Abstract
The enzyme isopentenyl pyrophosphate (IPP) isomerase catalyzes the reversible isomerization of IPP to produce dimethylallyl pyrophosphate, the initial substrate leading to the biosynthesis of carotenoids and many other long-chain isoprenoids. Expression of IPP isomerase, and of two enzymes specific to the carotenoid pathway (lycopene β-cyclase and β-carotene-C-4-oxygenase), was followed in the green unicellular alga Haematococcus pluvialis after exposure to high illumination. This alga uniquely accumulates carotenoids in the cytoplasm and in late developmental stages turns deep-red in color because of accumulation of ketocarotenoids in the cytosol. The carotenoid/chlorophyll ratio increased 3-fold in wild type and 6-fold in a precocious carotenoid-accumulating mutant (Car-3) within 24 h after increasing the illumination from 20 to 150 μmol photon m−2⋅s−1. Two cDNAs encoding IPP isomerase in Haematococcus, ipiHp1 and ipiHp2, were identified. Although otherwise highly similar (95% identity overall), the predicted sequence of ipiHp1 contained a 12-aa region not found in that of ipiHp2. This was reflected by a size difference between two polypeptides of 34 and 32.5 kDa, both of which reacted with an antibody to the product of ipiHp1. We suggest that the 32.5-kDa form is involved with the carotenoid accumulation in the cytoplasm, since the 32.5-kDa polypeptide was preferentially up-regulated by high light preceding the carotenoid increase and only this form was detected in red cysts.
Carotenoids are essential in the normal development of photosynthetic membranes. When carotenoid synthesis is inhibited with an herbicide such as norflurazon, the greening of seedlings is prevented (1). In fact, the absence of cyclic carotenoids is eventually lethal in oxygenic photosynthetic organisms because protection for the photosynthetic apparatus is lacking. In photosynthetic membranes, carotenoids serve as antenna pigments for light harvesting and also in the dissipation of light energy, thereby protecting plants from excess light (2). Attractive coloration in daffodil and marigold petals and in red tomato and pepper fruits are examples in which carotenoid accumulation is of economic value. Similarly, a few green algae can accumulate notable amounts of the red-colored ketocarotenoid astaxanthin, which is valued as a natural colorant for certain fish and crustaceans. In higher plants the carotenoids are synthesized within plastids and accumulate within specialized plastids (chromoplasts). By contrast, in some green algae such as Haematococcus pluvialis the carotenoids (mostly astaxanthin) accumulate in the cytoplasm. The accumulation of carotenoids in the cytosol raises the possibility that there is an extraplastidic site of carotenoid biosynthesis in Haematococcus. Alternatively, carotenoids synthesized in the chloroplast may be exported and accumulate in the cytoplasm.
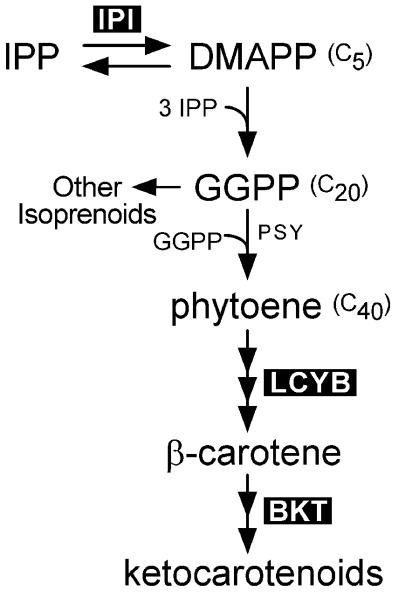
In algae and plants the five-carbon compound IPP and its allylic isomer, dimethylallyl pyrophosphate (DMAPP), serve as substrates for the synthesis of carotenoids and other isoprenoids, including such compounds as sterols and growth factors, and in the phytol moiety of chlorophyll and some quinones (3–5). The starting point for assembly of the C40 backbone of carotenoids is the isomerization of IPP to DMAPP (Fig. 1). Sequential addition of three IPP molecules to DMAPP yields the C20 geranylgeranyl pyrophosphate (GGPP). The condensation of two GGPP molecules forms the C40 phytoene, which is the first committed step of carotenoid synthesis. Coloration appears after a series of desaturation reactions that lengthen the conjugated series of carbon–carbon double bonds. Subsequent cyclization of the end groups to bicyclic compounds and their modification by the addition of hydroxyl or ketol groups give rise to the various yellow, pink, orange, and red carotenoid pigments found in nature (6, 7). The carotenoid biosynthetic pathway is an ancient one. Eubacteria synthesize a wide variety of carotenoids structurally similar to those in plants and algae, but the biosynthetic pathways have greatly diverged as suggested by the relatively low similarity of the genes encoding the enzymes of carotenoid synthesis (6, 8).
Figure 1.
Pathway of ketocarotenoid biosynthesis from IPP/DMAPP. Expression of the enzymes indicated by black boxes, IPP isomerase (IPI), lycopene β-cyclase (LYCB), and β-carotene-C-4-oxygenase (BKT), were studied. GGPP, geranylgeranyl pyrophosphate; PSY, phytoene synthase.
Although IPP isomerase (IPI in Fig. 1) catalyzes the interconversion of IPP and DMAPP, the equilibrium is in favor of DMAPP production, and suggestions have been made that this isomerization reaction is a rate-limiting step for isoprenoid biosynthesis (9, 10). IPP isomerase activity was stimulated by light in differentiating chloroplasts of maize etioplasts, unlike the enzyme activity of GGPP synthase and farnesyl pyrophosphate synthase and the carotenoid-committed enzyme phytoene synthase (9).
