Abstract
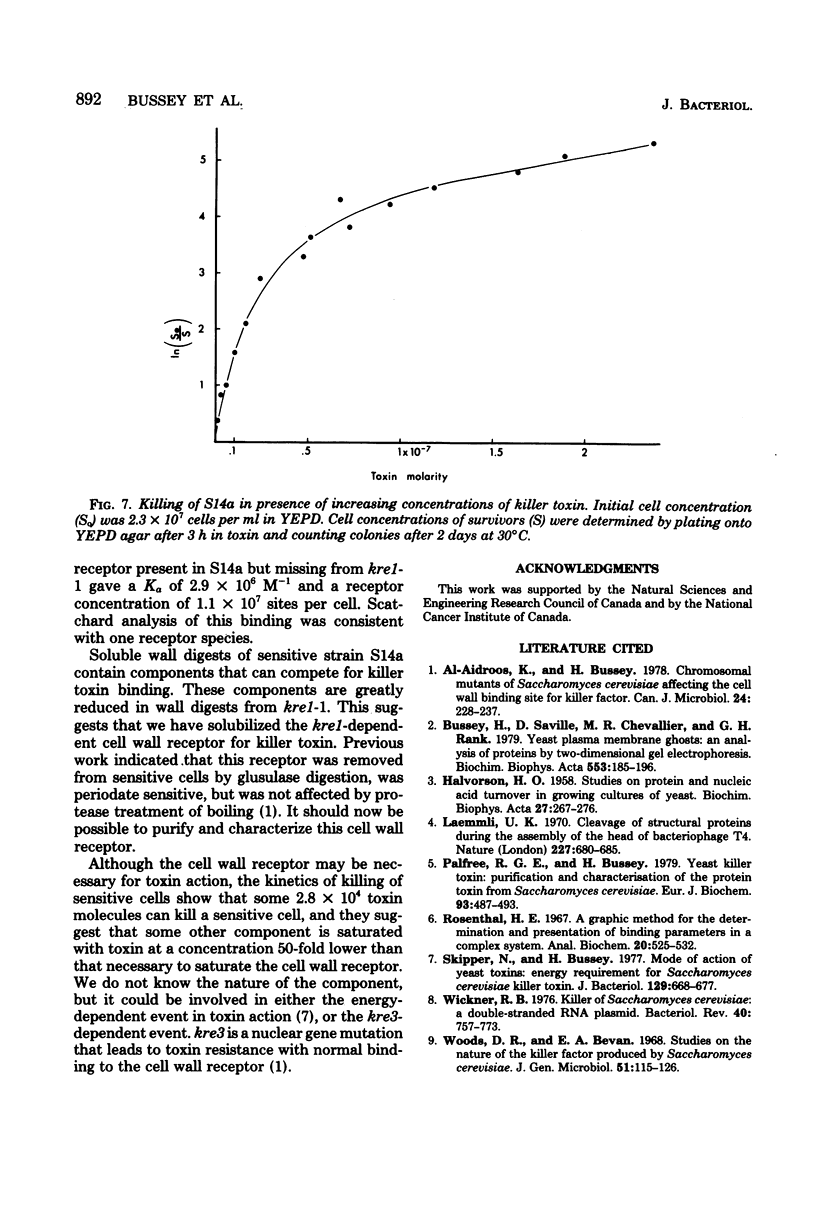
35S-labeled killer toxin protein bound to cells of sensitive Saccharomyces cerevisiae S14a. Strains that were resistant to toxin through mutation in the nuclear genes kre1 kre2 bound toxin only weakly. Non-radioactive toxin competed effectively with 35S-labeled toxin for binding to S14a, but did not compete significantly in the binding to mutant kre1-1. This implied that binding to kre1-1 was nonspecific. A Scatchard analysis of the specific binding to S14a gave a linear plot, with an association constant of 2.9 x 10(6) M-1 and a receptor number of 1.1 x 10(7) per cell. Killer toxin receptors were solubilized from the cell wall by zymolyase digestion. Soluble, non-dialyzable cell wall digest from S14a competed with sensitive yeast cells for 35S-labeled toxin binding and reduced toxin-dependent killing of a sensitive strain. Wall digest from kre1-1 competed only weakly for toxin binding with sensitive cells and caused little reduction of toxin-dependent killing. Although the abundant (1.1 x 10(7) per cell) receptor appeared necessary for toxin action, as few as 2.8 x 10(4) toxin molecules were necessary to kill a sensitive cell of S14a. The kinetics killing of S14a suggested that some component was saturated with toxin at a concentration 50-fold lower than that needed to saturate the wall receptor.
