Abstract
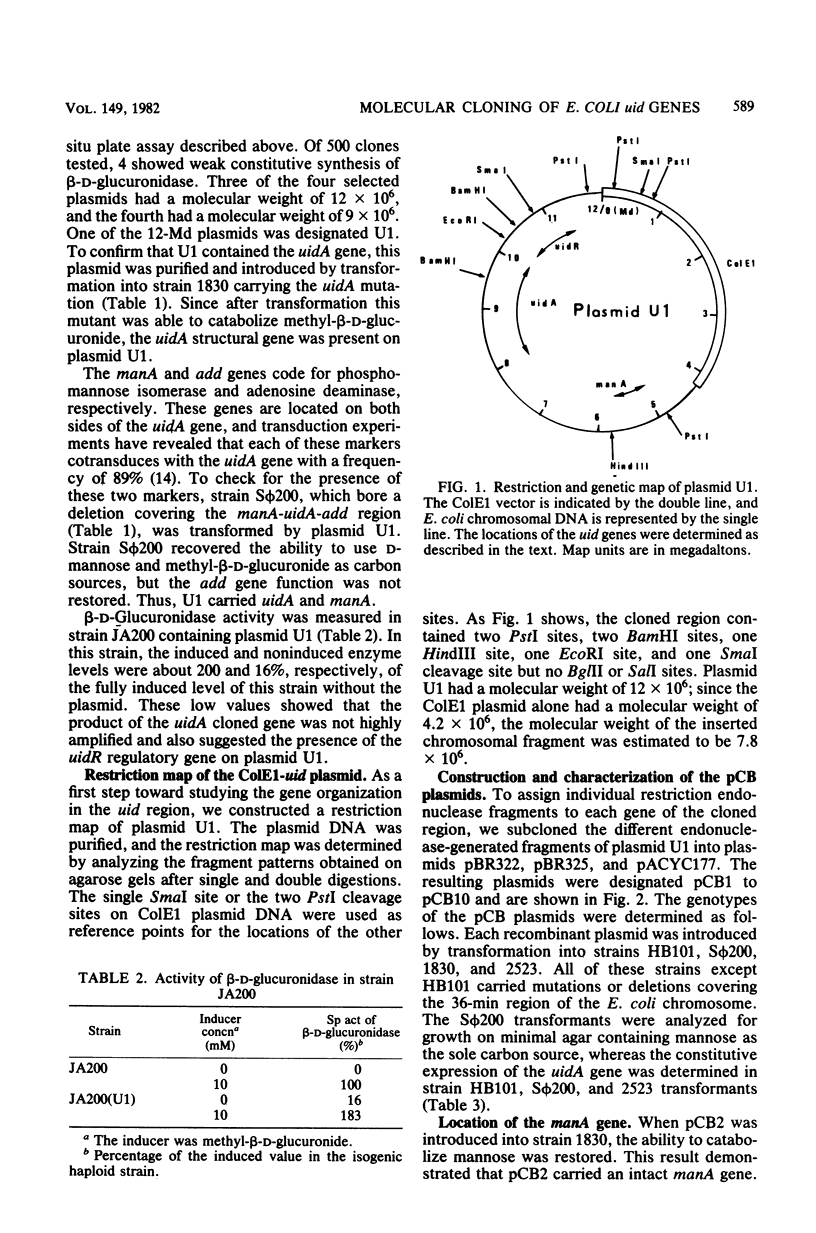
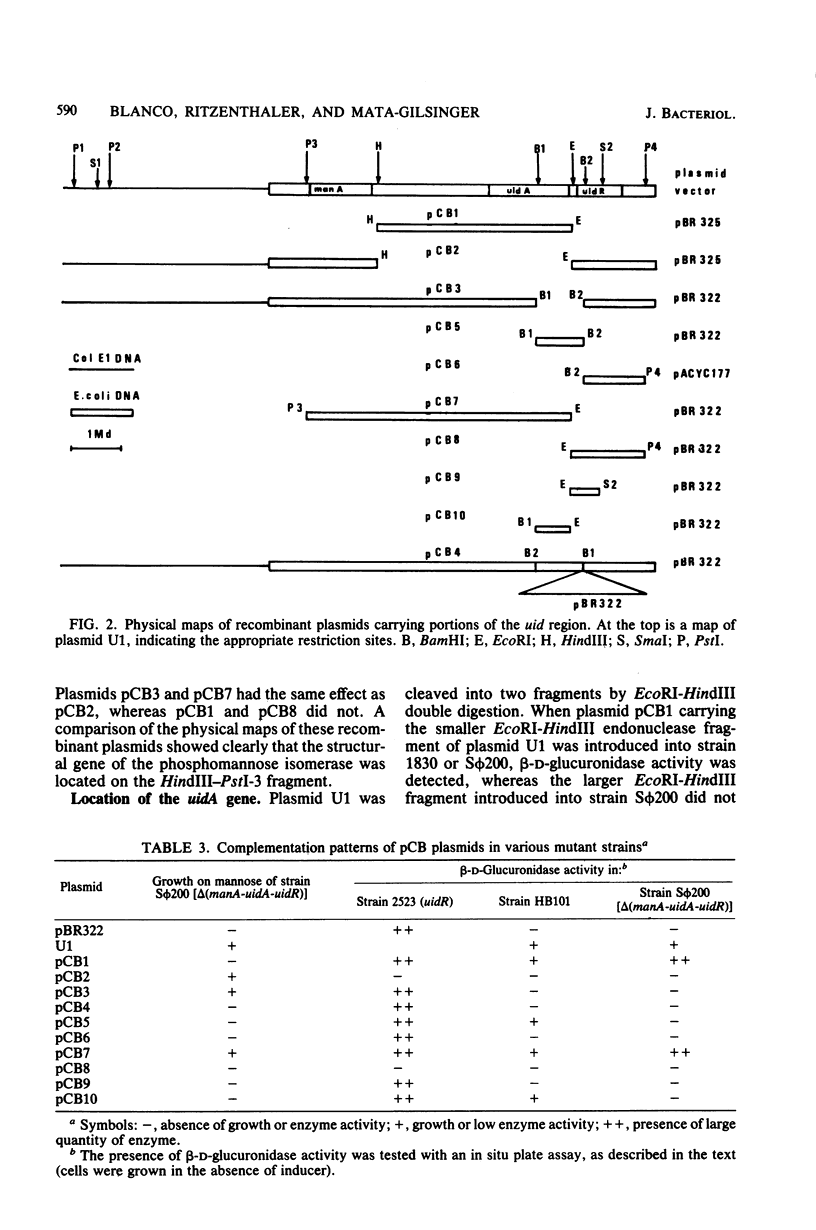
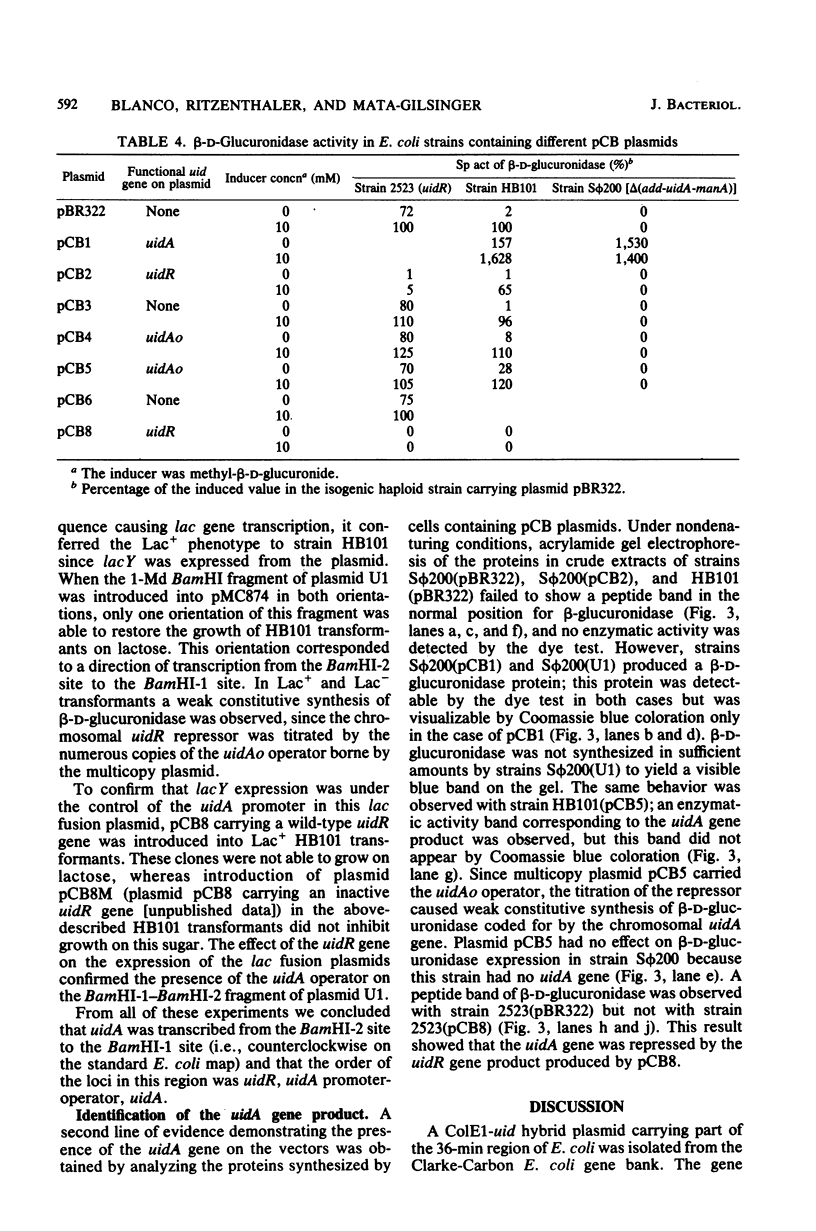
The two genes of the Escherichia coli K-12 uid region (the structural gene uidA and the regulatory gene uidR) were isolated on a ColE1-uid hybrid plasmid from the bank of Clarke and Carbon. We made a restriction map of this region and correlated it with the genetic map by subcloning the uid restriction fragments into plasmids pBR322, pBR325, and pACYC177. In these plasmids, amplification of the products of the uidA and uidR genes occurred. The enzyme coded for by uidA was identified by polyacrylamide gel electrophoresis of crude extracts from strains containing the uidA plasmid. A 1-megadalton EcoRI-BamHI segment contained the uidAo operator, and the direction of transcription of the uidA gene was determined. The restriction analysis also suggested that the order of the loci in this region is manA, uidA, uidAo, uidR.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barker D. F., Kuhn J., Campbell A. M. Sequence and properties of operator mutations in the bio operon of Escherichia coli. Gene. 1981 Jan-Feb;13(1):89–102. doi: 10.1016/0378-1119(81)90046-9. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds J., Barrett C. Characterization of Escherichia coli mutants tolerant to bacteriocin JF246: two new classes of tolerant mutants. J Bacteriol. 