, CDH (EC 1.1.99.1) activity was determined on toluene-permeabilized cells by monitoring the production of [1,2-14C]glycine betaine aldehyde as described in Landfald and Strøm (14). For betaine aldehyde dehydrogenase, BADH (EC 1.2.1.8), choline sulfatase, CHS (EC 3.1.6.6), and malate dehydrogenase, MDH (EC 1.1.1.37) assays, the cells were disrupted by two passages through a French press (8,500 lb/in2), followed by centrifugation at 12,000 × g for 10 min at 4°C to remove cell debris. BADH and MDH activities were determined by measuring the NADH production on a spectrophotometer at 340 nm as described previously (10, 37). CHS activity was determined by monitoring the [14C]choline formation from [1,2-14C]choline-O-sulfate or [methyl-14C]phosphorylcholine. No CDH activity (which would further metabolize choline) was present in the supernatant of the extract as this enzyme is bound to the cell membrane. The assay mixture contained 10 μl 14C-labeled substrate (105 dpm), 10 μl 20 mM cold substrate, and 180 μl cell extract. The reaction mix was incubated for 30 min at 30°C and stopped by heating for 5 min at 80°C. The sample was evaporated, resuspended in 10 μl H2O, and analyzed by electrophoresis (60 min, 40 V/cm) on Whatman 3MM paper presoaked in 0.75 M formic acid, pH 2.0. The electrophoregram was submitted to autoradiography, and the production of [14C]choline was measured by quantifying the spots by scintillation counting. Competition assay mixture (200 μl) contained [1,2-14C]choline-O-sulfate (105 dpm), 0.1 mM choline-O-sulfate, 100 mM Tris⋅HCl, pH7.6, and 1 or 10 mM of the tested competitor. Sulfatase activity was determined by using 4-nitrocatechol sulfate (Sigma) as described previously (38).
Protein concentrations of cell extracts were determined by the method of Lowry et al. (39) with BSA as standard.
Nucleotide Sequence Accession Number.
The DNA sequence of the S. meliloti betIC genes has been submitted to the GenBank database and assigned accession number U39940.
RESULTS
Phenotypic Analysis of the bet Region.
In previous work, a S. meliloti Tn5 mutant (LTS23-1020) lacking choline dehydrogenase activity was identified by its inability to grow on choline as sole carbon and nitrogen source and its incapacity to use this compound as osmoprotectant (18). A cosmid clone was isolated by complementation from a pRK290 genomic library containing wild-type S. meliloti 102F34 DNA. The betA gene encoding choline dehydrogenase was identified and sequenced, as was the betB gene encoding betaine aldehyde dehydrogenase located upstream of betA.
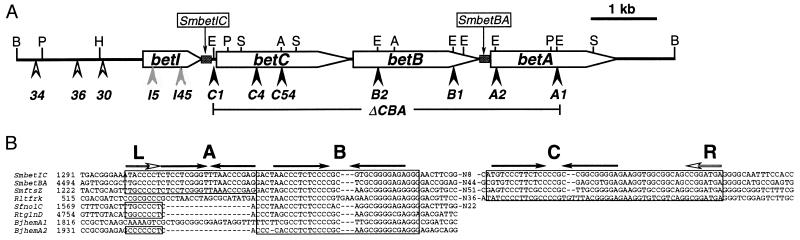
To identify the region required for choline metabolism, we carried out site-directed mutagenesis of the 8.6-kb BglII fragment cloned in pCHO341 by insertion of the Ω interposon in the EcoRI restriction sites and by saturation Tn5 mutagenesis of the plasmid. Twenty-three Tn5 insertions were mapped physically within the BglII fragment by restriction analysis. Complementation assay of seven Tn5 insertions located upstream of betB and the five Ω insertions revealed that only the five Tn5 insertions that mapped upstream of the first EcoRI site (I5, 30, 34, 36, and I45) were able to restore osmoprotection by choline in LTS23-1020 (Fig. 1A). This result suggested that the bet region necessary for choline metabolism extended at least 2 kb upstream of the betBA genes and hinted the presence of one or more extra genes. Five Ω (betA1, betA2, betB1, betB2, and betC1) and five Tn5 (betC4, betI5, 36, betI45, and betC54) insertions were recombined into the 102F34 wild-type genome to confirm the complementation phenotype. The mutant strains obtained with the noncomplementing mutations (betC1, betC4, betC54, betB1, betB2, betA1, and betA2) were unable to use choline as osmoprotectant. Surprisingly, the two insertions, betI5 and betI45, that complemented LTS23–1020 showed reduced osmoprotection by choline, and only insertion 36 retained a wild-type phenotype. No BADH and CDH activity could be detected in the mutant strains UNA156(betB2), UNA208(betC4) and UNA137(betCBA) (Table 1), which suggested that the insertions located upstream of betBA have a polar effect on the expression of these two genes. The mutant UNA130 (betA2) had no detectable CDH activity but retained wild-type BADH level (Table 1), which also supports the presence of an operon in the region spanning insertions betC1 to betA2. Low BADH and CDH activities were found in UNA210 (betI5) and could explain the leaky phenotype of the mutant (Table 1), which likely resulted from an incomplete polar effect of the betI∷Tn5 insertion on betCBA transcription, because introducing a plasmid-located wild-type copy of betI in UNA210 did not improve osmoprotection by choline (data not shown).
