Abstract
As reported before, the metabolic activity of nucleus basalis neurons is reduced significantly in Alzheimer patients. Because the apolipoprotein E (ApoE) ɛ4 genotype is a major risk factor for Alzheimer’s disease (AD), we determined whether the decrease in metabolic activity in nucleus basalis neurons in AD is ApoE-type dependent. The size of the Golgi apparatus (GA) was determined as a measure of neuronal metabolic activity in 30 controls and 41 AD patients with a known ApoE genotype by using an image analysis system in the nucleus basalis of Meynert. A polyclonal antibody directed against MG-160, a sialoglycoprotein of the GA, was used to visualize this organelle. There was a very strong reduction in the size of the GA in the nucleus basalis of AD patients. Furthermore, a strong and significant extra reduction in the size of the GA was found in the nucleus basalis neurons of AD patients with either one or two ApoE ɛ4 alleles compared with Alzheimer patients without ApoE ɛ4 alleles. Our data show that the decreased activity of nucleus basalis neurons in AD is ApoE ɛ4 dependent and suggest that ApoE ɛ4 participates in the pathogenesis of AD by decreasing neuronal metabolism.
Alzheimer’s disease (AD) is the most common cause of dementia in elderly. This disorder is characterized by progressive memory loss, other cognitive impairments, and by neuropathological lesions, i.e., neuritic plaques, neurofibrillary tangles, and neuropil threads (1, 2). Epidemiological and molecular genetic studies have revealed that the genetic variation in apolipoprotein E (ApoE) is an important risk factor for AD (3–6).
Human ApoE is a 37-kDa protein encoded by a four-exon gene of 3.6 kb in length located on the long arm of chromosome 19. ApoE polymorphism consists of three types, i.e., ApoE ɛ2, ApoE ɛ3, and ApoE ɛ4, which results in six different ApoE phenotypes in the population (6). ApoE ɛ3, the most common isoform, has a cysteine at residue 112 and an arginine at residue 158, whereas ApoE ɛ4 has an arginine at both sites. ApoE ɛ2 has a cysteine at both sites. ApoE ɛ2, ɛ3, and ɛ4 have allele frequencies of ≈0.08, 0.78, and 0.14, respectively (7, 8). The inheritance of one or two ApoE ɛ4 alleles increases the risk of AD and decreases the age of onset of this disease (9), whereas ApoE ɛ2 appears to reduce the risk of AD and increase the age of onset (10). The ApoE ɛ4/4 genotype is associated with a mean age of AD onset of 60–70 yr in most populations studied. Few ApoE ɛ4/4 individuals reach the age of 90 yr without developing AD (11–14). The presence of ApoE ɛ4 has a direct impact on amyloid accumulation, neurofibrillary tangle formation, neurotrophin receptor loss (15), and cholinergic deficits (15–18).
The suggestion that reduced neuronal activity in AD brains may by itself be a crucial hallmark for AD (19, 20) raised questions on the nature of the relationship between AD pathology and neuronal activity. In a series of studies, we established that plaques, tangles, and decreased neuronal activity as determined by the size of the Golgi apparatus (GA) occur independently from each other in various brain areas of AD patients (20–23). The nucleus basalis of Meynert (NBM) is neuropathologically severely affected in AD and also shows severely decreased neuronal activity (20). As a measure of neuronal metabolic activity that can be applied to formalin-fixed paraffin-embedded postmortem material, we used the size of the GA. It has been shown that all newly synthesized proteins destined for fast axonal transport are processed through the GA (24) and that the GA is involved in many physiological posttranslational modifications including the transport and targeting of a variety of proteins destined for secretion, the plasma membrane, and lysosomes (25, 26). Therefore, the decreased size of the neuronal GA reflects an impairment of protein processing. Because ApoE ɛ4 is one of the major risk factors for AD, in the present study we examined whether there is a relationship between the reduction of the size of the GA and the size of NBM neurons and the type of ApoE in AD patients. In each Alzheimer patient the ApoE genotype was determined, the GA of the NBM neurons visualized by immunocytochemistry, and the size of the organelle measured by image analysis.
Indeed, a clearly reduced neuronal activity of NBM neurons was found in AD brains compared with that of nondemented controls. Moreover, a similar extra decreased neuronal activity of NBM neurons was found in the presence of ApoE ɛ4 alleles in AD, which suggests that ApoE ɛ4 alleles participate in the pathogenesis of AD by decreasing neuronal metabolic rate.
MATERIALS AND METHODS
Tissue Collection.
