Abstract
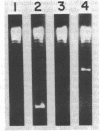
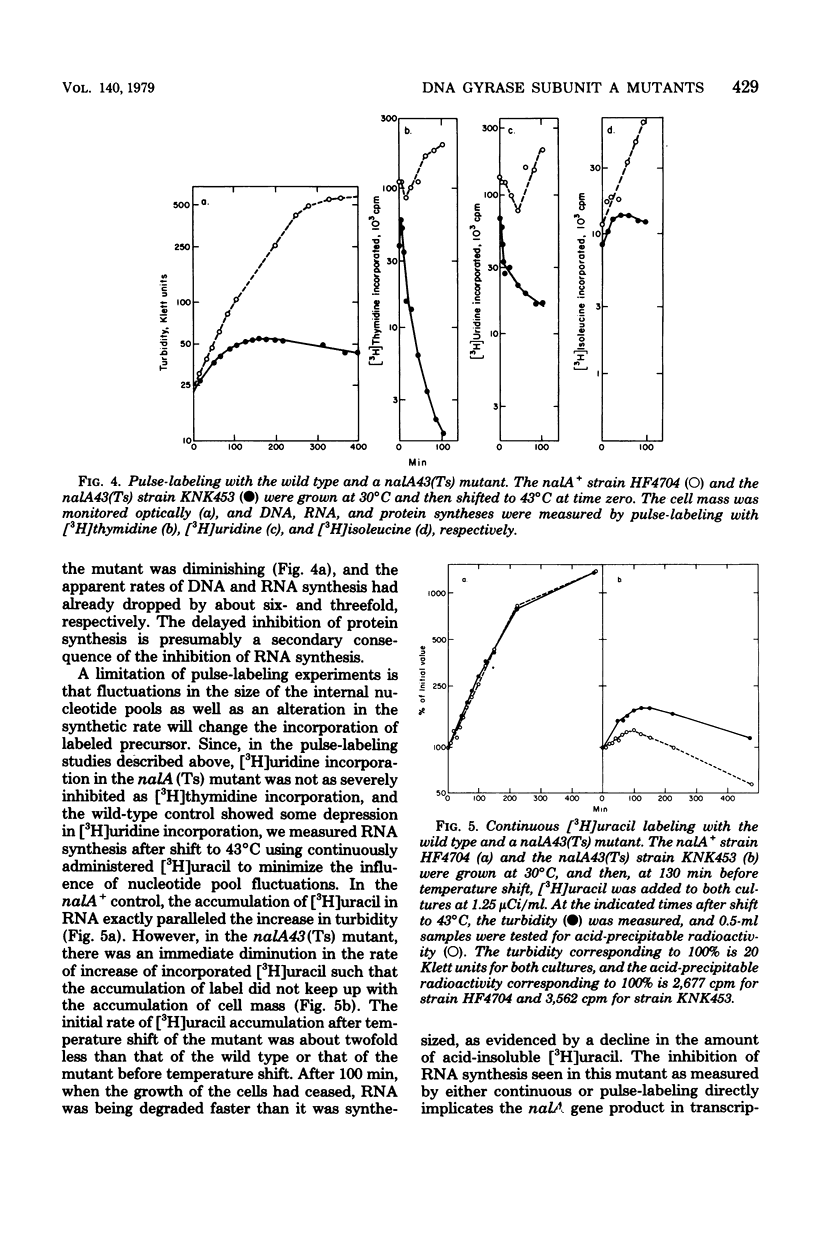
Temperature-sensitive nalA mutants of Escherichia coli have been used to investigate the structure and functions of deoxyribonucleic acid (DNA) gyrase. Extracts of one such mutant (nalA43) had thermosensitive DNA gyrase subunit A activity but normal gyrase subunit B activity, proving definitively that nalA is the structural gene for subunit A. Extracts of a second nalA (Ts) mutant (nalA45) had a 50-fold deficiency of gyrase subunit A activity. The residual DNA supertwisting was catalyzed by the mutant DNA gyrase rather than by a novel supertwisting enzyme. The nalA45(Ts) extract was also deficient in the nalidixic acid target, which is defined as the protein necessary to confer drug sensitivity to in vitro DNA replication directed by a nalidixic acid-resistant mutant extract. Thus, gyrase subunit A and the nalidixic acid target are one and the same protein, the nalA gene product. Shift of the nalA43(Ts) mutant to a nonpermissive temperature resulted in a precipitous decline in the rate of [3H]thymidine incorporation, demonstrating an obligatory role of the nalA gene product in DNA replication. The rates of incorporation of [3H]uridine pulses and continuously administered [3H]uracil were quickly reduced approximately twofold upon temperature shift of the nalA43(Ts) mutant, and therefore some but not all transcription requires the nalA gene product. The thermosensitive growth of bacteriophages φX174 and T4 in the nalA43(Ts) host shows that these phages depend on the host nalA gene product. In contrast, the growth of phage T7 was strongly inhibited by nalidixic acid but essentially unaffected by the nalA43(Ts) mutation. The inhibition of T7 growth by nalidixic acid was, however, eliminated by temperature inactivation of the nal43 gene product. Therefore, nalidixic acid may block T7 growth by a corruption rather than a simple elimination of the nalidixic acid target. Possible mechanisms for such a corruption are considered, and their relevance to the puzzling dominance of drug sensitivity is discussed.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austin S., Scaife J. A new method for selecting RNA polymerase mutants. J Mol Biol. 1970 Apr 14;49(1):263–267. doi: 10.1016/0022-2836(70)90394-3. [DOI] [PubMed] [Google Scholar]
- Botchan P., Wang J. C., Echols H. Effect of circularity and superhelicity on transcription from bacteriophagelambda DNA. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3077–3081. doi: 10.1073/pnas.70.11.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J. Strand breakage by the DNA untwisting enzyme results in covalent attachment of the enzyme to DNA. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3800–3804. doi: 10.1073/pnas.74.9.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzarelli N. R. The mechanism of action of inhibitors of DNA synthesis. Annu Rev Biochem. 1977;46:641–668. doi: 10.1146/annurev.bi.46.070177.003233. [DOI] [PubMed] [Google Scholar]
- De Wyngaert M., Hinkle D. C. Involvement of DNA gyrase in replication and transcription of bacteriophage T7 DNA. J Virol. 1979 Feb;29(2):529–535. doi: 10.1128/jvi.29.2.529-535.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Snyder M. Superhelical Escherichia coli DNA: relaxation by coumermycin. J Mol Biol. 1978 Apr 5;120(2):145–154. doi: 10.1016/0022-2836(78)90061-x. [DOI] [PubMed] [Google Scholar]