Figure 1.
(A) Physical and genetic map of the pROX341 plasmid containing the bet genes of S. meliloti. The insertions with the allele number are shown by arrowheads below the map. The arrowhead content indicates a minus (solid), leaky (gray), or wild-type (open) choline growth phenotype. The position of the two mosaic elements SmbetIC and SmbetBA is indicated by shaded boxes. Restriction sites shown are ApaI (A), BglII (B), EcoRI (E), HindIII (H), PstI (P), and SmaI (S). (B) Sequence alignment of the four mosaic elements and the four palindromes that were identified in Rhizobiaceae. The numbers on the left show the position of the elements within the published sequences. The conserved left and right domains are boxed, and the three palindromes (A, B, and C) are indicated by solid arrows. The flanking left (L) and right (R) heptamers are shown by open arrows. Only the 5-bp flanking sequence of the central region is given with the number of remaining nucleotides (N). Sm, S. meliloti; Rlt, R. leguminosarum bv. trifolii; Sf, S. fredii; Rt, R. tropicii; Bj, B. japonicum.
Table 1.
Choline growth phenotype and enzyme activities in bet mutants
| Strain (genotype) | Growth* | Enzyme activity†
|
||
|---|---|---|---|---|
| CDH | BADH | MDH | ||
| 102F34R (wild type) | 100 | 113 ± 22 | 84 ± 17 | 214 ± 8 |
| UNA130 (betA2∷Ω) | NG | ND | 61 ± 12 | 214 ± 3 |
| UNA156 (betB2∷Ω) | NG | ND | ND | 213 ± 2 |
| UNA208 (betC4∷Tn5) | NG | ND | ND | 185 ± 31 |
| UNA210 (betI5∷Tn5) | 20 | 18 ± 2 | 3 ± 1 | 186 ± 20 |
| UNA137 (betCBAΔΩ) | NG | ND | ND | 190 ± 7 |
NG, no growth improvement compared with LAS + 0.5 M NaCl (<10% wild-type growth). ND, not detected (CDH, <10 nmol/min per mg; BADH, <1 nmol/min per mg).
The ability of the strains to use choline as osmoprotectant was determined in LAS + 0.5 M NaCl + 1 mM choline. Values represent the percentage of wild-type growth.
Cells were grown in MCAA + choline. Enzyme activity was determined from triplicate assays and given in nmol/min per mg of protein ± SEM.
Molecular Characterization of the bet Locus.
The sequence analysis of a HindIII-ApaI fragment localized upstream of betB revealed two nonoverlapping ORFs encoding proteins of, respectively, 203 and 512 aa, designed as BetI and BetC and transcribed in the same direction as betB and betA (Fig. 1A). The betI gene starts with the usual initiation codon ATG while betC appears to start with the rare initiation codon GTG, and both were preceded by a putative ribosome-binding site (RBS) (5′-GAGGAG-N7-ATG-3′ and 5′-GAGGAA-N7-GTG-3′ for betI and betC, respectively). The Tn5 insertions were mapped within the betI coding region after residues 28 (I5) and 175 (I45) and within the betC coding region after residues 301 (C4) and 434 (C54) (Fig. 1A). The first EcoRI site (betC1∷Ω) of the BglII fragment maps between the RBS and the initiation codon of betC. The stop codon of betC is located only 1 nt upstream of the betB start codon, suggesting that the two genes are transcribed together. The betI and betC genes are separated by 167 nt while betA is located 211 nt downstream of betB. In E. coli, the gaps between the betIBA genes never exceed a few nucleotides (40). A comparison analysis by alignment of the betIC and betBA intergenic regions identified an element characterized by two conserved domains of 62 and 50 bp flanking a central region variable in sequence and length (Fig. 1B). The left domain is formed by two large palindromes of 21 (A) and 26 (B) nt while the right domain contains one palindrome of 26 nt (C). The B and C inverted repeats are almost identical with only one base change, suggesting that they originated by duplication. The external heptamers of the left (L) and right (R) domain also form an inverted repeat flanking the entire element. A GenBank search of this region detected two more copies of the element, one in S. meliloti downstream of ftsZ (accession no. L25440) and another downstream of frk in Rhizobium leguminosarum bv. trifolii (accession no. U08434) (Fig. 1B). The latter did not contain the first palindrome (A). The search also identified three other sequences with the palindrome B alone, downstream of nolC in S. fredii (accession no. L03521), downstream of glnD in R. tropicii (accession no. U47030), and in duplicated form downstream of hemA in B. japonicum (accession no. M16751) (Fig. 1B). These results suggest that this palindrome is a repeat element that can form a more complex structure leading to the mosaic element found in the S. meliloti bet genes.