Brains from 30 nondemented controls ranging in age from 29 to 94 (58.4 ± 3; mean ± SEM) and 41 Alzheimer patients ranging in age from 40 to 98 yr (72.8 ± 2) were obtained at autopsy (see Table 1 for clinico-pathological information). One AD patient was ApoE ɛ2/3 (40-yr-old), 15 were of the ApoE ɛ3/3 genotype (69.9 ± 3.1 yr), 16 of the ApoE ɛ3/4 genotype (75.3 ± 3.6 yr), and 9 of the genotype ɛ4/4 (73.2 ± 3.1 yr). AD was diagnosed clinically based on National Institute of Neurological and Communication Disorders and Stroke–Alzheimer’s Disease and Related Disorders Association criteria, and the diagnosis “probable AD” was established by excluding other causes of dementia (27). The patients had a global deterioration scale of 4–7 for severity of dementia (ref. 28; Table 1). The patients suffered between 3 and 14 yr of dementia before death (Table 1). The clinical diagnosis AD was neuropathologically confirmed by systematic study of the following structures: the temporal superior gyrus, orbital gyrus, cingulate gyrus, hippocampus, striatum, thalamus, and locus coeruleus. The diagnosis was confirmed on the basis of the presence of a large number of plaques, tangles, and dystrophic neurites. The integrity of controls also was established by such a systemic neuropathological examination. The hypothalamus containing the NBM was dissected out and fixed in 4% formaldehyde in PBS (pH 7.4) at room temperature, for ≈1 mo. The fixed hypothalamus was dehydrated in graded ethanols, embedded in paraffin, and cut serially in 6-μm coronal sections. For anatomical orientation, every fiftieth section was mounted on chrome-aluminum sulfate-coated glass slides, deparaffinized, hydrated, and stained with thionine (0.5%).
Table 1.
Clinco-pathological information on controls and Alzheimer patients with different ApoE genotypes
| No. | Diagnosis | Age | Bw, g | PMD, hr | Fix, d | GDS | ApoE Genotype | Disease duration, mo |
|---|---|---|---|---|---|---|---|---|
| 30 | Control | 58.4 ± 3.5 | 1,265 ± 30 | 17.42 ± 3.35 | 49 ± 15 | — | — | — |
| 1 | Alzheimer | 40 | 1,410 | 2.83 | 28 | 7 | 2/3 | 60 |
| 15 | Alzheimer | 69.9 ± 3.1 | 1,071.3 ± 38.87 | 4.01 ± 0.24 | 32.5 ± 2.8 | 6.2 ± 0.26 | 3/3 | 90.0 ± 12 |
| 16 | Alzheimer | 75.3 ± 3.6 | 1,168.7 ± 52.72 | 4.13 ± 0.20 | 31.0 ± 2.8 | 6.5 ± 0.12 | 3/4 | 96.0 ± 8 |
| 9 | Alzheimer | 73.2 ± 3.1 | 1,180.2 ± 48.64 | 4.55 ± 0.38 | 29.7 ± 2.1 | 6.33 ± 0.44 | 4/4 | 105.3 ± 10 |
No, Number of patients; PMD, postmortem delay (in hours); fix, fixation time (in days); BW, brain weight (in grams); GDS, Global Deterioration Scale (28).
Immunocytochemistry.
A polyclonal antibody was raised against immunoaffinity purified MG-160, a sialoglycoprotein of the medial cisternae of the rat neuronal GA. The specificity of this antibody for GA membranes had been established previously by light and ultrastructural immunocytochemistry (29–31). For the optimal retrieval of the MG-160 antigen, we used microwave oven heating before incubation with the primary antibody (32, 33).
For an extensive description of the immunocytochemical procedures, see Salehi et al. (20). In brief, the sections were incubated with anti-MG-160 (first antibody) and diluted 1:800 for 1 hr at room temperature followed by an overnight incubation at 4°C. Subsequently, the sections were washed in tris-buffered saline and incubated with biotinylated goat-anti-rabbit Ig (second antibody: Vector Laboratories) at a dilution of 1:500 for 1 hr at room temperature and finally with avidin-biotin-horseradish peroxidase (Vector Laboratories) at a dilution of 1:1500 for 1 hr at room temperature. 3′,3′-Diaminobenzidine-tetrahydrochloride (Sigma) was used as chromogen. Staining enhancement was obtained by adding ammonium nickel sulfate (2.2 mg/ml). The sections were then dehydrated in graded ethanol and xylene and coverslipped with Entellan (Merck).
Nucleus Basalis.
The NBM, or Ch4 area according to Mesulam’s nomenclature (34), provides the major cholinergic innervation for the cerebral cortex and the amygdala (34, 35). In the human brain, in addition to choline acetyltransferase, Ch4 neurons also express acetylcholinesterase, calbindin-d28k, the tyrosine kinase receptors A, B, and C, and the low affinity neurotrophin receptors (36–38). The NBM is severely affected in AD. Originally it was presumed that massive neuronal death is one of the major hallmarks of AD, in the NBM and other regions (39–41). However, recent studies have indicated that neuronal atrophy rather than cell loss is the main phenomenon in the NBM in AD (20, 42, 43).