1973 Nov;116(2):885–892. doi: 10.1128/jb.116.2.885-892.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyneker H. L., Shine J., Goodman H. M., Boyer H. W., Rosenberg J., Dickerson R. E., Narang S. A., Itakura K., Lin S., Riggs A. D. Synthetic lac operator DNA is functional in vivo. Nature. 1976 Oct 28;263(5580):748–752. doi: 10.1038/263748a0. [DOI] [PubMed] [Google Scholar]
- Jochimsen B., Nygaard P., Vestergaard T. Location on the chromosome of Escherichia coli of genes governing purine metabolism. Adenosine deaminase (add), guanosine kinase (gsk) and hypoxanthine phosphoribosyltransferase (hpt). Mol Gen Genet. 1975 Dec 30;143(1):85–91. doi: 10.1007/BF00269424. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Marians K. J., Wu R., Stawinski J., Hozumi T., Narang S. A. Cloned synthetic lac operator DNA is biologically active. Nature. 1976 Oct 28;263(5580):744–748. doi: 10.1038/263744a0. [DOI] [PubMed] [Google Scholar]
- Mata M., Delstanche M., Robert-Baudouy J. Isolation of specialized transducing bacteriophages carrying the structural genes of the hexuronate system in Escherichia coli K-12: exu region. J Bacteriol. 1978 Feb;133(2):549–557. doi: 10.1128/jb.133.2.549-557.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novel G., Didier-Fichet M. L., Stoeber F. Inducibility of beta-glucuronidase in wild-type and hexuronate-negative mutants of Escherichia coli K-12. J Bacteriol. 