Full text
PDF
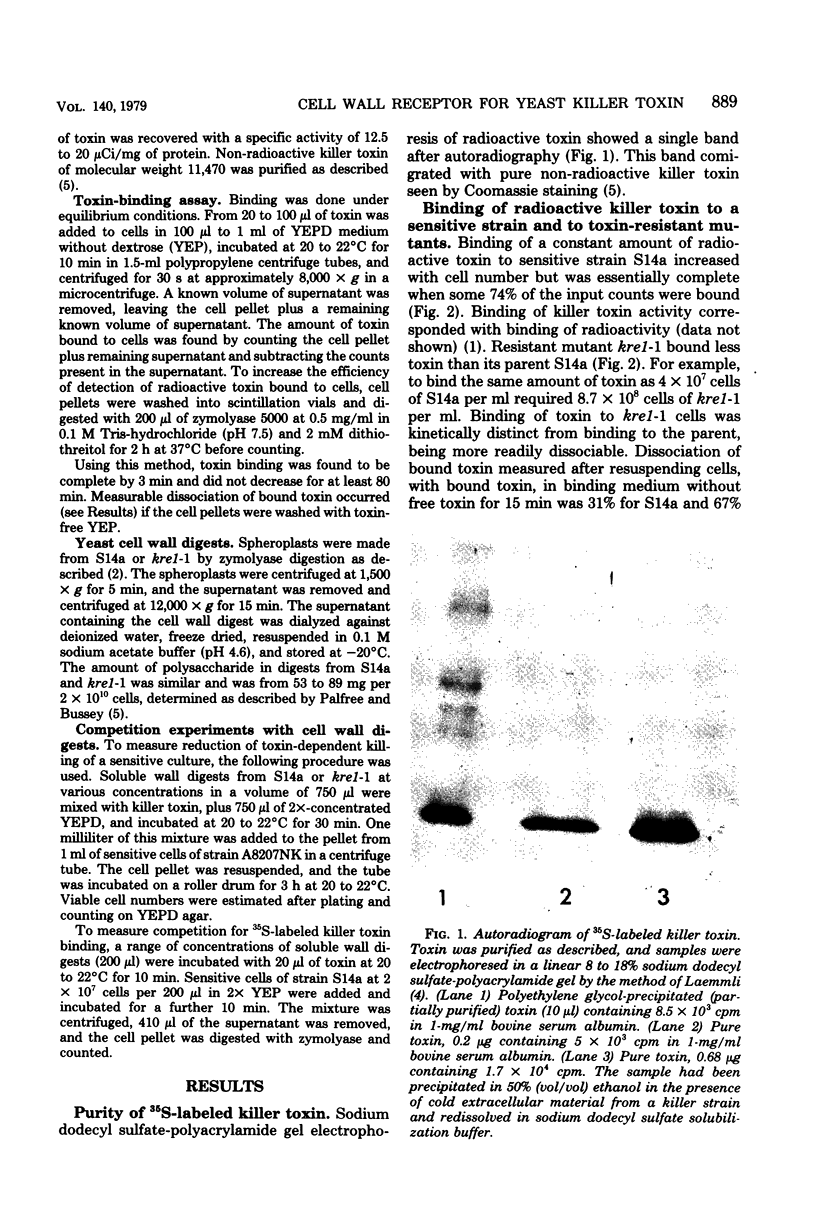
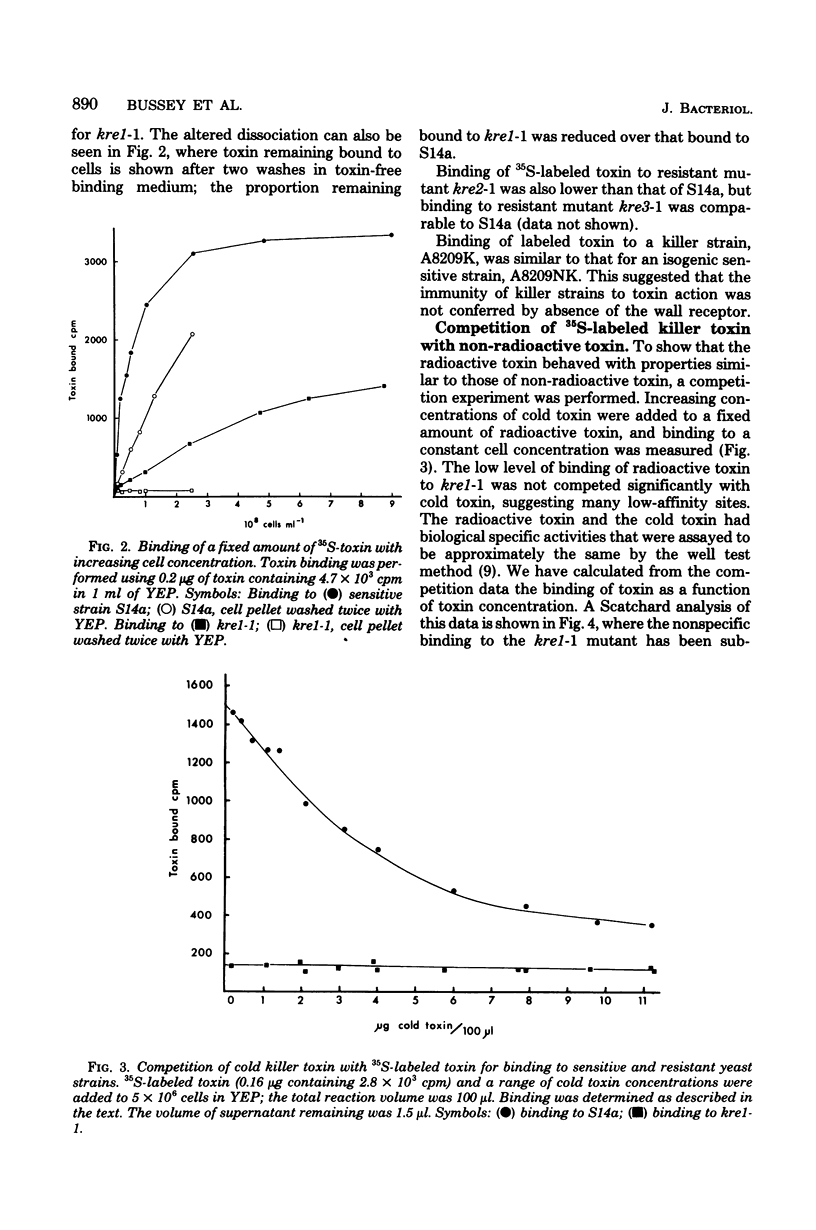
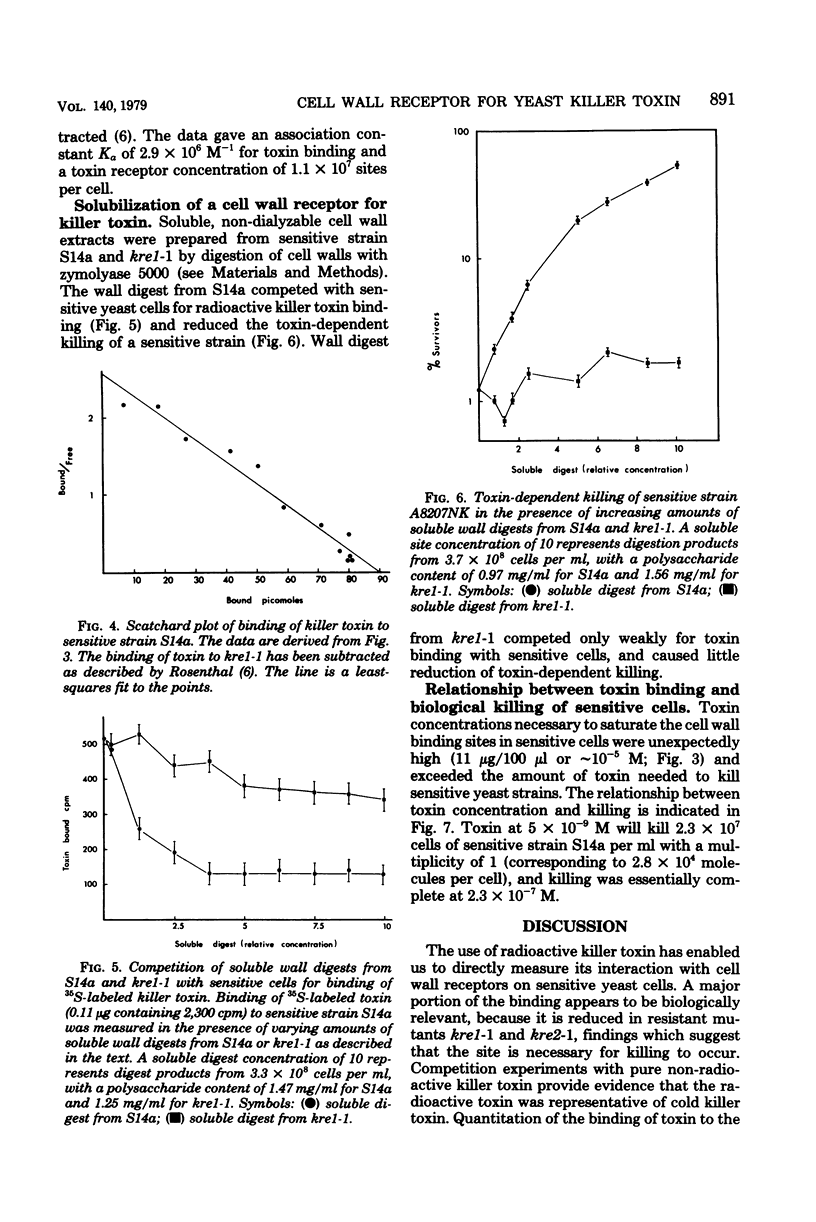
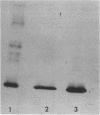
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Al-Aidroos K., Bussey H. Chromosomal mutants of Saccharomyces cerevisiae affecting the cell wall binding site for killer factor. Can J Microbiol. 1978 Mar;24(3):228–237. doi: 10.1139/m78-041. [DOI] [PubMed] [Google Scholar]
- Bussey H., Saville D., Chevallier M. R., Rank G. H. Yeast plasma membrane ghosts. An analysis of proteins by two-dimensional gel electrophoresis. Biochim Biophys Acta. 1979 May 17;553(2):185–196. doi: 10.1016/0005-2736(79)90224-4. [DOI] [PubMed] [Google Scholar]