Morphometry.
Because the NBM is a very extensive cell system, the measurements were performed in a standardized part of the NBM, i.e., in the medial subdivision of Ch4 (34). NBM-containing sections were selected on the basis of a standardized location of the fornix, anterior commissure, optic tract, and supraoptic nucleus (20).
Measurement of the GA surface was performed by using an IBAS image analysis system [Kontron AT-based system]. The image analysis system was connected to a Bosch TYK9B TV camera equipped with a chalnycon tube mounted on a Zeiss microscope. The microscope was equipped with planapo-objectives. All measurements were performed by using a 560-nm filter, which coincides with the maximum absorption of the diaminobenzidine/nickel sulfate precipitate in the sections. For an extensive description of the analysis procedures, see Salehi et al. (20).
Statistical Methods.
The differences in the mean values of the neuronal GA and mean cellular profile area among AD groups with different ApoE genotypes were tested by using the Kruskal–Wallis test and Mann–Whitney U test. To test the correlation between different parameters such as fixation time, postmortem delay, and age of the subjects and the mean size of the GA, the Pearson’s correlation coefficient was used. A P-value <0.05 was considered to be significant.
APOE Genotyping.
ApoE genotyping was performed on frozen tissue from the cerebellum of the Alzheimer patients. The genotype of each extracted DNA sample was determined by PCR amplification by using the primers 5′-ATAAATATAAAATATAAATAACAGAATTCGCCCCGGCCTGGTACAC-3′ and 5′-TAAGCTTGGCACGGCTGTCCAAGGA-3′. Then the PCR product was digested by Cfol, and fragments were separated by electrophoresis in a 5% agarose gel (44).
RESULTS AND DISCUSSION
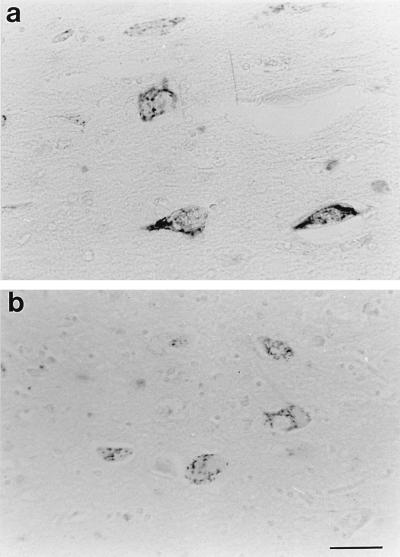
The immunocytochemical visualization of the GA in the NBM neurons revealed a cytoplasmatic staining with a perinuclear distribution in both neurons and glia, consistent with the location of the GA (Fig. 1A–B). Qualitative microscopic analysis showed that the area occupied by the GA in the cytoplasm of NBM neurons in control subjects was clearly larger than that of Alzheimer patients. Furthermore, the GA size in Alzheimer patients with ApoE genotype ɛ3/3 (Fig. 1A) was generally larger than that of Alzheimer patients with ApoE genotype ɛ3/4 (Fig. 1B) or ɛ4/4. In addition to this reduction in GA size in Alzheimer patients with ApoE genotype ɛ3/4 or ɛ4/4, a reduction in the cell profile area was observed in these patients as compared with Alzheimer patients with ApoE genotype ɛ3/3.
Figure 1.
Immunocytochemical staining of the Golgi apparatus in Alzheimer patients with ApoE genotype ɛ3/3 and ApoE genotype ɛ3/4. Note the reduction in Golgi size in the nucleus basalis neurons of Alzheimer patients with ApoE genotype ɛ3/4 (B) vs. the Alzheimer patients with ApoE genotype ɛ3/3 (A). (Bar = 30 μm.)
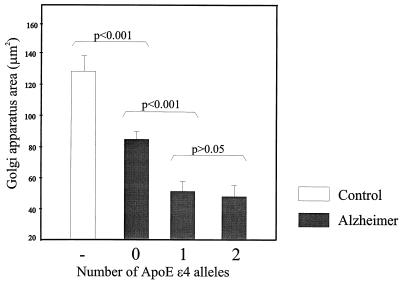
Image analysis confirmed the qualitative microscopical impression. The mean area of the GA in control subjects (127.8 ± 10.4 μm2; mean GA size ± SEM) was significantly (P < 0.001) larger than that of Alzheimer patients (63.7 ± 4.3 μm2). As far as the Alzheimer group was concerned, the Kruscal–Wallis test showed significant (P = 0.002) differences in GA size and mean profile area among the three groups of patients with different ApoE genotypes (Fig. 2). Alzheimer patients with ApoE genotype ɛ3/3 had a mean GA size of 85.1 ± 5.2 μm2, whereas Alzheimer patients with ApoE genotype ɛ3/4 had a mean GA size of 51.9 ± 5.9 μm2. The patients with ApoE genotype ɛ4/4 had a mean GA size of 48.8 ± 8.1 μm2. The mean GA size of Alzheimer patients with ApoE genotype ɛ3/4 or ɛ4/4 was significantly (P < 0.001) lower than the mean GA size of Alzheimer patients with ApoE genotype ApoE ɛ3/3 (Fig. 2). The size of the GA of Alzheimer patients with ApoE genotype ɛ4/4 was not different (P = 0.760) from that of Alzheimer patients with ApoE genotype ɛ3/4. The cell profile area of Alzheimer patients with ApoE genotype ɛ3/4 (254.1 ± 17.0 μm2) or 4/4 (187.3 ± 23.0 μm2) was significantly lower (P <0.001) than that of Alzheimer patients with ApoE genotype ɛ3/3 (346.1 ± 12.1 μm2).