Analysis of the Encoded Proteins.
A blastp search of GenBank revealed that BetI showed homology to regulatory proteins of the TetR/AcrR family (41). These regulators of approximately 200 aa in length are characterized by a conserved N-terminal region that contains a helix–turn–helix motif involved in binding to their DNA target sequence (42). Only with one member of this family, the BetI repressor of E. coli, did the homology (33.5% identity) extend to the entire protein (Fig. 2A). E. coli BetI has been shown to regulate the expression of the betIBA operon depending on the choline concentration (16, 43).
Figure 2.
clustalv alignment of the deduced amino acid sequence for S. meliloti BetI (A) and BetC (B). The BetI regulator (SmBetI) is compared with its homologue in E. coli (EcBetI). The choline sulfatase (SmBetC) is compared with the phosphonate monoester hydrolase of Burkholderia caryophilli (BcPehA), the hypothetical YidJ protein of E. coli (EcYidJ), and the arylsulfatases of Pseudomonas aeruginosa (PaAtsa), E. coli (EcAslA), and Klebsiella pneumoniae (KpAtsA). Boxes numbered 1–6 indicate conserved domains in all six sequences. Strictly conserved residues are shown in black, and residues conserved in at least four sequences are shown in gray.
A similar analysis of BetC, which is not present in the E. coli bet operon, revealed weak but significant homology to a family of hydrolases that comprises mostly sulfatases. The highest homology was found with a phosphonate monoester hydrolase from Burkholderia caryophilli (27% identity, GenBank accession no. U44852), human iduronate sulfatase and arylsulfatase (26% identity, SwissProt accession nos. P22304 and P51689), a hypothetical sulfatase-like protein from E. coli (24% identity, SwissProt accession no. P31447), and arylsulfatases from Pseudomonas aeruginosa, E. coli, and Klebsiella pneumoniae (23% identity, SwissProt accession nos. P51691, P25549, and P20713). An alignment of the characterized prokaryotic member of this family allowed the identification of six conserved domains separated by regions with high variability in length (Fig. 2B). The domains 1, 2, 3, 5, and 6 are also conserved in eukaryotic sulfatases, with the same strictly conserved residues (44).
Importance of Choline Precursors in S. meliloti Nutrition and Osmotolerance.
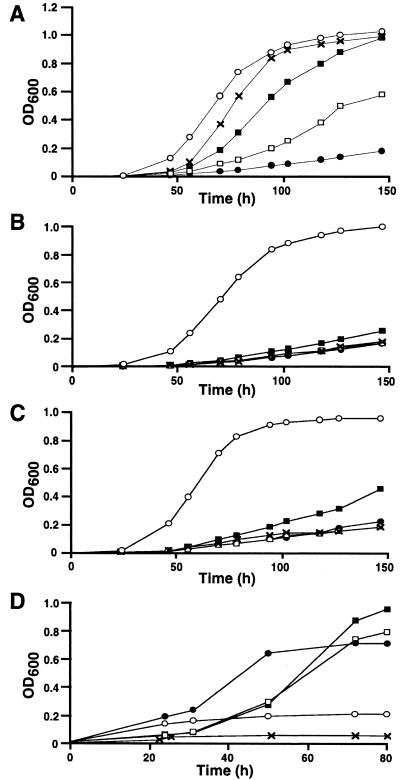
The above result prompted us to search for the substrate hydrolyzed by BetC. The transformation of choline to glycine betaine requires only the enzymes encoded by betA and betB (10). As glycine betaine is the final product of the pathway, the likely function of BetC would be the transformation of an unknown precursor into choline. We examined the ability of wild-type S. meliloti and several bet mutants to utilize phosphorylcholine and choline-O-sulfate, two possible precursors of choline, as osmoprotectants. In a wild-type background (Fig. 3A), choline-O-sulfate is capable of restoring growth in LAS medium containing a high salt concentration (0.5 M NaCl) much more efficiently than phosphorylcholine. The mutant strains UNA156 (betB2∷Ω) and UNA208 (betC4∷Tn5) were compared with the wild-type parental strain 102F34R (Fig. 3 B and C). Whereas the UNA156 mutant grew well when glycine betaine was used as osmoprotectant, choline, choline-O-sulfate, and phosphorylcholine could not improve growth in the presence of 0.5 M NaCl. The mutant UNA208 had a similar phenotype for choline and phosphorylcholine, but choline-O-sulfate showed some osmoprotection, albeit much weaker than in the wild-type strain. These results indicate that S. meliloti does not use these precursors as osmolytes per se, but are metabolized to glycine betaine via choline.
