Abstract
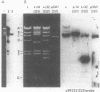
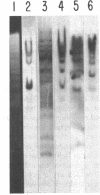
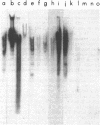
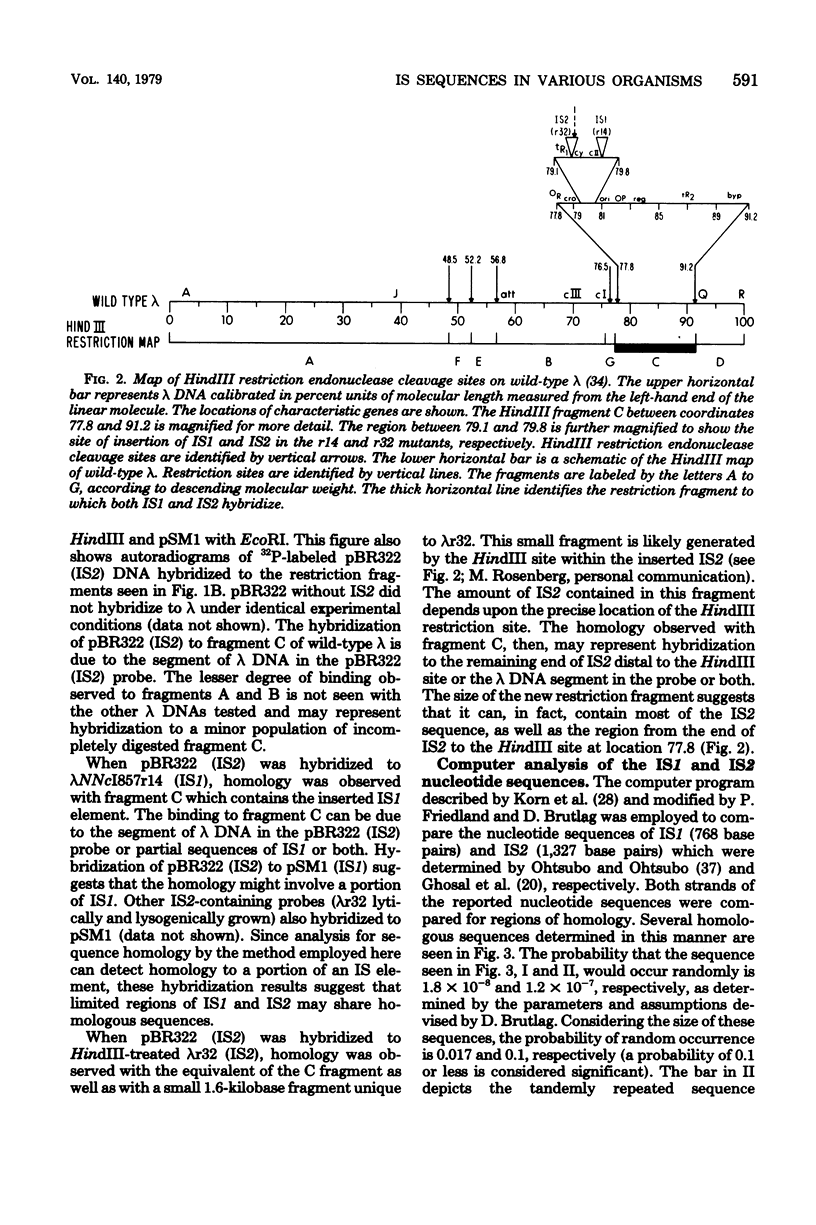
Plasmid and phage deoxyribonucleic acid (DNA) harboring bacterial insertion sequence (IS) elements IS1, IS2, and IS5 were characterized and used as probes to detect homologous sequences in various procaryotic and eucaryotic genomes. The hybridization method used permits the detection of sequences partially homologous to the elements. Hybridization of the IS-containing probes to each other revealed a region of limited homology between IS1 and IS2. Homologous sequences were then detected by computer analysis of the published IS1 and IS2 nucleotide sequences. The homologous sequence contains a tandemly repeated tetranucleotide sequence which resembles the repeated sequence at the hot spot for spontaneous mutations in the lacI gene (P. J. Farabaugh, U. Schmeissner, M. Hofer, and J. Miller, J. Mol. Biol. 126:847–863, 1978). Homology between the IS elements and various genomes was determined by hybridizing labeled DNA containing IS1, IS2, and IS5 sequences to Southern blots of chromosomal DNA cleaved with restriction endonucleases. IS1 and IS5 appear limited to the enteric bacteria, whereas IS2 sequences can also be detected in Pseudomonas putida, Pseudomonas aeruginosa, and Serratia marcescens. Bacteria which appear not to possess extrachromosomal elements, e.g., Caulobacter crescentus, did not show homology with any insertion sequences tested. In addition, sequences homologous to IS1, IS2, or IS5 were not detected in Saccharomyces cerevisiae, Dictyostelium discoideum, or calf thymus DNA.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed A. Mechanism of reversion of the gal3 mutation of Escherichia coli. Mol Gen Genet. 1975;136(3):243–253. doi: 10.1007/BF00334019. [DOI] [PubMed] [Google Scholar]
- Allet B., Bukhari A. I. Analysis of bacteriophage mu and lambda-mu hybrid DNAs by specific endonucleases. J Mol Biol. 1975 Mar 15;92(4):529–540. doi: 10.1016/0022-2836(75)90307-1. [DOI] [PubMed] [Google Scholar]
- Berg D. E., Drummond M. Absence of DNA sequences homologous to transposable element Tn5 (Kan) in the chromosome of Escherichia coli K-12. J Bacteriol. 1978 Oct;136(1):419–422. doi: 10.1128/jb.136.1.419-422.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F. R., Fiandt M., Hass K. K., Twose P. A., Szybalski W. Deletions and insertions in the immunity region of coliphage lambda: revised measurement of the promoter-startpoint distance. Virology. 1974 Dec;62(2):458–471. doi: 10.1016/0042-6822(74)90407-3. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Johnsrud L., Miller J. H. DNA sequence at the integration sites of the insertion element IS1. Cell. 1978 Mar;13(3):411–418. doi: 10.1016/0092-8674(78)90315-x. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Chow L. T. Sequence arrangements of the Escherichia coli chromosome and of putative insertion sequences, as revealed by electron microscopic heteroduplex studies. J Mol Biol. 1977 Jul 15;113(4):611–621. doi: 10.1016/0022-2836(77)90225-x. [DOI] [PubMed] [Google Scholar]
- Cohen S. N. Transposable genetic elements and plasmid evolution. Nature. 1976 Oct 28;263(5580):731–738. doi: 10.1038/263731a0. [DOI] [PubMed] [Google Scholar]