- HALVORSON H. Studies on protein and nucleic acid turnover in growing cultures of yeast. Biochim Biophys Acta. 1958 Feb;27(2):267–276. doi: 10.1016/0006-3002(58)90333-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Palfree R. G., Bussey H. Yeast killer toxin: purification and characterisation of the protein toxin from Saccharomyces cerevisiae. Eur J Biochem. 1979 Feb 1;93(3):487–493. doi: 10.1111/j.1432-1033.1979.tb12847.x. [DOI] [PubMed] [Google Scholar]
- Rosenthal H. E. A graphic method for the determination and presentation of binding parameters in a complex system. Anal Biochem. 1967 Sep;20(3):525–532. doi: 10.1016/0003-2697(67)90297-7. [DOI] [PubMed] [Google Scholar]
- Skipper N., Bussey H. Mode of action of yeast toxins: energy requirement for Saccharomyces cerevisiae killer toxin. J Bacteriol. 1977 Feb;129(2):668–677. doi: 10.1128/jb.129.2.668-677.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. Killer of Saccharomyces cerevisiae: a double-stranded ribonucleic acid plasmid. Bacteriol Rev. 1976 Sep;40(3):757–773. doi: 10.1128/br.40.3.757-773.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D. R., Bevan E. A. Studies on the nature of the killer factor produced by Saccharomyces cerevisiae. J Gen Microbiol. 1968 Apr;51(1):115–126. doi: 10.1099/00221287-51-1-115. [DOI] [PubMed] [Google Scholar]