Figure 3.
Osmoprotection growth phenotype of S. meliloti strains 102F34R (A), UNA156 (B), and UNA208 (C) grown in LAS + 0.5 M NaCl (•) supplemented with 1 mM of glycine betaine (○), choline (X), choline-O-sulfate (■), or phosphorylcholine (□). Growth phenotype (D) of S. meliloti strain 102F34R in sulfur-free M9 medium (NaCl, 0) supplemented with 10 mM of glycine betaine + MgSO4 (•), glycine betaine (○), choline-O-sulfate + MgSO4 (■), or choline-O-sulfate (□), and of strain UNA208 in the same M9 medium supplemented with choline-O-sulfate + MgSO4 (X).
The ability of choline-O-sulfate to act as sole carbon and nitrogen sources was tested in low osmolarity (NaCl, 0) M9 medium (Fig. 3D). Growth of the wild-type strain was supported by choline-O-sulfate but less efficiently than with glycine betaine while a betC mutant (UNA208) was not able to use choline-O-sulfate for growth. Choline-O-sulfate also was assayed as a potential sole sulfur source for growth. While 102F34R required sulfate in the medium when growing with glycine betaine as the sole carbon source, the absence of additional sulfate did not reduce significantly the growth when choline-O-sulfate is present (Fig. 3D). These results are consistent with the ability of S. meliloti to use choline-O-sulfate as the sole carbon, nitrogen, and sulfur sources for growth via the glycine betaine biosynthetic pathway.
betC Encodes a Choline Sulfatase.
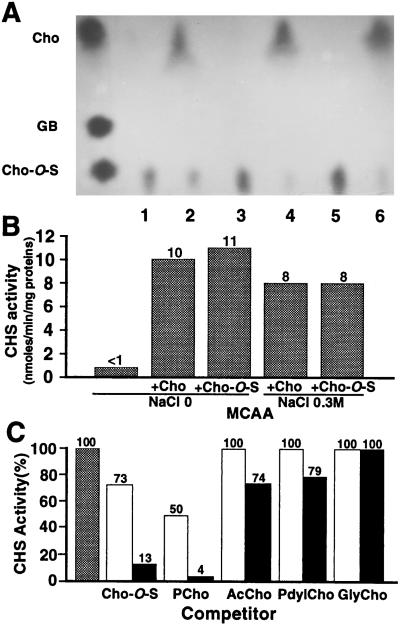
To demonstrate that choline-O-sulfate is converted into choline, enzymatic analysis was performed with wild-type cells grown in MCAA medium alone or supplemented with choline or choline-O-sulfate. Cell extracts were incubated with 14C-choline-O-sulfate (7 mM) as described in Materials and Methods, and the reaction mixtures were subjected to paper electrophoresis (Fig. 4A). 14C-choline was produced when the cells were grown in presence of choline or choline-O-sulfate, but not in MCAA alone (Fig. 4). The presence of a high-salt concentration (0.3 M NaCl) in the growth medium slightly reduced the choline sulfatase activity (Fig. 4B). The S. meliloti choline sulfatase was found to be partially inhibited by phosphate as the presence of 50 mM KH2PO4 in the reaction mix reduced the measured activity by approximately 65% (data not shown). Since choline sulfatase of lower eukaryotes has been shown to be induced when sulfate is absent, S. meliloti cells grown in sulfur-free M9-mannitol medium using methionine as the sulfur source were assayed for activity. No 14C-choline was produced (Fig. 4A, lane 3) which indicates that choline sulfatase was induced only by the substrate or by choline. The assay of extracts obtained from the mutant strains UNA156(betB2∷Ω) and UNA208(betC4∷Tn5) grown in MCAA supplemented with choline-O-sulfate clearly showed that the inducible choline sulfatase activity was encoded by the betC gene (Fig. 4A, lanes 5 and 6).