The current study was prompted by the fact that little is known about the compartmentation of the biosynthetic enzymes leading to carotenoids and about the effect of light on carotenoid synthesis in green algae and plants. In a single-celled alga such as Haematococcus it is simpler to follow the expression of light-stimulated isoprenoid synthetic enzymes and carotenoids in the same cell in contrast to higher plants, where carotenoids usually accumulate in specialized organs and in multicellular maturing tissues. Also, we had found previously that carotenoid production was enhanced in bacteria upon addition of a gene for IPP isomerase (11). We investigated the expression of three types of enzymes (Fig. 1), including IPP isomerase, lycopene β-cyclase, and β-carotene-C-4-oxygenase, in Haematococcus that were transferred from low to high light. The expression of these enzymes was correlated with the accumulation of carotenoids in wild-type Haematococcus as well as in a mutant (Car-3) with enhanced pigment accumulation. Two highly similar cDNAs for IPP isomerase were found, unlike in animals (12), yeast (13), and bacteria (14), where only one gene (or cDNA) has been reported, suggesting that the different gene products in the single-celled Haematococcus may reflect a requirement for this enzyme in different cell compartments. Two polypeptides, whose molecular weights closely correspond to those predicted by the cDNA sequences, were found to be immunoreactive to antisera raised against a recombinant gene product. Increased light intensity stimulated expression of both IPP isomerase genes and was positively correlated with carotenoid accumulation. However, expression of the two polypeptides differed, with one being relatively constant while the other increased in concert with the increase in the carotenoid level. This correlation suggests a direct relationship of one of the IPP isomerase levels with the cytoplasmic carotenoid accumulation in Haematococcus.
MATERIALS AND METHODS
Cell Growth, Light Treatment, and Mutagenesis.
Axenic cultures of Haematococcus pluvialis Flotow (Göttingen Culture Collection) were grown in a mineral liquid medium with acetate (15, 16). One-liter batches were grown in wide-bottomed Fernbach flasks (3-liter capacity) under white light provided by fluorescent lamps (Sylvania Electric Products, Fall River, MA; F48T12/D/VHO, 115W). Illumination at 20 μmol photon m−2⋅s−1 for 14 h at 22°C was followed by 10 h of darkness at 18°C. To enhance carotenoid production, log phase cultures grown as above were exposed to constant illumination of 150 μmol photon m−2⋅s−1 (22°C) and samples (50 ml) were collected at selected time points and harvested in a table-top clinical centrifuge.
Mutagenesis and Mutant Selection.
Cells from a log phase culture (400 ml at 2.0 × 105 cells/ml) were harvested by centrifugation (22°C at 3,000 × g for 5 min). The pelleted cells were rinsed twice in 200 ml sterile deionized water, resuspended in 20 ml sterile sodium phosphate buffer (0.1 M, pH 7), and incubated with ethyl methyl sulfonate at a final concentration of 0.2 M for 1 h with stirring on an illuminated stirrer. After dilution with 180 ml sterile deionized water, centrifugation, and two subsequent rinses in the growth medium lacking acetate, cells were incubated in the acetate-minus medium in darkness for 12 h and grown at 40 μmol photon m−2 ⋅s−1 (white light) for 3 days. To enrich for carotenoid-accumulating mutants, cultures were placed under constant blue-light illumination (250 μmol photon m−2⋅s−1, Sylvania Electric Products, F48T12/246/VHO) for 3 days. Subsequently, 50-ml aliquots were harvested by centrifugation and spread on 20 agar plates (2.5 ml per plate; growth medium plus 1% agar) and grown under low-intensity white light (15 μmol photon m−2⋅s−1). After 2 weeks, each brown colony selected from a background of green colonies under a microscope was streaked on growth medium in 1% agar for subsequent analysis. Four stable mutants (Car 1–4) were selected on the basis of their carotenoid accumulation, growth characteristics, and cell morphology. Of these, Car-3 was investigated further since its growth rate approximated that of wild type.
Construction of λ cDNA Expression Library and Screening for Clones Encoding IPP Isomerase.
An H. pluvialis cDNA library was constructed according to the instructions in Stratagene’s ZAP cDNA synthesis kit. The mRNA was extracted from cells grown at 80 μmol photon m−2⋅s−1 and exposed to white light (250 μmol photon m−2⋅s−1) for 9 h with the expectation that this treatment would induce the expression of genes involved in carotenoid biosynthesis. Double-stranded cDNAs were produced, selected, and ligated with uni-ZAP XR. The λ phage library was packaged with Gigapack II Gold packing extract. Approximately 1.5 × 106 pfu were obtained in the original library. The entire library was amplified once on 30 large agar plates.
The λ phage cDNA library was excised in vivo, using Stratagene’s ExAssist/SOLR system, to produce a phagemid cDNA library. Screening of the phagemid library was as described in detail in Cunningham et al. (17) and Sun et al. (11). Presumptive cDNA clones encoding ipiHp, identified (see Results) on the basis of enhanced accumulation of carotenoid pigments in Escherichia coli, were inoculated individually in liquid Luria–Bertani medium containing ampicillin and chloramphenicol, and the plasmid DNA was isolated by using a procedure described by Del Sal et al. (18).
Antisera Production and Purification.