1974 Oct;120(1):89–95. doi: 10.1128/jb.120.1.89-95.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novel G., Novel M. Mutants d'Escherichia coli K 12 affectés pour leur croissance sur méthyl-beta-D-glucuronide: localisation of gène de structure de la beta-D-glucuronidase (uid A. Mol Gen Genet. 1973;120(4):319–335. [PubMed] [Google Scholar]
- Novel M., Novel G. Regulation of beta-glucuronidase synthesis in Escherichia coli K-12: constitutive mutants specifically derepressed for uidA expression. J Bacteriol. 1976 Jul;127(1):406–417. doi: 10.1128/jb.127.1.406-417.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novel M., Novel G. Regulation of beta-glucuronidase synthesis in Escherichia coli K-12: pleiotropic constitutive mutations affecting uxu and uidA expression. J Bacteriol. 1976 Jul;127(1):418–432. doi: 10.1128/jb.127.1.418-432.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzenthaler P., Mata-Gilsinger M., Stoeber F. Construction and expression of hybrid plasmids containing Escherichia coli K-12 uxu genes. J Bacteriol. 1980 Sep;143(3):1116–1126. doi: 10.1128/jb.143.3.1116-1126.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzenthaler P., Mata-Gilsinger M., Stoeber F. Molecular cloning of Escherichia coli K-12 hexuronate system genes: exu region. J Bacteriol. 