Figure 4.
(A) Autoradiography after electrophoresis of cell extracts incubated 30 min with 14C-choline-O-sulfate. Cell extracts were obtained from wild-type strain 102F34R grown in MCAA (lane 1), MCAA + choline (lane 2), MCAA + choline-O-sulfate (lane 4), and sulfur-free M9 with 0.1% (wt/vol) methionine (lane 3). Cell extracts from mutants UNA208 (lane 5) and UNA156 (lane 6) were obtained after growth in MCAA + choline-O-sulfate. (B) Choline sulfatase activity determined from wild-type 102F34R cell extracts grown under different conditions. Each value was obtained from duplicate assays. (C) Competition assay with 10- (open) and 100-fold (solid) excess competitor compared with choline-O-sulfate using wild-type cell extract grown in MCAA + choline. Shaded column, no competitor. Cho, choline; Cho-O-S, choline-O-sulfate; GB, glycine betaine; Pcho, phosphorylcholine; AcCho, acetylcholine; PdylCho, phosphatidylcholine; GlyCho, glycerophosphatidylcholine.
The substrate specificity of the enzyme was analyzed by determining the ability of different choline esters to compete with choline-O-sulfate as substrate. Of four compounds tested, only phosphorylcholine showed significant competition (Fig. 4C). Acetylcholine and phosphatidylcholine inhibited choline-O-sulfate hydrolysis by less than 26% when supplied in a 100-fold excess while glycerophosphatidylcholine had no visible effect. The ability of the enzyme to hydrolyze phosphorylcholine was confirmed by using 14C-labeled substrate, but the measured activity (1.7 nmol/min per mg protein) is low compared with choline-O-sulfate (10 nmol/min per mg protein).
DISCUSSION
The molecular analysis of the bet locus in S. meliloti identified four genes, betICBA, organized in an operon. In addition to the previously described betBA genes encoding, respectively, betaine aldehyde dehydrogenase and choline dehydrogenase (18), the betI gene also shares homology to the betI gene of E. coli (40). Its product is likely to be involved in the regulation of the S. meliloti bet operon although this has not yet been demonstrated experimentally. The S. meliloti bet locus is characterized by the presence of an extra gene, betC, located between betI and betB, absent from the E. coli bet locus. Interestingly, the bet genes of S. meliloti are separated by significantly large intergenic regions of, respectively, 167 and 211 bp between betIC and betBA. A highly conserved domain within these two regions has also been found in other rhizobia sequences and shows a complex secondary structure (Fig. 1B) characteristic of mosaic elements (45). Two such elements named RIME1 and RIME2 have been identified previously in S. meliloti and other rhizobia (30), but the sequence identified in the bet locus differs by several points: (i) three conserved palindromes are present in the element instead of two, (ii) two of the palindromes are highly homologous, and (iii) the central domain is very variable in sequence and size while the RIME are conserved over their entire length. The presence of single copies of one of the palindrome in rhizobial genomes (Fig. 1B) suggests that the complex structure of the bet mosaic elements was formed by duplication, and incorporation of different motifs, as it is the case for the enterobacteria bacterial-interspersed mosaic elements (BIME), which were formed from duplication of the REP palindromic unit (46). The reason for the presence of two conserved mosaic elements within the S. meliloti bet operon is unknown. Although the phenotypical data obtained with the bet mutants indicate that they form a single transcriptional unit, previously determined enzyme activity does suggest that some degree of differential regulation occurs between the betaine aldehyde dehydrogenase and the choline dehydrogenase (10), and these mosaic elements could be involved in posttranscriptional regulation of the bet operon.