A cDNA fragment encoding part of IPP isomerase (ipiHp1) of Haematococcus (starting at amino acid residue 77 as in Fig. 3) was used to produce a polypeptide that was used to generate polyclonal antibody production in rabbits. The cDNA fragment was excised from the cloning vector [pBluescript SK(−), Stratagene] by digestion with the restriction enzymes BamHI and XhoI, and then directionally cloned into the BamHI-XhoI-digested expression vector pET28b (Novagen) to fuse the ipiHp to a polyhistidine tag (to facilitate later purification of the expressed protein). The resulting plasmid was introduced into E. coli strain BL21(DE3). Cell extracts, upon isopropyl β-d-thiogalactosidase induction, showed a prominent band in the 33-kDa region after SDS/PAGE, which was purified on an affinity chromatography column by using a pTrcHis Xpress purification kit (Invitrogen) and separation on 12% SDS/PAGE. The excised gel band, ground to a powder in liquid nitrogen and mixed with complete Freund’s adjuvant, served as antigen. For purification of antisera, affinity columns were prepared by coupling the specific fusion protein in 0.1 M ALD solution (1 M NaCNBH3, Sterogene Bioseparations, Carlsbad, CA) to Actigel ALD (6 h, 20C°) and exhaustive rinsing with 15 mM phosphate/0.5 M NaCl, pH 7.0. The affinity-purified antibody fractions were eluted with ActiSep and buffer and stored at 4°C. For lycopene β-cyclase antigen we used the EcoRI-PvuII fragment, encompassing about 2/3 of the coding region of an Arabidopsis lycopene β-cyclase cDNA (17), and cloned it into the pET28b vector to produce a His6-tagged fusion protein of about 45 kDa.
Figure 3.
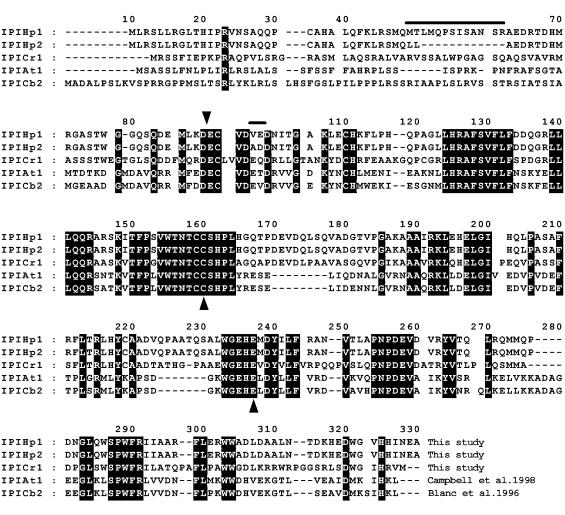
Amino acid sequence alignment of IPP isomerases predicted by cDNAs of Haematococcus pluvialis (IPIHp1 and IPIHp2) and Chlamydomonas reinhardtii (IPICr1) are compared with sequences from the flowering plant Clarkia brewerii (IPICb2) (29) and Arabidopsis thaliana (IPIAt1) (30). The sequences were aligned and shaded by using the program clustalw (http://dot.imgen.bcm.tmc.edu:9331/multialign) and shaded by using genedoc (http://www.concentric.net/∼Ketchup/gddl.htm). The dark shading with white letters indicates amino acid identity for the aligned residue for all five proteins whereas the lightly shaded sequences represent amino acid identity for four of five sequences. The lines above the sequences indicates differences in the predicated amino acid sequences of IPIHp1 and IPIHp2. The inverted arrowhead (▾) above IPIHp1 designates the location of a truncation from the N terminus. Residues required for catalytic activity (31) are marked by upright arrowheads (▴).
Haematococcus Sample Preparation.
Pelleted cells, rinsed in deionized water, were resuspended in a microfuge tube in 0.5–1.0 ml buffer (250 mM sucrose/25 mM Hepes⋅KOH/3 mM DTT/1 mM benzamidine/5 mM ɛ-amino-n-caproic acid, pH 7) and broken by grinding with a micropestle and small pieces of glass microfiber filter (Whatman GF/D), followed by storage at −80°C.
Western Blotting.
Aliquots of the broken cells were mixed with an equal volume of 2× sample buffer [100 mM Tris⋅HCl, pH 6.8/20% glycerol (vol/vol)], 4% SDS, and 10% 2-mercaptoethanol) and boiled for 5 min. Samples of 1.2 μg of Chl were loaded per lane and polypeptides were separated on a 12% SDS/PAGE gel (20) and transferred to a poly(vinylidene difluoride) membrane (Millipore) according to the method of Peluso and Rosenberg (20). Membranes were blocked in 5% (wt/vol) nonfat dry milk in TBS buffer (50 mM Tris⋅HCl/200 mM NaCl, pH 7.4) for 1 h (21). Primary (and secondary) antibodies were in 1% milk in TBS with 0.1% Tween 20 (vol/vol) at pH 7.4 in the following dilution: anti-IPP isomerase and anti-lycopene cyclase at 1:500, and anti-rubisco (to tobacco, which was a kind gift from the laboratory of H. Bohnert) at 1:10,000, for 1 h. After washing the membranes with TBS plus 0.1% Tween 20, horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit, Jackson ImmunoResearch) was applied at a concentration of 1:5,000 for 1 h. After washing, membranes were developed by using SuperSignal Substrate (Pierce).
Reverse Transcription–PCR Analysis of Gene Expression.
Total RNA was isolated by using the TRI reagent (Molecular Research Center, Cincinnati). The synthesis of first-strand cDNA was performed following the procedure in the Preamplification system of GIBCO. Total RNA was quantified by spectrometry and the intensity of the two rRNA bands, relative to standards, on agarose gels. For each sample, a serial dilution (1:2:4) of total RNA was used for cDNA synthesis to ascertain linearity of amplification. Amplification of the message was done by conventional PCR (94°C, 1 min; 58°C, 1 min; 72°C, 2 min) with several repeats. The two specific primers for ipiHp1 were 5′-AGCCCAGCATCTCAGCCAATC-3′ and 5′-CGAGCCATTTTACCTGACCTT-3′; for ipiHp2, 5′-AGCTGATGCTGAAGGACGAGT-3′ and 5′-AGAGAGACGATGAATATGATGC-3′; and for the β-carotene-C-4-oxygenase, 5′-ACTGGTTGCCTGTGTCCGAAG-3′ and 5′-CAGTGCAGGTCAAAGTGGTAGC-3′ (22).