1981 Jan;145(1):181–190. doi: 10.1128/jb.145.1.181-190.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SISTROM W. R. On the physical state of the intracellularly accumulates substrates of beta-galactoside-permease in Escherichia coli. Biochim Biophys Acta. 1958 Sep;29(3):579–587. doi: 10.1016/0006-3002(58)90015-5. [DOI] [PubMed] [Google Scholar]
- Spudich J. A., Horn V., Yanofsky C. On the production of deletions in the chromosome of Escherichia coli. J Mol Biol. 1970 Oct 14;53(1):49–67. doi: 10.1016/0022-2836(70)90045-8. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J Mol Biol. 1973 Sep 15;79(2):237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]
- Sykes R. B., Nordström K. Microiodometric determination of beta-lactamase activity. Antimicrob Agents Chemother. 1972 Feb;1(2):94–99. doi: 10.1128/aac.1.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin B. E., Nordström K. Plasmid incompatibility and control of replication: copy mutants of the R-factor R1 in Escherichia coli K-12. J Bacteriol. 1975 Nov;124(2):641–649. doi: 10.1128/jb.124.2.641-649.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin B. E., Nordström K. R plasmid gene dosage effects in Escherichia coli K-12: copy mutants of the R plasmic R1drd-19. Plasmid. 1977 Nov;1(1):1–7. doi: 10.1016/0147-619x(77)90003-8. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]