Figure 2.
Graph showing the size of the mean Golgi apparatus in controls and Alzheimer patients with ApoE genotype ɛ3/3 compared with Alzheimer patients with ApoE genotype ɛ3/4 and ɛ4/4. Note the clear reduction in the size of Golgi apparatus in Alzheimer patients with one or two ApoE ɛ4 alleles, compared with Alzheimer patients without ApoE ɛ4 alleles. There is no significant difference (P = 0.760) in Golgi apparatus size between Alzheimer patients with one ApoE ɛ4 allele and two ɛ4 alleles.
Using the Pearson’s test, no significant correlation could be found between fixation time postmortem delay, disease duration, and age of the subjects and the mean GA size either in controls or in Alzheimer patients with different ApoE genotypes. Moreover, no significant correlation could be found between fixation time or postmortem delay of the subjects and the cell profile area of the either controls or Alzheimer patients with different ApoE genotypes. Nor was the cell profile area dependent on the age of the subjects (Table 2). Furthermore, there were no significant differences in age (P = 0.296), brain weight (P = 0.249), postmortem delay (P = 0.507), fixation time (P = 0.872), global deterioration scale (P = 0.652), or duration of the disease (P = 0.451) among the three groups of AD patients (Table 1) with either one or two or without any ApoE ɛ4 alleles.
Table 2.
P-value of correlation* between the GA size, cell profile area and clinico-pathological parameters of the subjects used in this study
| Group | Parameter | Age | PMD | Fix | Bw | Disease duration | GDS |
|---|---|---|---|---|---|---|---|
| Controls | GA area | 0.126 | 0.087 | 0.989 | 0.834 | — | — |
| Controls | Cell area | 0.342 | 0.104 | 0.747 | 0.199 | — | — |
| Alzheimer | GA area | 0.733 | 0.228 | 0.279 | 0.834 | 0.377 | −0.105 |
| Alzheimer | Cell area | 0.441 | 0.432 | 0.172 | 0.337 | 0.406 | −0.127 |
PMD, postmortem delay; ∗, Pearson’s correlation test; Fix, fixation time; GDS, Global Deterioration Scale (28).
Mutations in different genes, including amyloid precursor protein, presenilin-1 and presenilin-2, are invariably associated with AD (45–48). However, the ApoE ɛ4 variant associated with AD also can be found in quite elderly, cognitively normal individuals (49), indicating that ApoE ɛ4 should be considered as a risk factor for AD. Furthermore, polymorphism in specific regions of the ApoE gene might be of importance for the occurrence of AD (50).
The mechanism of involvement of ApoE in AD is not known. It was hypothesized (i) that ApoE affects the deposition of amyloid by changing the clearance, the deposition or the generation of β-amyloid, or (ii) interacts with the microtubule-associated protein τ, in that ApoE ɛ4 does not protect τ from aggregation into neurofibrillary tangles. In addition, a role of ApoE in neuronal repair and degeneration has been suggested (51–53), and it was proposed that ApoE ɛ4 might be less able to support neuronal survival during neurodegeneration than ApoE ɛ2 or ɛ3. The prevalence of ApoE indeed correlated with β-amyloid accumulation (3, 5, 55) and neurofibrillary tangle formation (55, 56) supporting the possibility of a causal relationship between ApoE and the development of AD neuropathology. Moreover, the role of ApoE in the cascade of cholesterol and phospholipid transport and re-uptake and the involvement of these mechanisms in repair in brain aging and AD (57) has been taken into consideration. It was proposed that cholesterol, released during the breakdown of the synaptic terminal, is transported by ApoE to neurons undergoing reinnervation (57). Because the transport of cholesterol and other lipoproteins play a central role in synaptogenesis, it is possible that Alzheimer patients differing in their ApoE phenotype also differ in their capacities of synaptogenesis. Recent in vitro data have shown that in the presence of a lipid source, ApoE ɛ3 enhances and ApoE ɛ4 inhibits neurite outgrowth in neuronal cell cultures (58). The results of the present study suggest a new mechanism for the involvement of ApoE ɛ4 because it was found that the neurons of Alzheimer patients with ApoE genotype ɛ3/4 or ɛ4/4 are less metabolically active than those of Alzheimer patients with ApoE genotype ApoE ɛ3/3. Interestingly, we could not find a significant difference in the size of the GA between Alzheimer patients with ApoE genotype ɛ4/4 and those with ɛ3/4. This result indicates that the relationship between the size of the GA and the number of ApoE ɛ4 alleles is not dose-dependent and that there may be a threshold effect in the NBM, as was found for the cholinergic deficit in the NBM (18).