Pigment Determination.
Pigment extractions were performed under dim room light. Chlorophyll content was estimated in 90% acetone extracts by using the equation of Jeffrey and Humphey (23). Carotenoids were estimated by the equation of Jaspers (24) (mg/liter = 4.1E467 − 0.0435Chla − 0.367Chlb), using the absorption maximum at 476 nm instead of 450 nm, to allow for the somewhat longer wavelength absorption of astaxanthin.
RESULTS
High Light Enhances Carotenoid Production in Haematococcus.
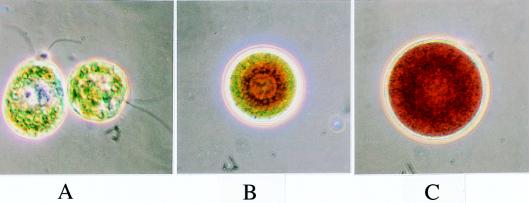
A unique developmental feature of the unicellular alga Haematococcus pluvialis is the transition from a green to a red stage (Fig. 2 A–C) with an accompanying accumulation of ketocarotenoids in the cytoplasm. An increase in cell size and an encystment stage of red cells (Fig. 2C) are common phenomena under natural conditions and are reproduced readily in the laboratory. The accumulation of carotenoids is a function, to some degree, of the illumination (Table 1). An increase in total carotenoid content in Haematococcus can be taken as a measure of carotenoid accumulation outside the chloroplast. The appearance of carotenoid-colored bodies can be followed by light microscopy and the corresponding electron-dense globules by electron microscopy (16, 25, 26).
Figure 2.
Developmental sequence of Haematococcus pluvialis cells from the green stage (A), to extraplastidic carotenoid accumulation and increase in cell size (B), to the enlarged red-cyst stage (C). (×1,600.)
Table 1.
Carotenoid-to-chlorophyll ratios increase with time of exposure to 150 μmol photon m−2⋅s−1
| Time, h | Car/Chl
|
|
|---|---|---|
| Wild type | Mutant Car-3 | |
| 0 | 0.13 | 0.15 |
| 6 | 0.17 | 0.24 |
| 12 | 0.22 | 0.28 |
| 24 | 0.40 | 0.87 |
| 48 | 0.51 | 1.10 |
Log-phase cultures grown at 20 μmol photon m−2⋅s−1 white light were exposed to 150 μmol photons m−2⋅s−1 for the time indicated in the left column. Pigment determination is on mol-to-mol basis.
Carotenoid accumulation in low light (<60 μmol photon m−2⋅s−1) tends not to occur under nutrient replete conditions, but carotenoid-rich, red-colored cells with thick walls (cysts) are found in nature often under high-light and low-nutrient status. Whereas it is commonly assumed that carotenoid accumulation is obligately linked with cyst formation, this is not the case. For example, the mutant Car-3 is one of several mutants, produced by chemical mutagenesis and selection under high-blue-light conditions, whose carotenoid-rich cells remain in the vegetative state. Initially, the mutant Car-3 (growing at near wild-type rates) has the same Car/Chl ratio as wild type, but it is precocious in carotenoid accumulation induced by high-light exposure. Within 24 h of the inducing illumination it shows more than a 2-fold increase in the Car/Chl ratio relative to the wild type (Table 1). These results led us to undertake a further comparison of the time course of expression of IPP isomerase and that of several enzymes directly involved in carotenoid biosynthesis in the wild type and the Car-3 mutant.
Identification of Two IPP Isomerase cDNAs in Haematococcus.
Since IPP isomerase is the first enzyme required for formation of many isoprenoids, and having observed previously that carotenoid production was enhanced by increasing the copy number of IPP isomerase genes in E. coli (11), we sought cDNAs encoding IPP isomerase by screening a cDNA library of Haematococcus. We used the “carotenoid color-reporting system” with the pAC-LYC plasmid construct previously employed in our laboratory (27) to screen E. coli colonies for enhanced carotenoid production and selected five deep-pink clones from the cDNA library that conferred a deep-pink phenotype, indicative of enhanced lycopene accumulation. From the sequences we identified two (ipiHp1 and ipiHp2, Hp designation for Haematococcus pluvialis) cDNAs as IPP isomerases by comparison with sequences of IPP isomerase from yeast (13) and other plants (28). Although highly similar, the two Haematococcus cDNA sequences almost certainly represent two distinct genes; first, because an alignment of the two shows 17 differences (out of 886 bases) spread widely throughout the coding region. Second, major dissimilarities are present in the 3′ end untranslated region, and, most significantly, a 36-base region (169–205) in ipiHp1 has no counterpart in the ipiHp2 sequence (not shown).