- Falco S. C., Zivin R., Rothman-Denes L. B. Novel template requirements of N4 virion RNA polymerase. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3220–3224. doi: 10.1073/pnas.75.7.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer E., Wolf H., Hantke K., Parmeggiani A. Elongation factor Tu resistant to kirromycin in an Escherichia coli mutant altered in both tuf genes. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4341–4345. doi: 10.1073/pnas.74.10.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOSS W. A., DEITZ W. H., COOK T. M. MECHANISM OF ACTION OF NALIDIXIC ACID ON ESCHERICHIA COLI.II. INHIBITION OF DEOXYRIBONUCLEIC ACID SYNTHESIS. J Bacteriol. 1965 Apr;89:1068–1074. doi: 10.1128/jb.89.4.1068-1074.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J. I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Itoh T., Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hane M. W., Wood T. H. Escherichia coli K-12 mutants resistant to nalidixic acid: genetic mapping and dominance studies. J Bacteriol. 1969 Jul;99(1):238–241. doi: 10.1128/jb.99.1.238-241.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Peebles C. L., Sugino A., Cozzarelli N. R. Purification of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1773–1777. doi: 10.1073/pnas.75.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkle D. C., Richardson C. C. Bacteriophage T7 deoxyribonucleic acid replication in vitro. Requirements for deoxyribonucleic acid synthesis and characterization of the product. J Biol Chem. 1974 May 10;249(9):2974–2980. [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. I. Involvement of DNA gyrase in bacteriophage T7 DNA replication. Nature. 1977 Nov 3;270(5632):78–80. doi: 10.1038/270078a0. [DOI] [PubMed] [Google Scholar]
- Iwakura Y., Ishihama A., Yura T. RNA polymerase mutants of Escherichia coli. Streptolydigin resistance and its relation to rifampicin resistance. Mol Gen Genet. 1973 Mar 1;121(2):181–196. doi: 10.1007/BF00277531. [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N., McEntee K., Geballe A. P., Cozzarelli N. R. Lambda transducing phages for the nalA gene of Escherichia coli and conditional lethal nalA mutations. Mol Gen Genet. 1978 Nov 29;167(2):129–137. doi: 10.1007/BF00266906. [DOI] [PubMed] [Google Scholar]
- LEDERBERG J. Streptomycin resistance; a genetically recessive mutation. J Bacteriol. 1951 May;61(5):549–550. doi: 10.1128/jb.61.5.549-550.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- McCarthy D. Gyrase-dependent initiation of bacteriophage T4 DNA replication: interactions of Escherichia coli gyrase with novobiocin, coumermycin and phage DNA-delay gene products. J Mol Biol. 1979 Jan 25;127(3):265–283. doi: 10.1016/0022-2836(79)90329-2. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., O'Dea M. H., Gellert M. DNA gyrase: subunit structure and ATPase activity of the purified enzyme. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5960–5963. doi: 10.1073/pnas.75.12.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Cozzarelli N. R. Site-specific cleavage of DNA by E. coli DNA gyrase. Cell. 1979 May;17(1):175–184. doi: 10.1016/0092-8674(79)90305-2. [DOI] [PubMed] [Google Scholar]