The most striking finding of this work was the presence of a novel gene, betC, in the S. meliloti bet operon. The BetC protein shared significant homology to sulfatases and was identified as a choline sulfatase by enzymatic assays (Fig. 4). Choline sulfatase, first identified in Pseudomonas nitroreducences (47), has been characterized in P. aeruginosa (48) and in filamentous fungi (49). The S. meliloti enzyme seemed to have more characteristics similar to the choline sulfatase from Pseudomonas enzyme than from fungi. The enzyme from Aspergillus was inhibited completely by 10 μM PO43− (50) while in S. meliloti and Pseudomonas some activity was retained at 50 mM PO43− (47). As choline-O-sulfate acts as a sulfur storage compound in fungi, the enzyme, like other sulfatases, is activated only during sulfur deficiency (49). In S. meliloti and Pseudomonas, however, choline sulfatase is induced by the substrate that is used as a carbon, nitrogen, and/or sulfur source for growth (Fig. 3) (22). Of the other choline esters tested, only phosphorylcholine showed competition with choline-O-sulfate (Fig. 4C) and was hydrolyzed, although the phosphatase activity of the enzyme was less than 20% of the sulfatase activity. Phosphorylcholine seemed to be a strong competitor, suggesting that the enzyme had a high affinity for this substrate, but this may also be because of a slower reaction (the enzyme is bound longer to the substrate) or an indirect effect caused by PO43− ions, which were shown previously to inhibit choline sulfatase activity. Phosphorylcholine was also a competitor for the P. aeruginosa choline sulfatase, but was not hydrolyzed (48). It is worth noticing that this strain has a periplasmic phosphorylcholine phosphatase that is lacking in S. meliloti (51). The reduced osmoprotection conferred by phosphorylcholine (Fig. 3) thus may be correlated to the low phosphatase activity observed. Interestingly, choline, the product of the reaction, was also an inducer of choline sulfatase synthesis in S. meliloti (Fig. 4), which would be in agreement with the bet genes forming an operon whose unique function is to synthesize glycine betaine. Likewise, no significant hydrolysis of 4-nitrocatechol sulfate in cell extracts from 102F34R cultures in sulfate-rich medium (data not shown) indicates that the enzyme is not an arylsulfatase. In addition to its role in sulfur storage in fungi and growth metabolite in some bacteria, choline-O-sulfate can act as a compatible solute in response to osmotic stress and is accumulated as such by plant species of the Plumbaginaceae family (19). Bacteria such as E. coli also could use choline-O-sulfate as osmoprotectant, but less efficiently than glycine betaine (19). Unlike S. meliloti, enterobacteria that are lacking choline sulfatase activity cannot metabolize choline-O-sulfate, and this may be linked to the inability of these bacteria to use choline or glycine betaine except as osmoprotectant. S. meliloti, however, does not use choline-O-sulfate as osmoprotectant per se since this compound cannot significantly restore growth in salt-stressed bet mutants (Fig. 3C).
The relationship between compounds of plant root exudates and compounds found in root is poorly understood, but many root compounds are probably exuded under environmental stresses encountered in soil (52). Root extracts of alfalfa, the host plant of S. meliloti, contain many amino acids, organic acids, numerous carbohydrates (7), and betaines such as stachydrine (proline betaine) and trigonelline (53). In addition, the natural occurrence of choline and derivatives is widespread in higher plants. In Phaseolus vulgaris, the choline pool ranged from 3.7 μmol/g fresh weight in leaves to 2.2 μmol/g fresh weight in roots whereas acetylcholine level was much lower (54). In alfalfa shoots, the level of choline was found to be increased by 100% under salt stress (55). The existence of high levels of choline-O-sulfate mainly in roots of Plumbaginaceae such as Limonium vulgare grown in saline conditions is well established (19). Furthermore, this compound accounted for 5–15% of sulfate in Zea mays, Hordeum vulgare, and Helianthus annuus (21). However, in mesophyte plants, choline-O-sulfate levels usually are low (<1 μmol/g dry weight) and much lower than the levels of free choline and its esters, phosphoryl-, glycerophosphoryl-, and phosphatidylcholine (56). Nevertheless, all these compounds eventually may reach soil bacteria though exudation or decomposition of plant material. Any of these compounds present in sufficient concentration could serve as an energy substrate for the growth of rhizosphere microbes. This concept is of particular interest in the case of S. meliloti because, besides the genes involved in catabolism of major compounds (amino acids, organic acids, carbohydrates), various strains contain genes that permit catabolism of stachydrine, trigonelline, and carnitine (57). Data presented here indicate that in addition to choline, choline-O-sulfate and phosphorylcholine also can be used by S. meliloti either as an energy substrate or as an osmoprotectant after subsequent conversion into glycine betaine. Given the competition among soil bacteria for plant carbon sources, it is tempting to argue that S. meliloti is better adapted than other soil bacteria for using substrate or osmoprotectant. Indeed, it has the unique capacity to use choline-O-sulfate as an energy source, whereas B. subtilis, for example, can use this compound only to cope effectively with high osmolarity (E. Bremer, personal communication). Altogether, these data form the basis for detailed investigations on the regulation of the bet locus of S. meliloti in free-living cells and also in planta at different symbiotic stages.