For the longer cDNA, a polypeptide (IPIHp1) of 305 aa with a molecular weight of 34,535 and a pI of 6.04 is predicted (Fig. 3). The second cDNA encodes a product, IPIHp2, with 297 aa residues, a molecular weight of 33,189, and a pI of 5.94. This second polypeptide lacks a 12-aa region present near the N terminus of IPIHp1, and several other differences in the predicted polypeptides are indicated in Fig. 3. For example, in the flanking region of the 12-aa region, the MT in IPIHp1 is substituted by LL in IPIHp2, and (progressing toward the C terminus) another difference has VE in IPIHp1 replaced by AD in IPIHp2. With a 95% sequence identity the two gene products of Haematococcus are closely related to each other, but quite different from known higher-plant IPIs. An IPI predicted from a cDNA from the related green alga Chlamydomonas reinhardtii more closely resembles the Haematococcus IPI isomerases, but even here their sequence identity is only 55% (Fig. 3) vs. 42% sequence identity for Haematococcus and those of higher plants such as Clarkia or Arabidopsis (28–30). Nevertheless, the alignment in Fig. 3 highlights a number of identical and highly conserved regions that are common to both green algae and higher plants. Among these are two critical residues C and E (upright arrowheads in Fig. 3), which, by mutational analysis in yeast, have been found to be essential for catalytic activity (31). Also, upon truncating the first 76-aa residues of the N terminus of ipiHp1 (inverted arrowhead in Fig. 3) we found a reduction in carotenoid accumulation in E. coli, suggesting another site important for maximal activity but not essential for catalysis. The predicted amino acid sequence of IPIHp2 is virtually identical to the predicted gene product pHP11 from another Haematococcus strain (NIES-144) (32), differing only by five conservative substitutions over the entire predicted amino acid sequence.
Accumulation of IPP Isomerase mRNA Is Up-Regulated by Increasing Light Intensity and Precedes Enhanced Accumulation of mRNA for the β-Carotene C-4-Oxygenase.
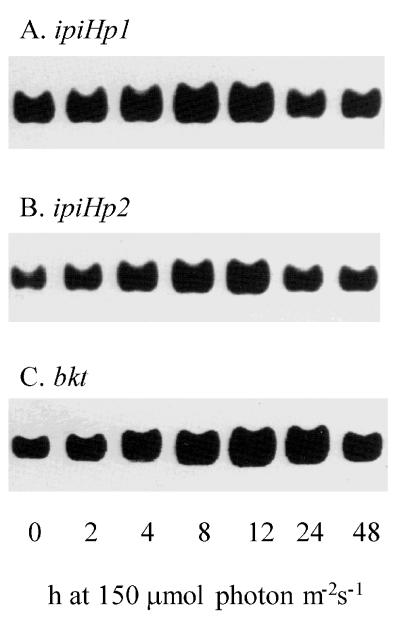
The RNA gel blot in Fig. 4 A and B establishes that the steady-state level of mRNA for the IPP isomerase genes (ipiHp1 and ipiHp2) is up-regulated by increasing the light intensity from 20 to 150 μmol photon m−2⋅s−1. After a 4-h lag the message level increased to a maximum at 8 h, but declined by 24 h with no further increase by 48 h. The early increase of the message occurs before the increase of the Car/Chl ratio (Table 1), which would be expected for production of carotenoids requiring IPP/DMAPP as initial substrates. Although the increase in the Car/Chl ratio occurred earlier in the Car-3 mutant than in wild type, there was not a notable difference in the pattern of steady-state message levels over the same time course.
Figure 4.
Increase of steady-state mRNA for IPP isomerases ipiHp1 (A) and ipiHp2 (B) in response to exposure to high light intensity precedes that for a β-carotene-C-4-oxygenase gene (C) as shown by reverse transcription–PCR for cells of Haematococcus wild type. Times of exposure to 150 μmol photon m−2⋅s−1, after from 20 μmol photon m−2⋅s−1, are indicated at the bottom.
Formation of the red-colored astaxanthin and canthaxanthin occurs in the late stages of carotenoid biosynthesis and is catalyzed by a β-carotene-C-4-oxygenase. A pair of primers (as in Materials and Methods) was produced to the sequence determined by Kajiwara et al. (22) to test the expression of the β-carotene-C-4-oxygenase gene (bkt). The maximal steady-state level of the oxygenase message was not evident until 12 h after exposure to high light (Fig. 4C), which is substantially later than the maximal level of message for the IPP isomerase and consistent with later synthesis of ketocarotenoids in cell development.
Differential Expression of IPP Isomerase Polypeptides and Their Increase in the Car-3 Mutant Are Correlated with Enhanced Carotenoid Accumulation.
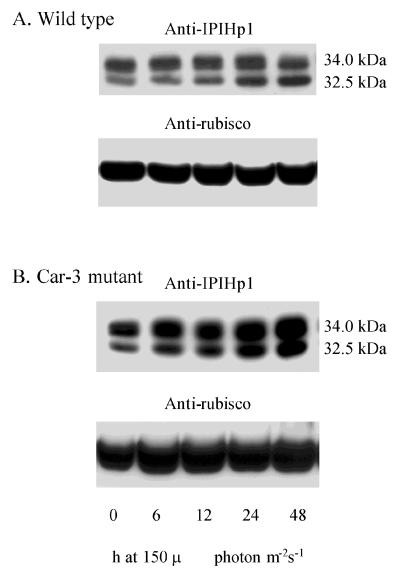
Differential expression over time of two IPP isomerases is suggested by the immunoblot in Fig. 5A with a polyclonal antibody to the IPIHp1 recombinant polypeptide. Polypeptides of 34 and 32.5 kDa were recognized by this antiserum that correspond closely to those predicted (34.5 and 33.2 kDa) from the cDNA sequences. Thus, we consider it highly probable that the 34- and 32.5-kDa polypeptides are the respective gene products of ipiHp1 and ipiHp2. Further, an antiserum produced to the 12-aa region present only in IPIHp1 (Fig. 3) recognizes only the upper band (not shown).
Figure 5.