- Nomura M., Engbaek F. Expression of ribosomal protein genes as analyzed by bacteriophage Mu-induced mutations. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1526–1530. doi: 10.1073/pnas.69.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles C. L., Higgins N. P., Kreuzer K. N., Morrison A., Brown P. O., Sugino A., Cozzarelli N. R. Structure and activities of Escherichia coli DNA gyrase. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):41–52. doi: 10.1101/sqb.1979.043.01.008. [DOI] [PubMed] [Google Scholar]
- Puga A., Tessman I. Mechanism of transcription of bacteriophage S13. II. Inhibition of phage-specific transcription by nalidixic acid. J Mol Biol. 1973 Mar 25;75(1):99–108. doi: 10.1016/0022-2836(73)90531-7. [DOI] [PubMed] [Google Scholar]
- Ryan M. J. Coumermycin A1: A preferential inhibitor of replicative DNA synthesis in Escherichia coli. I. In vivo characterization. Biochemistry. 1976 Aug 24;15(17):3769–3777. doi: 10.1021/bi00662a020. [DOI] [PubMed] [Google Scholar]
- Sanzey B. Modulation of gene expression by drugs affecting deoxyribonucleic acid gyrase. J Bacteriol. 1979 Apr;138(1):40–47. doi: 10.1128/jb.138.1.40-47.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Kubo M., Imamoto F. Promoter-specific inhibition of transcription by antibiotics which act on DNA gyrase. Nature. 1978 Oct 5;275(5679):420–423. doi: 10.1038/275420a0. [DOI] [PubMed] [Google Scholar]
- Snyder M., Drlica K. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J Mol Biol. 1979 Jun 25;131(2):287–302. doi: 10.1016/0022-2836(79)90077-9. [DOI] [PubMed] [Google Scholar]
- Sparling P. F., Modolell J., Takeda Y., Davis B. D. Ribosomes from Escherichia coli merodiplods heterozygous for resistance to streptomycin and to spectinomycin. J Mol Biol. 1968 Nov 14;37(3):407–421. doi: 10.1016/0022-2836(68)90111-3. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L. Letters to the editor: Novobiocin-a specific inhibitor of semiconservative DNA replication in permeabilized Escherichia coli cells. J Mol Biol. 1975 Jul 25;96(1):201–205. doi: 10.1016/0022-2836(75)90191-6. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L. REPLICAtion of small plasmids in extracts of Escherichia coli: requirement for both DNA polymerases I and II. Mol Gen Genet. 1976 Dec 8;149(2):151–158. doi: 10.1007/BF00332883. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L. Replication of Escherichia coli DNA in vitro: inhibition by oxolinic acid. Eur J Biochem. 1976 Mar 1;62(3):491–497. doi: 10.1111/j.1432-1033.1976.tb10183.x. [DOI] [PubMed] [Google Scholar]
- Sugino A., Higgins N. P., Brown P. O., Peebles C. L., Cozzarelli N. R. Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4838–4842. doi: 10.1073/pnas.75.10.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumida-Yasumoto C., Hurwitz J. Synthesis of phiX174 viral DNA in vitro depends on phiX replicative form DNA. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4195–4199. doi: 10.1073/pnas.74.10.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumida-Yasumoto C., Yudelevich A., Hurwitz J. DNA synthesis in vitro dependent upon phiX174 replicative form I DNA. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1887–1891. doi: 10.1073/pnas.73.6.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketo A., Watanabe H. Effect of nalidixic acid on the growth of bacterial viruses. J Biochem. 1967 Apr;61(4):520–522. doi: 10.1093/oxfordjournals.jbchem.a128579. [DOI] [PubMed] [Google Scholar]