Acknowledgments
This work was funded by the European Communities Biotech Program, as part of the Project of Technological Priority 1993–1997 (BIO2CT930400, D.L.R.), and the Centre National de la Recherche Scientifique. E.B. received a doctoral fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche. We are grateful to T. M. Finan, N. T. Keen, J. Frey, and J. A. Pocard for providing bacterial strains and plasmids.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U39940).
References
- 1.Le Rudulier D, Strøm A R, Dandekar A M, Smith L T, Valentine R C. Science. 1984;224:1064–1068. doi: 10.1126/science.224.4653.1064. [DOI] [PubMed] [Google Scholar]
- 2.Csonka L N. Microbiol Rev. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Csonka L N, Hanson A D. Annu Rev Microbiol. 1991;45:569–606. doi: 10.1146/annurev.mi.45.100191.003033. [DOI] [PubMed] [Google Scholar]
- 4.Rhodes D, Hanson A D. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:357–384. [Google Scholar]
- 5.Miller K J, Wood J M. Annu Rev Microbiol. 1996;50:101–136. doi: 10.1146/annurev.micro.50.1.101. [DOI] [PubMed] [Google Scholar]
- 6.Long S R. Cell. 1989;56:203–214. doi: 10.1016/0092-8674(89)90893-3. [DOI] [PubMed] [Google Scholar]
- 7.Fougère F, Le Rudulier D, Streeter J G. Plant Physiol. 1991;96:1228–1236. doi: 10.1104/pp.96.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Rudulier D, Bernard T. FEMS Microbiol Rev. 1986;39:67–72. [Google Scholar]
- 9.Bernard T, Pocard J A, Perroud B, Le Rudulier D. Arch Microbiol. 1986;143:359–364. [Google Scholar]
- 10.Smith L T, Pocard J A, Bernard T, Le Rudulier D. J Bacteriol. 1988;170:3142–3149. doi: 10.1128/jb.170.7.3142-3149.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tani Y, Mori N, Ogata K, Yamada H. Agric Biol Chem. 1979;43:815–820. [Google Scholar]
- 12.Rozwadowski K L, Khachatourians G G, Selvaraj G. J Bacteriol. 1991;173:472–478. doi: 10.1128/jb.173.2.472-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagasawa T, Mori N, Tani Y, Ogata K. Agric Biol Chem. 1976;40:2077–2084. [Google Scholar]
- 14.Landfald B, Strøm A R. J Bacteriol. 1986;165:849–855. doi: 10.1128/jb.165.3.849-855.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rathinasabapathi B, Burnet M, Russell B L, Gage D A, Liao P C, Nye G J, Scott P, Golbeck J H, Hanson A D. Proc Natl Acad Sci USA. 1997;94:3454–3458. doi: 10.1073/pnas.94.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamark T, Røkenes T P, McDougall J, Strøm A R. J Bacteriol. 1996;178:1655–1662. doi: 10.1128/jb.178.6.1655-1662.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boch J, Kempf B, Schmid R, Bremer E. J Bacteriol. 1996;178:5121–5129. doi: 10.1128/jb.178.17.5121-5129.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pocard J A, Vincent N, Boncompagni E, Smith L T, Poggi M C, Le Rudulier D. Microbiology. 1997;143:1369–1379. doi: 10.1099/00221287-143-4-1369. [DOI] [PubMed] [Google Scholar]
- 19.Hanson A D, Rathinasabapathi B, Chamberlin B, Gage D A. Plant Physiol. 1991;97:1199–1205. doi: 10.1104/pp.97.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanson A D, Rathinasabapathi B, Rivoal J, Burnet M, Dillon M O, Gage D A. Proc Natl Acad Sci USA. 1994;91:306–310. doi: 10.1073/pnas.91.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nissen P, Benson A A. Science. 1961;134:1759. doi: 10.1126/science.134.3492.1759. [DOI] [PubMed] [Google Scholar]
- 22.Harada T. Biochim Biophys Acta. 1964;81:193–196. [Google Scholar]
- 23.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. p. 431. [Google Scholar]
- 24.Finan T M, Kunkel B, DeVos G F, Signer E R. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevens C M, Vohra P. J Amer Chem Soc. 1955;77:4935–4936. [Google Scholar]
- 26.Stumpf D K. Plant Physiol. 1984;75:273–274. doi: 10.1104/pp.75.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarosh O K, Charles T C, Finan T M. Mol Microbiol. 1989;3:813–823. doi: 10.1111/j.1365-2958.1989.tb00230.x. [DOI] [PubMed] [Google Scholar]