Immunologically detected levels of IPP isomerase of Haematococcus cells grown under 20 μmol photon m−2⋅s−1 (0 h) and subsequently exposed to 150 μmol photon m−2⋅s−1 (6–48 h). Each lane of the SDS/polyacrylamide gel was loaded with 1.2 μg chlorophyll. Two polypeptides of 34 kDa and 32.5 kDa were recognized when membranes were probed with an antibody to the ipiHp1 gene product in wild-type cells (A) and in Car-3 mutant cells (B). Comparable samples were probed with anti-rubisco (large subunit) in bottom lanes, and bound antibody was visualized by enhanced chemiluminescence.
There is a notable increase of the 32.5-kDa relative to the 34-kDa immunoreactive band over the first 24 h on exposure to 150 μmol photon m−2⋅s−1. This increase corresponds to the visual accumulation of carotenoid globules outside the chloroplast and thus leads to the consideration that expression of the two isomerase genes is differentially regulated and that the proteins are perhaps targeted to different cell compartments.
If IPP/DMAPP is a rate-limiting substrate for polyisoprenoid synthesis, as has been postulated (9), then increasing the level of IPP isomerase may result in enhanced carotenoid production. In fact, enhanced carotenoid accumulation was observed in E. coli after insertion of the ipiHp1 gene into plasmids containing the carotenoid gene cluster (resulting in plasmid pAC-BETA-04) (11). We previously had taken advantage of this phenotype in the identification and functional analysis of a β-carotene hydroxylase from Arabidopsis (11). Also, a similar construct made with the gene equivalent to ipiHp2, and an isomerase gene from the yeast Phaffia rhodozyma, led to enhanced carotenoid accumulation in E. coli (32). In the mutant Car-3, which accumulates carotenoids at an earlier stage than wild type, a higher IPP isomerase content might be expected. Indeed, in Fig. 5 it is evident that the level of IPP isomerase is greater in Car-3 than in wild type. Such a positive correlation between IPP isomerase expression and enhanced carotenoid accumulation level in Car-3 is concordant with the demonstrated carotenoid increase in E. coli upon introduction of a cDNA-encoding IPP isomerase (11).
Lycopene β-Cyclase Expression Increases only in Car-3.
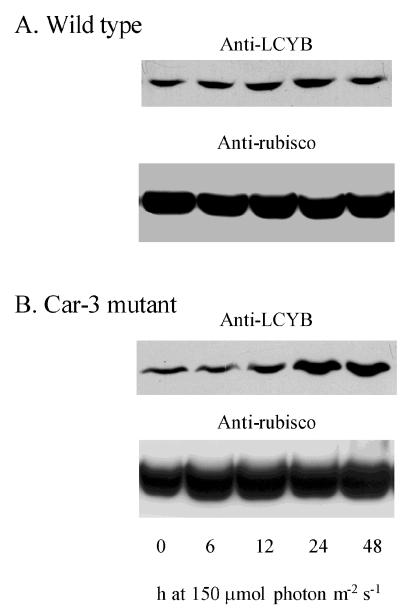
β-carotene is a bicyclic compound formed from the linear lycopene via the action of the β-cyclase enzyme. Its formation is a major branch point in carotenoid synthesis, and it is a precursor for canthaxanthin and astaxanthin, which are the red-colored di-ketocarotenoid end products in Haematococcus (22, 33). An antibody produced to a fusion protein of the β-cyclase (of Arabidopsis) (17) strongly reacted with a polypeptide with an apparent molecular mass of ca. 50 kDa on SDS/PAGE (Fig. 6) in Haematococcus. The 50-kDa size closely corresponds to that observed in Arabidopsis (not shown) and that inferred from the predicted polypeptide sequence (6). There is no apparent increase in lycopene β-cyclase in wild-type cells during the developmental transition from green to red cells. However, in Car-3 there is an increase in the level after a 24-h exposure to 150 μmol photon m−2⋅s−1 and is maintained without decline at 48 h (Fig. 5B). Interestingly, there is a notable decline by 48 h of the β-carotene C-4-oxygenase message (Fig. 4C).
Figure 6.
Immunoblot to determine level of lycopene β-cyclase. An increase is observed by 24 h in Car-3 mutant cells (B), which also have a much greater carotenoid accumulation than in wild-type cells (A). Each lane of the SDS/polyacrylamide gel was loaded with 1.2 μg chlorophyll. Membranes were probed with an antibody prepared to a recombinant protein of a gene encoding lycopene β-cyclase (LYCB) of Arabidopsis (as in methods of ref. 17). Protein level of rubisco was used as a control, and bound antibody was visualized by enhanced chemiluminescence.
DISCUSSION
Our interest in the biosynthetic pathway of carotenoids and the isoprenoid precursors that provide the substrate for this pathway (6) has led us to study this pathway in a unicellular alga where carotenoid accumulation can be manipulated readily by changes in light intensity. In Haematococcus, the massive accumulation of carotenoids in the cytoplasm can lead to deep-red-colored cells, but the chloroplast remains green and never becomes a chromoplast, unlike in higher plants where the photosynthetic lamellae disappear during carotenoid accumulation in the stroma.
The role of carotenoid accumulation in the unicellular alga Haematococcus is not understood, although various potential photoprotective roles have been proposed (34–36). In such a unicellular photoautotroph, dependent as it is on photosynthesis for its continued growth and survival, it is difficult to imagine that enhanced accumulation of carotenoids would not be beneficial, since carotenoid production requires redirection of a substantial amount of the cells’ carbon resources into synthesis of isoprenoids. Carotenoid accumulation in Haematococcus is inducible by a variety of environmental stress conditions, including nutrient deprivation and high illumination (34). Thus, the accumulation of carotenoids in this alga is one facet of a general response to stress conditions that presumably is of advantage to its survival. We conjecture that the accumulation of ketocarotenoids plays a role in minimizing the oxidation of lipids. An antioxidant role of carotenoids has been shown in the yeast Phaffia (37) and proposed for Haematococcus (38). Further, Sprey (39) found a close correlation between increasing lipid content and accumulation of β-carotene, astaxanthin, and canthaxanthin in oil globules of Haematococcus. Carotenoids thus would be of primary benefit to the cells by inhibition of lipid peroxidation (40) within membranes and also in protecting the storage lipids that Haematococcus relies on as an energy source under photosynthesis-limited conditions.