We have shown in the present study that the presence of one or two ApoE ɛ4 alleles is accompanied by a decreased metabolic activity of neurons as indicated by the diminished size of the GA, which was interpreted in our studies as an indicator of reduced neuronal metabolic activity. However, it may well be that the GA itself is a direct target of ApoE ɛ4 alleles. In support of this possibility, Verde et al. (59) have shown that ATP depletion blocks the export of proteins from the endoplasmic reticulum to the intermediate compartment of the Golgi complex, which may lead to GA shrinkage (59).
Our present study clearly confirms our previous work (20) that in AD patients the GA (63.70 ± 4.3) is approximately one-half the size of that in controls (127.83 ± 10.4). One of the questions that we are currently trying to address is that whether the number of ApoE ɛ4 alleles in controls is related also to the GA size. To do so, we are optimizing a method to determine the number of ApoE ɛ4 alleles in the paraffin-embedded tissue of the controls. However, an extremely large collection would be needed to obtain a group of neuropathologically confirmed control cases with ApoE ɛ4 alleles. No case with ApoE ɛ4/4 genotype was found in the collection of the Netherlands Brain Bank out of 117 nondemented controls.
One of the most consistent neurochemical abnormalities in AD brains is a cholinergic deficit, as evidenced by a loss of choline acetyltransferase and acetylcholinesterase in the neocortex and hippocampus, and a degeneration of cholinergic neurons in the NBM (20, 39). Recent studies have indicated that Alzheimer patients carrying an ApoE ɛ4 allele have a more severe cholinergic deficit in the hippocampus, temporal cortex (18), and frontal cortex (15). The reduced activity of choline acetyltransferase in the temporal cortex reported by Poirier et al. (18) in Alzheimer patients was found to be similar in patients with either one or two ApoE ɛ4 alleles, just as we found a similar reduction in metabolic rate in the NBM of these two patient groups. All these data support the notion that ApoE may play a crucial role in the metabolic activity of the cholinergic neurons of the NBM. Our data suggest that ApoE ɛ4 may be a risk factor for the development of AD by reducing neuronal metabolism. It may be presumed that a lower protein synthetic ability of the NBM neurons in AD predisposes them to neurodegeneration (19).
An enhanced dose-dependent burden of β-amyloid has been observed in ApoE ɛ4 carriers (55, 56, 60). It has been shown that inhibition of energy metabolism can influence amyloid precursor protein processing, leading to decreased secretion of nonamyloidogenic fragments of amyloid precursor protein (61–63). One may suggest, therefore, that ApoE ɛ4 alleles increase the burden of amyloid by inhibition of the energy metabolism. On the other hand, we found no clear relationship between the GA size and the distance of neurons from neuritic plaques (A.S., C. W. Pool, M.M., R. Ravid, N. K. Gonatas, and D.F.S., unpublished data), pleading against such a local inhibitory effect of amyloid on neuronal metabolism.
The proposal that ApoE influences the metabolic activity of neurons may have clinical implications because it supports the idea that in cholinergic treatment of Alzheimer patients the ApoE genotype should be taken into account. The differential responses to drugs with cholinergic enhancing effects, such as tacrine of AD individuals with different ApoE types could be explained by our data. This possibility is supported by the study of Poirier et al. (18), who showed that patients without ɛ4 alleles responded significantly better than AD patients carrying the ApoE ɛ4 alleles to a 30-wk treatment with tacrine.
The next question in our study will be what the mechanism is by which ApoE subtypes may influence the rate of metabolism.
Acknowledgments
Would like to express our appreciation to Prof. N. K. Gonatas for providing the MG-160 antibody and critical reading of our manuscript. Human brain tissue was obtained from the Netherlands Brain Bank, Amsterdam (coordinator Dr. R. Ravid). Neuropathological diagnosis of Alzheimer’s disease was performed by Dr. W. Kamphorst and Prof. P. van der Valk of the Pathological Institute of the Free University, Amsterdam. We also wish to thank U. Unmehopa and B. Fisser (Netherlands Institute for Brain Research, Amsterdam) for their contribution to the processing of the brain tissue. This work was supported by a grant from the Van den Houten Foundation and Internationale Stichting Alzheimer Onderzoek (ISAO; to A.S.).