- Contreras I., Shapiro L., Henry S. Membrane phospholipid composition of Caulobacter crescentus. J Bacteriol. 1978 Sep;135(3):1130–1136. doi: 10.1128/jb.135.3.1130-1136.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Deonier R. C., Davidson N. The sequence organization of the integrated F plasmid in two Hfr strains of Escherichia coli. J Mol Biol. 1976 Nov 5;107(3):207–222. doi: 10.1016/s0022-2836(76)80002-2. [DOI] [PubMed] [Google Scholar]
- Deonier R. C., Hadley R. C. Distribution of inverted IS-length sequences in the E. coli K12 genome. Nature. 1976 Nov 11;264(5582):191–193. doi: 10.1038/264191a0. [DOI] [PubMed] [Google Scholar]
- Deonier R. C., Hadley R. G., Hu M. Enumeration and identification of IS3 elements in Escherichia coli strains. J Bacteriol. 1979 Mar;137(3):1421–1424. doi: 10.1128/jb.137.3.1421-1424.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Fiandt M., Szybalski W., Malamy M. H. Polar mutations in lac, gal and phage lambda consist of a few IS-DNA sequences inserted with either orientation. Mol Gen Genet. 1972;119(3):223–231. doi: 10.1007/BF00333860. [DOI] [PubMed] [Google Scholar]
- Ghosal D., Saedler H. DNA sequence of the mini-insertion IS2--6 and its relation to the sequence of IS2. Nature. 1978 Oct 19;275(5681):611–617. doi: 10.1038/275611a0. [DOI] [PubMed] [Google Scholar]
- Ghosal D., Sommer H., Saedler H. Nucleotide sequence of the transposable DNA-element IS2. Nucleic Acids Res. 1979 Mar;6(3):1111–1122. doi: 10.1093/nar/6.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D. IS1 insertion generates duplication of a nine base pair sequence at its target site. Cell. 1978 Mar;13(3):419–426. doi: 10.1016/0092-8674(78)90316-1. [DOI] [PubMed] [Google Scholar]
- Hu S., Ohtsubo E., Davidson N. Electron microscopic heteroduplex studies of sequence relations among plasmids of Escherichia coli: structure of F13 and related F-primes. J Bacteriol. 1975 May;122(2):749–763. doi: 10.1128/jb.122.2.749-763.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Otsubo E., Davidson N., Saedler H. Electron microscope heteroduplex studies of sequence relations among bacterial plasmids: identification and mapping of the insertion sequences IS1 and IS2 in F and R plasmids. J Bacteriol. 1975 May;122(2):764–775. doi: 10.1128/jb.122.2.764-775.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Reichardt K., Botstein D. Inversions and deletions of the Salmonella chromosome generated by the translocatable tetracycline resistance element Tn10. J Mol Biol. 1979 Jan 5;127(1):89–115. doi: 10.1016/0022-2836(79)90461-3. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Translocatable elements in procaryotes. Cell. 1977 May;11(1):11–23. doi: 10.1016/0092-8674(77)90313-0. [DOI] [PubMed] [Google Scholar]
- Korn L. J., Queen C. L., Wegman M. N. Computer analysis of nucleic acid regulatory sequences. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4401–4405. doi: 10.1073/pnas.74.10.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCCLINTOCK B. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- Malamy M. H., Fiandt M., Szybalski W. Electron microscopy of polar insertions in the lac operon of Escherichia coli. Mol Gen Genet. 1972;119(3):207–222. doi: 10.1007/BF00333859. [DOI] [PubMed] [Google Scholar]
- Malamy M. H. Frameshift mutations in the lactose operon of E. coli. Cold Spring Harb Symp Quant Biol. 1966;31:189–201. doi: 10.1101/sqb.1966.031.01.027. [DOI] [PubMed] [Google Scholar]