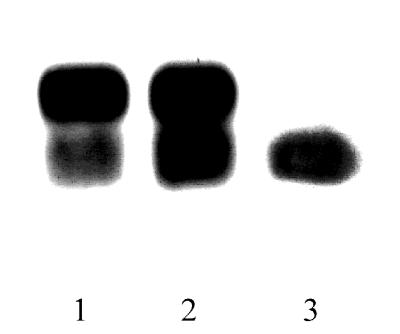
It is of interest to consider our results on the differential expression of the two IPP isomerase polypeptides, i.e., the increased expression of the 32.5-kDa form relative to the 34-kDa form, in a physiologically relevant framework. If the 32.5-kDa form is responsible for ensuring the substrate supply for cytoplasmically localized carotenoids, one would expect it to be maintained in red cyst cells. In fact, the 32.5-kDa form alone was found in completely red cultures while the 34-kDa form had decreased below the level of detection on immunoblots (Fig. 7). The fact that Kajiwara et al. (32) found the cDNA only for the 32.5-kDa form of IPP isomerase can be regarded as further support for the observed differential expression, since red cysts (probably enriched in mRNA for this smaller form) were used for cDNA preparation.
Figure 7.
In the red-cyst stage the 32.5-kDa form of IPP isomerase is readily observed but the 34-kDa form is no longer detected. Each lane of the SDS/polyacrylamide gel contained 1.2 μg of chlorophyll. Lane 1, SDS extract from green cells (20 μmol photon m−2⋅s−1, 6 h); lane 2, SDS extract from cells in an early stage of carotenoid accumulation (150 μmol photon m−2⋅s−1, 24 h); lane 3, SDS extract from red cysts. Membranes were probed with the antibody to the ipiHp1 gene product and visualized with the enhanced chemiluminescence assay.
The inferred sizes of the IPP isomerases of plants and algae (Fig. 3) are larger than those of mammals (12), yeast (13), and Rhodobacter (41). The increased lengths of about 70-aa residues are largely extensions at the N termini, but lack recognizable targeting sequences for different cell compartments. Whereas, it is generally assumed that IPP is synthesized in the cytoplasm (4, 5), Paton et al. (12) recently showed that IPP isomerase is targeted to peroxisomes in mammals. Consistent with our results is the expectation that in Haematococcus the 32.5-kDa form of IPP isomerase is targeted to the cytoplasm, and that the 34-kDa form is targeted to the chloroplast. However, our current sequences do not provide a definitive targeting sequence.
With the antibody to the lycopene β-cyclase of Arabidopsis we detected only one polypeptide band in Haematococcus (Fig. 6), consistent with our findings in higher plants. Its location is unknown in the cell as is that of β-carotene-C-4-oxygenase. If other β-cyclases do not exist in this alga, then the β-carotene (probable precursor for ketocarotenoids) must somehow be exported from the chloroplast. The sequences of β-carotene-C-4-oxygenase cDNAs from different Haematococcus strains by Lotan and Hirschberg (33) and Kajiwara et al. (22) are very similar but not identical, allowing for the possible existence of multiple carotene oxygenases in Haematococcus.
In conclusion, our results provide support for the hypothesis that IPP isomerase catalyzes a crucial rate-limiting step in IPP/DMAPP production for the biosynthesis of carotenoids. In the green alga Haematococcus the carotenoids accumulate in the cytoplasm, unlike in higher plants, in which chromoplasts are the site of carotenoid accumulation. Thus, this organism provides a novel experimental system for investigating localization and regulation of carotenoid production. Given that the expression of one of two IPP isomerases is differentially up-regulated by higher light intensity, we suggest that the 32.5-kDa form is responsible for carotenoid accumulation in the cytoplasm.
Acknowledgments
This work was supported partially by a grant from the Binational Agricultural Development Fund (Project number US-2006-91R), by the National Science Foundation (NSF award number 9631257), and by the Maryland Agricultural Experiment Station (E.G.).