ABBREVIATIONS
- AD
Alzheimer’s disease
- ApoE
apolipoprotein E
- GA
Golgi apparatus
- NBM
nucleus basalis of Meynert
References
- 1.Trojanowski J Q, Schmidt M L, Shin R-W, Bramblett G T, Goedert M, Lee V M-Y. Clin Neurosci. 1993;1:184–191. [Google Scholar]
- 2.Trojanowski J Q, Schmidt M L, Shin R W, Bramblett T T, Rao D, Lee V M-Y. Brain Pathol. 1993;3:45–54. doi: 10.1111/j.1750-3639.1993.tb00725.x. [DOI] [PubMed] [Google Scholar]
- 3.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, Ikeda M, Chi H, Lin C, Li C, Holman K, et al. Nature (London) 1995;29:754–760. doi: 10.1038/376775a0. , 1995. [DOI] [PubMed] [Google Scholar]
- 4.Levy-Lahad E, Bird T D. Ann Neurol. 1996;40:829–840. doi: 10.1002/ana.410400604. [DOI] [PubMed] [Google Scholar]
- 5.Roses A D. J Neuropathol Exp Neurol. 1994;53:429–437. doi: 10.1097/00005072-199409000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Poirier J, Natbantoglu J, Guillaume D, Bertrand P. Pathobiology of Alzheimer’s Disease. New York: Academic; 1995. pp. 225–245. [Google Scholar]
- 7.Mahley R W. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 8.Ordovas J M, Litwack-Klein L, Wilson P W, Schaefer M M, Schaefer E J. J Lipid Res. 1987;28:371–380. [PubMed] [Google Scholar]
- 9.Corder E H, Saunders A M, Strittmatter W J, Schmechel D E, Gaskell P C, Small G W, Roses A D, Haines J L, Pericak-Vance M A. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 10.Talbot C, Lendon C, Craddock N, Shears S, Morris J C, Goate A. Lancet. 1994;343:1432–1433. doi: 10.1016/s0140-6736(94)92557-7. [DOI] [PubMed] [Google Scholar]
- 11.Saunders A M, Schmader K, Breitner J C, Benson M D, Brown W T, Goldfarb L, Goldgaber D, Manwaring M G, Szymanski M H, McCown N. Lancet. 1993;342:710–711. doi: 10.1016/0140-6736(93)91709-u. [DOI] [PubMed] [Google Scholar]
- 12.Corder E H, Saunders A M, Risch N J, Strittmatter W J, Schmechel D E, Gaskell P C, Jr, Rimmler J B, Locke P A, Conneally P M, Schmader K E. Nat Genet. 1994;7:180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- 13.Schächter F, Faure-Delanef L, Guénot F, Rouger H, Froguel P, Lesueur-Ginot L, Cohen D. Nat Genet. 1994;6:29–32. doi: 10.1038/ng0194-29. [DOI] [PubMed] [Google Scholar]
- 14.Rebeck G W, Perls T T, West H L, Sodhi P, Lipsitz L A, Hyman B T. Neurology. 1994;144:1513–1516. doi: 10.1212/wnl.44.8.1513. [DOI] [PubMed] [Google Scholar]
- 15.Arendt T, Schindler C, Brückner M K, Eschrich K, Bigl V, Zedlick D, Marcova L. J Neurosci. 1997;17:516–529. doi: 10.1523/JNEUROSCI.17-02-00516.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirier J. Trends Neurosci. 1994;17:525–530. doi: 10.1016/0166-2236(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 17.Poirier J, Nalbantaglu J, Guillaume D, Bertrand P. In: Neurofibrillary Tangles and Amyloid Protein in Alzheimer’s Disease. Goate A, Ashall F, editors. New York: Academic; 1994. pp. 525–530. [Google Scholar]
- 18.Poirier J, Delisle M-C, Quirion R, Aubert I, Farlow M, Lahiri D, Hui S, Bertrand P, Nalbantoglu J, Gilfix B, Gauthier S. Proc Natl Acad Sci USA. 1995;92:12260–12264. doi: 10.1073/pnas.92.26.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swaab D F. Neurobiol Aging. 1991;12:317–324. doi: 10.1016/0197-4580(91)90008-8. [DOI] [PubMed] [Google Scholar]
- 20.Salehi A, Lucassen P J, Pool C W, Gonatas N K, Ravid R, Swaab D F. Neuroscience. 1994;59:871–880. doi: 10.1016/0306-4522(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 21.Salehi A, Van de Nes J A P, Hofman M A, Gonatas N K, Swaab D F. Brain Res. 1995;678:29–39. doi: 10.1016/0006-8993(95)00138-g. [DOI] [PubMed] [Google Scholar]