- Mosharrafa E., Pilacinski W., Zissler J., Fiandt M., Szybalski W. Insertion sequence IS2 near the gene for prophage lambda excision. Mol Gen Genet. 1976 Aug 10;147(1):103–109. doi: 10.1007/BF00337943. [DOI] [PubMed] [Google Scholar]
- Murray K., Murray N. E. Phage lambda receptor chromosomes for DNA fragments made with restriction endonuclease III of Haemophilus influenzae and restriction endonuclease I of Escherichia coli. J Mol Biol. 1975 Nov 5;98(3):551–564. doi: 10.1016/s0022-2836(75)80086-6. [DOI] [PubMed] [Google Scholar]
- Nevers P., Saedler H. Transposable genetic elements as agents of gene instability and chromosomal rearrangements. Nature. 1977 Jul 14;268(5616):109–115. doi: 10.1038/268109a0. [DOI] [PubMed] [Google Scholar]
- Nisen P. D., Kopecko D. J., Chou J., Cohen S. N. Site-specific DNA deletions occurring adjacent to the termini of a transposable ampicillin resistance element (Tn3). J Mol Biol. 1977 Dec 25;117(4):975–978. doi: 10.1016/s0022-2836(77)80008-9. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S. S., Brorein W. J., Jr, Dunsmuir P., Rubin G. M. Transposition of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell. 1979 Jun;17(2):415–427. doi: 10.1016/0092-8674(79)90168-5. [DOI] [PubMed] [Google Scholar]
- Ptashne K., Cohen S. N. Occurrence of insertion sequence (IS) regions on plasmid deoxyribonucleic acid as direct and inverted nucleotide sequence duplications. J Bacteriol. 1975 May;122(2):776–781. doi: 10.1128/jb.122.2.776-781.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak B. Gal mRNA initiated within IS2. Mol Gen Genet. 1976 Dec 8;149(2):135–143. doi: 10.1007/BF00332881. [DOI] [PubMed] [Google Scholar]
- Reif H. J., Saedler H. IS1 is involved in deletion formation in the gal region of E. coli K12. Mol Gen Genet. 1975;137(1):17–28. doi: 10.1007/BF00332538. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Saedler H., Heiss B. Multiple copies of the insertion-DNA sequences IS1 and IS2 in the chromosome of E. coli K-12. Mol Gen Genet. 1973 May 9;122(3):267–277. doi: 10.1007/BF00278602. [DOI] [PubMed] [Google Scholar]
- Saedler H., Reif H. J., Hu S., Davidson N. IS2, a genetic element for turn-off and turn-on of gene activity in E. coli. Mol Gen Genet. 1974;132(4):265–289. doi: 10.1007/BF00268569. [DOI] [PubMed] [Google Scholar]
- Saedler H., Starlinger P. 0 degree mutations in the galactose operon in E. coli. I. Genetic characterization. Mol Gen Genet. 1967;100(2):178–189. doi: 10.1007/BF00333604. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Starlinger P., Saedler H. IS-elements in microorganisms. Curr Top Microbiol Immunol. 1976;75:111–152. doi: 10.1007/978-3-642-66530-1_4. [DOI] [PubMed] [Google Scholar]
- Starlinger P., Saedler H., Rak B., Tillmann E., Venkov P., Waltschewa L. mRNA distal to polar nonsense and insertion mutation in the gal operon of E. coli. Mol Gen Genet. 1973 May 9;122(3):279–286. doi: 10.1007/BF00278603. [DOI] [PubMed] [Google Scholar]
- Strobel E., Dunsmuir P., Rubin G. M. Polymorphisms in the chromosomal locations of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell. 1979 Jun;17(2):429–439. doi: 10.1016/0092-8674(79)90169-7. [DOI] [PubMed] [Google Scholar]
- Wood N. B., Rake A. V., Shapiro L. Structure of Caulobacter deoxyribonucleic acid. J Bacteriol. 1976 Jun;126(3):1305–1315. doi: 10.1128/jb.126.3.1305-1315.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]