ABBREVIATIONS
- Car
carotenoids
- Chl
chlorophyll
- DMAPP
dimethylallyl pyrophosphate
- GGPP
geranylgeranyl pyrophosphate
- IPI
isopentenyl pyrophosphate isomerase
- IPP
isopentenyl pyrophosphate
Footnotes
References
- 1.Sandmann G, Böger P. In: Target Sites of Herbicide Action. Böger P, Sandmann G, editors. Boca Raton, FL: CRC; 1989. pp. 25–44. [Google Scholar]
- 2.Demmig-Adams B, Adams W W., III . In: Carotenoids in Photosynthesis. Young A J, Britton G, editors. London: Chapman and Hall; 1993. pp. 206–252. [Google Scholar]
- 3.Gray J C. Adv Bot Res. 1987;14:25–91. [Google Scholar]
- 4.Chappell J. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:521–547. [Google Scholar]
- 5.McGarvey D J, Croteau R. Plant Cell. 1995;7:1015–1026. doi: 10.1105/tpc.7.7.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham F X, Jr, Gantt E. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:557–583. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- 7.Sandmann G. Eur J Biochem. 1994;223:7–24. doi: 10.1111/j.1432-1033.1994.tb18961.x. [DOI] [PubMed] [Google Scholar]
- 8.Armstrong G A. Annu Rev Microbiol. 1997;51:629–659. doi: 10.1146/annurev.micro.51.1.629. [DOI] [PubMed] [Google Scholar]
- 9.Albrecht M, Sandmann G. Plant Physiol. 1994;105:529–534. doi: 10.1104/pp.105.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos-Valdivia A C, van der Heijden R, Verpoorte R. Nat Products Rep. 1997;14:591–603. doi: 10.1039/np9971400591. [DOI] [PubMed] [Google Scholar]
- 11.Sun Z, Gantt E, Cunningham F X., Jr J Biol Chem. 1996;271:24349–24352. doi: 10.1074/jbc.271.40.24349. [DOI] [PubMed] [Google Scholar]
- 12.Paton V G, Shackelford J E, Krisans S K. J Biol Chem. 1997;272:18945–18950. doi: 10.1074/jbc.272.30.18945. [DOI] [PubMed] [Google Scholar]
- 13.Anderson M S, Muchlbacher M, Street I P, Proffitt J, Poulter C D. J Biol Chem. 1989;264:19169–19175. [PubMed] [Google Scholar]
- 14.Hahn F M, Xuan J W, Chambers A F, Poulter C D. Arch Biochem Biophys. 1996;332:30–34. doi: 10.1006/abbi.1996.0312. [DOI] [PubMed] [Google Scholar]
- 15.Bednarik D P, Hoober J K. Arch Biochem Biophys. 1985;240:369–379. doi: 10.1016/0003-9861(85)90042-6. [DOI] [PubMed] [Google Scholar]
- 16.Tan S, Cunningham F X, Jr, Youmans M, Grabowski B, Sun Z, Gantt E. J Phycol. 1995;31:897–905. [Google Scholar]
- 17.Cunningham F X, Jr, Pogson B, Sun Z, McDonald K A, DellaPenna D, Gantt E. Plant Cell. 1996;8:1613–1626. doi: 10.1105/tpc.8.9.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del Sal G, Manfioletti G, Schneider C. Nucleic Acids Res. 1988;16:9878. doi: 10.1093/nar/16.20.9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Peluso R W, Rosenberg G H. Anal Biochem. 1987;162:389–398. doi: 10.1016/0003-2697(87)90409-x. [DOI] [PubMed] [Google Scholar]
- 21.Mustardy L, Cunningham F X, Jr, Gantt E. Plant Physiol. 1990;94:334–340. doi: 10.1104/pp.94.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kajiwara S, Kakizono T, Saito T, Kondo K, Ohtani T, Nishio N, Nagai S, Misawa N. Plant Mol Biol. 1995;29:343–352. doi: 10.1007/BF00043657. [DOI] [PubMed] [Google Scholar]
- 23.Jeffrey S W, Humphey G F. Biochem Physiol Pflanzen. 1975;167:191–194. [Google Scholar]
- 24.Jaspers E M W. Physiol Plant. 1965;18:933–940. [Google Scholar]
- 25.Lang N. J Phycol. 1968;4:12–19. doi: 10.1111/j.1529-8817.1968.tb04670.x. [DOI] [PubMed] [Google Scholar]
- 26.Santos M F, Mesquita J F. Cytologia. 1984;49:215–228. [Google Scholar]
- 27.Cunningham F X, Jr, Sun Z, Chamovitz D, Hirschberg J, Gantt E. Plant Cell. 1994;6:1107–1121. doi: 10.1105/tpc.6.8.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanc V M, Pichersky E. Plant Physiol. 1995;108:855–856. doi: 10.1104/pp.108.2.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanc V M, Mullin K, Pichersky E. Plant Physiol. 1996;111:652. [Google Scholar]
- 30.Campbell M, Hahn F M, Poulter C D, Leustek T. Plant Mol Biol. 1998;36:323–328. doi: 10.1023/a:1005935516274. [DOI] [PubMed] [Google Scholar]
- 31.Street I P, Coffman H R, Baker J A, Poulter C D. Biochemistry. 1994;33:4212–4217. doi: 10.1021/bi00180a014. [DOI] [PubMed] [Google Scholar]
- 32.Kajiwara S, Fraser P D, Kondo K, Misawa N. Biochem J. 1997;324:421–426. doi: 10.1042/bj3240421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lotan T, Hirschberg J. FEBS Lett. 1995;364:125–128. doi: 10.1016/0014-5793(95)00368-j. [DOI] [PubMed] [Google Scholar]
- 34.Boussiba S, Vonshak A. Plant Cell Physiol. 1991;32:1077–1080. [Google Scholar]
- 35.Young Y Y R, Lee Y K. Phycologia. 1991;30:257–261. [Google Scholar]
- 36.Hagen C, Braune W, Björn L O. J Phycol. 1994;30:241–248. [Google Scholar]
- 37.Schroeder W A, Johnson E A. J Biol Chem. 1995;270:18374–18379. doi: 10.1074/jbc.270.31.18374. [DOI] [PubMed] [Google Scholar]
- 38.Hagen C, Braune W, Greulich F. J Photochem Photobiol B. 1993;20:153–160. [Google Scholar]
- 39.Sprey B. Protoplasma. 1970;71:235–250. doi: 10.1007/BF01279634. [DOI] [PubMed] [Google Scholar]
- 40.Frank H A, Cogdell R J. Photochem Photobiol. 1996;63:257–264. doi: 10.1111/j.1751-1097.1996.tb03022.x. [DOI] [PubMed] [Google Scholar]
- 41.Hahn H M, Baker J A, Poulter C D. J Bacteriol. 1996;178:619–624. doi: 10.1128/jb.178.3.619-624.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]