- 28.Frey J, Krisch H M. Gene. 1985;36:143–150. doi: 10.1016/0378-1119(85)90078-2. [DOI] [PubMed] [Google Scholar]
- 29.Ruvkun G B, Ausubel F M. Nature (London) 1981;289:85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- 30. Østerås M, Stanley J, Finan T M. J Bacteriol. 1995;177:5485–5494. doi: 10.1128/jb.177.19.5485-5494.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T E. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 32.Meade H M, Long S R, Ruvkun G B, Brown S E, Ausubel F M. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vieira J, Messing J. Methods Enzymol. 1987;153:3–34. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 34.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul S F, Gish W, Miller W, Meyers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Higgins D G, Bleasby A J, Fuchs R. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 37.Englard S, Siegal L. Methods Enzymol. 1969;13:99–100. [Google Scholar]
- 38.Beil S, Kehrli H, James P, Staudenmann W, Cook A M, Leisinger T, Kertesz M A. Eur J Biochem. 1995;229:385–394. doi: 10.1111/j.1432-1033.1995.0385k.x. [DOI] [PubMed] [Google Scholar]
- 39.Lowry O H, Rosebrough N J, Farr A L, Randall R J. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 40.Lamark T, Kaasen I, Eshoo M W, McDougall J, Strøm A R. Mol Microbiol. 1991;5:1049–1064. doi: 10.1111/j.1365-2958.1991.tb01877.x. [DOI] [PubMed] [Google Scholar]
- 41.Pan W, Spratt B G. Mol Microbiol. 1994;11:769–775. doi: 10.1111/j.1365-2958.1994.tb00354.x. [DOI] [PubMed] [Google Scholar]
- 42.Wissmann A, Baumeister R, Muller G, Hecht B, Pfleiderer K, Hillen W. EMBO J. 1991;10:4145–4152. doi: 10.1002/j.1460-2075.1991.tb04992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Røkenes T P, Lamark T, Strøm A R. J Bacteriol. 1996;178:1663–1670. doi: 10.1128/jb.178.6.1663-1670.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters C, Schmidt B, Rommerskirch W, Rupp K, Zuhlsdorf M, Vingron M, Meyer H E, Pohlmann R, von Figura K. J Biol Chem. 1990;265:3374–3381. [PubMed] [Google Scholar]
- 45.Lupski J R, Weinstock G M. J Bacteriol. 1992;174:4525–4529. doi: 10.1128/jb.174.14.4525-4529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilson E, Saurin W, Perrin D, Bachellier S, Hoffnung M. Nucleic Acids Res. 1991;19:1375–1383. doi: 10.1093/nar/19.7.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takebe I. J Biochem (Tokyo) 1961;50:245–255. doi: 10.1093/oxfordjournals.jbchem.a127440. [DOI] [PubMed] [Google Scholar]
- 48.Lucas J J, Burchiel S W, Segel I H. Arch Biochem Biophys. 1972;153:664–672. doi: 10.1016/0003-9861(72)90385-2. [DOI] [PubMed] [Google Scholar]
- 49.Spencer B, Hussey E C, Orsi B A, Scott J M. Biochem J. 1968;106:461–469. doi: 10.1042/bj1060461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott J M, Spencer B. Biochem J. 1968;106:471–477. doi: 10.1042/bj1060471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lucchini A E, Lisa T A, Domenech C H. Arch Microbiol. 1990;153:596–599. doi: 10.1007/BF00245271. [DOI] [PubMed] [Google Scholar]
- 52.Koshino H, Masaoka Y, Ichihara A. Phytochemistry. 1993;33:1075–1077. [Google Scholar]
- 53.Wood K V, Stringham K J, Smith D L, Volenec J J, Kerry L, Hendershot L, Jackson K A, Rich P J, Yang W-J, Rhodes D. Plant Physiol. 1991;96:892–897. doi: 10.1104/pp.96.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miura G A, Shih T M. Physiol Plant. 1984;61:417–421. [Google Scholar]
- 55.Wyn Jones R G, Storey R. In: The Physiology and Biochemistry of Drought Resistance in Plant. Paleg L G, Aspinall D, editors. Sydney: Academic; 1981. pp. 171–204. [Google Scholar]
- 56.Hanson A D, Gage D A. Aust J Plant Physiol. 1991;18:317–327. [Google Scholar]
- 57.Goldmann A, Boivin C, Fleury V, Message B, Lecoeur L, Maille M, Tepfer D. Mol Plant Microbe Interact. 1991;4:571–578. doi: 10.1094/mpmi-4-571. [DOI] [PubMed] [Google Scholar]