- 22.Salehi A, Heyn S, Gonatas N K, Swaab D F. Neurosci Lett. 1995;193:29–32. doi: 10.1016/0304-3940(95)11659-k. [DOI] [PubMed] [Google Scholar]
- 23.Salehi A, Ravid R, Gonatas N K, Swaab D F. J Neuropathol Exp Neurol. 1995;54:704–709. doi: 10.1097/00005072-199509000-00013. [DOI] [PubMed] [Google Scholar]
- 24.Hammerschlag R, Stone G C, Bolen F A, Lindsey J D, Ellisman M H. J Cell Biol. 1982;93:568–575. doi: 10.1083/jcb.93.3.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farquhar M G, Palade G E. J Cell Biol. 1981;91:77S–103S. doi: 10.1083/jcb.91.3.77s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farquhar M G. Annu Rev Cell Biol. 1985;1:447–488. doi: 10.1146/annurev.cb.01.110185.002311. [DOI] [PubMed] [Google Scholar]
- 27.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan E M. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 28.Reisberg B, Ferris S H, Leon De M J, Crook T. Am J Psychiatry. 1982;139:1136–1139. doi: 10.1176/ajp.139.9.1136. [DOI] [PubMed] [Google Scholar]
- 29.Gonatas J O, Mezitis S G, Stieber A, Fleischer B, Gonatas N K. J Biol Chem. 1989;264:646–653. [PubMed] [Google Scholar]
- 30.Croul S, Mezitis S G E, Stieber A, Chen Y, Gonatas J O, Goud B, Gonatas N K. J Histochem Cytochem. 1990;38:957–963. doi: 10.1177/38.7.2355176. [DOI] [PubMed] [Google Scholar]
- 31.Gonatas N K, Stieber A, Mourelatos Z, Chen Y, Gonatas J O, Appel S H, Hays A P, Hickey W F, Hauw J J. Am J Pathol. 1992;140:731–737. [PMC free article] [PubMed] [Google Scholar]
- 32.Shi S R, Key M E, Kalra K L. J Histochem Cytochem. 1991;39:741–748. doi: 10.1177/39.6.1709656. [DOI] [PubMed] [Google Scholar]
- 33.Lucassen P J, Ravid R, Gonatas N K, Swaab D F. Brain Res. 1993;632:105–113. doi: 10.1016/0006-8993(93)91144-h. [DOI] [PubMed] [Google Scholar]
- 34.Mesulam M M, Mufson E J, Levey A I, Wainer B H. Neuroscience. 1984;12:669–686. doi: 10.1016/0306-4522(84)90163-5. [DOI] [PubMed] [Google Scholar]
- 35.Mesulam M M, Mufson E J, Levey A, Wainer B H. J Comp Neurol. 1989;214:170–197. doi: 10.1002/cne.902140206. [DOI] [PubMed] [Google Scholar]
- 36.Geula C, Schatz C R, Mesulam M M. Neuroscience. 1993;54:461–476. doi: 10.1016/0306-4522(93)90266-i. [DOI] [PubMed] [Google Scholar]
- 37.Salehi A, Verhaagen J, Dijkhuizen P J, Swaab D F. Neuroscience. 1996;75:373–387. doi: 10.1016/0306-4522(96)00273-4. [DOI] [PubMed] [Google Scholar]
- 38.Kordower J H, Gash D M, Bothwell M, Hersh L, Mufson E J. Neurobiol Aging. 1989;10:67–74. doi: 10.1016/s0197-4580(89)80013-2. [DOI] [PubMed] [Google Scholar]
- 39.Etienne P, Robitaille Y, Wood P, Gauthier S, Nair N P V, Quirion R. Neuroscience. 1986;19:1279–1291. doi: 10.1016/0306-4522(86)90142-9. [DOI] [PubMed] [Google Scholar]
- 40.Whitehouse P J, Price D L, Clark A W, Coyle J T, DeLong M R. Ann Neurol. 1981;10:122–126. doi: 10.1002/ana.410100203. [DOI] [PubMed] [Google Scholar]
- 41.Whitehouse P J, Price D L, Struble R G. Science. 1982;215:1237–1239. [Google Scholar]
- 42.Rinne J O, Paljärvi L, Rinne U K. J Neurol Sci. 1987;79:67–76. doi: 10.1016/0022-510x(87)90260-7. [DOI] [PubMed] [Google Scholar]
- 43.Pearson R C A, Sofroniew M V, Cuello A C, Powell T P, Eckenstein F, Esiri M M, Wilcock G K. Brain Res. 1983;289:375–379. doi: 10.1016/0006-8993(83)90046-x. [DOI] [PubMed] [Google Scholar]
- 44.Crook R, Hardy J, Duff K J. J Neurosci Methods. 1994;53:125–127. doi: 10.1016/0165-0270(94)90168-6. [DOI] [PubMed] [Google Scholar]
- 45.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, Ikeda M, Lin C, Li G, Holman K. Nature (London) 1995;375:754–760. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 46.Rogaev E I, Sherrington R, Rogaeva E A, Levesque G, Ikeda M, Liang Y, Chi H, Lin G, Holman K, Tsuda T, et al. Nature (London) 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 47.Cruts M, Backhovens H, Wang S-Y, van Gassen G, Theuns J, De Jonghe C, Wehnert A, De Voecht J, De Winter G, Gras P, et al. Hum Mol Genet. 1995;4:2363–2371. doi: 10.1093/hmg/4.12.2363. [DOI] [PubMed] [Google Scholar]
- 48.Wasco W, Pettingell W P, Jondro P D, Schmidt S D, Gurubhagavatula S, Rodes L, Di Blast T. Nat Genet. 1995;1:848. doi: 10.1038/nm0995-848a. [DOI] [PubMed] [Google Scholar]
- 49.Rebeck G W, Perls T T, West H L, Sodhi P, Lipsitz L A, Hyman B T. Neurology. 1994;144:1513–1516. doi: 10.1212/wnl.44.8.1513. [DOI] [PubMed] [Google Scholar]
- 50.Bullido M J, Artiga M J, Recuero M, Sastre I, Garcia M A, Aldudo J, Lendon C, Han S W, Morris J C, Frank A, et al. Nat Genet. 1998;18:69–71. doi: 10.1038/ng0198-69. [DOI] [PubMed] [Google Scholar]
- 51.Poirier J, Davignon J, Bouthillier D, Kogan S, Bertrand P, Gauthier S. Lancet. 1993;342:697–699. doi: 10.1016/0140-6736(93)91705-q. [DOI] [PubMed] [Google Scholar]
- 52.Roses A D. J Neuropathol Exp Neurol. 1994;53:429–437. doi: 10.1097/00005072-199409000-00002. [DOI] [PubMed] [Google Scholar]
- 53.Masliah E, Mallory M, Alford M, Veinbergs I, Roses A D. Exp Neurol. 1995;136:107–122. doi: 10.1006/exnr.1995.1088. [DOI] [PubMed] [Google Scholar]
- 54.Strittmatter W J, Weisgraber K H, Goedert M, Saunders A M, Huang D, Corder E H, Dong L M, Jakes R, Alberts M J, Gilbert J R. Exp Neurol. 1994;125:163–171. doi: 10.1006/exnr.1994.1019. [DOI] [PubMed] [Google Scholar]
- 55.Strittmatter W J, Weisgraber K H, Huang D Y, Dong L M, Salvesen G S, Pericak-Vance M, Schmechel D, Saunders A M, Goldgaber D, Roses A D. Proc Natl Acad Sci USA. 1993;90:8098–8102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagy Z, Esiri M M, Jobst K A, Johanston C, Litchhfield S, Sim E, Smith A D. Neuroscience. 1995;69:757–761. doi: 10.1016/0306-4522(95)00331-c. [DOI] [PubMed] [Google Scholar]
- 57.Soininen H S, Riekkinen P J., Sr Trends Neurosci. 1996;19:224–228. doi: 10.1016/0166-2236(96)10027-8. [DOI] [PubMed] [Google Scholar]
- 58.Lovestone S, Anderton B H, Hartley C, Jensen T G, Jorgensen A L. NeuroReport. 1996;7:1005–1008. doi: 10.1097/00001756-199604100-00010. [DOI] [PubMed] [Google Scholar]
- 59.Verde C, Pascale M C, Martire G, Lotti L V, Torrisi M R, Helenius A, Bonatti S. Eur J Cell Biol. 1995;67:267–274. [PubMed] [Google Scholar]
- 60.Schmechel D E, Saunders A M, Strittmatter W J, Crain B J, Hulette C M, Joo S H, Pericak-Vance M A, Goldgaber D, Roses A D. Proc Natl Acad Sci USA. 1993;90:9649–9653. doi: 10.1073/pnas.90.20.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gasparini L, Racchi M, Benussi L, Curti D, Binetti G, Bianchetti A, Trabucchi M, Govoni S. Neurosci Lett. 1997;231:113–117. doi: 10.1016/s0304-3940(97)00536-3. [DOI] [PubMed] [Google Scholar]
- 62.Shi J, Xiang Y, Simkins J W. Brain Res. 1997;772:247–251. doi: 10.1016/s0006-8993(97)00827-5. [DOI] [PubMed] [Google Scholar]
- 63.Gabuzda D, Busciglio J, Chen L B, Matsudaira P, Yankner B A. J Biol Chem. 1994;269:13623–13628. [PubMed] [Google Scholar]