Abstract
The design and chemical synthesis of two de novo four-helix bundle proteins is described; each protein has two bound cofactors. Their construction from purified peptides is based on the modular assembly of different amphiphilic helices by chemoselective coupling to a cyclic peptide template. In the hydrophobic interior of the antiparallel four-helix bundle these proteins contain a heme in a binding pocket with two ligating histidine residues. A ruthenium-tris(bipyridine) complex is covalently bound to different positions at the hydrophilic side of one of the heme-binding helices. Laser-induced electron transfer across the varied distance through this helix has been studied and compared with a pathway analysis. The UV-visible, CD, and mass spectra are consistent with the structure and orientation predetermined by the template.
Keywords: synthetic protein/chemoselective coupling/ruthenium trisbipyridine/cytochrome b
Redox proteins represent the largest group of enzymes. Their cofactors or redox centers are embedded in structural domains with a wide variety of functions, including light-induced charge separation and electron transport. The size of a domain has come within reach of chemical methods by recent developments in peptide synthesis and chemoselective ligation to link unprotected peptides. The de novo design of redox proteins is an attractive approach to investigate the factors involved in electron transfer (ET) and to create proteins with novel functions. Synthetic peptides have been linked sequentially (1) or assembled by templates like transition metals (2), porphyrins (3), carbohydrates (4), and peptides (5). In particular, a cyclic peptide template with orthogonally protected amino acids provides selective ligation and a well-defined relative orientation of the peptide elements (6). Such a branched design of a template-assembled synthetic protein avoids the folding problem encountered in the association of linear peptides (7) and increases the stability of the folded structure (8). The binding of heme groups to bundles of amphiphilic linear helices has been extensively investigated (9, 10). For studies of the ET, different cofactors have been bound to a single helix (11) or bundles of two (12) and three helices (13). Light-induced ET through metalloproteins like cytochromes and myoglobins (14) has been investigated after modification of surface-exposed amino acid residues by ruthenium complexes. Electron tunneling through a protein (15) is a matter of discussion and has been approximated by a homogeneous barrier to ET in complexes like photosynthetic reaction centers (16). However, different regions of a given protein can indeed display different barriers to ET, as shown by an analysis of pathways through the protein (17). We have extended our model of cytochrome b termed MOP1 with a template-based antiparallel four-helix bundle with two different helices (18) to one with three different helices capable of light-induced ET. One of the two heme-binding amphiphilic helices carries besides the ligating histidine a cysteine at the hydrophilic side to which a ruthenium tris-bipyridine complex is bound. This helix has been designed to mediate the photoinitiated ET from the metal-to-ligand charge transfer triplet state of the ruthenium complex to the heme in the hydrophobic interior across different distances by variation of the position of the cysteine residue. The strategy to assemble synthetic proteins with variations in single peptide elements in a modular way offers the possibility to create families of proteins with specifically tuned functions. It shows the chemical synthesis of a nanomodule with high yield by controlled assembly of peptides and cofactor building blocks and represents a step toward artificial photoenzymes.
METHODS
Peptide Synthesis.
Chemicals for peptide synthesis were purchased from Perseptive Biosystems (Framingham, MA), and preloaded Fmoc-Gly-NovaSyn TGT resin was from Nova Biochem. All other chemicals of the highest available grade were obtained from Aldrich/Sigma or Merck. The amino acid sequences of the four helical peptides and the cyclic peptide template are given with the position of selectively cleavable cysteine protecting groups: 3-maleimidopropionyl (Mp)-H1, Mp-G-N-A-R-E-L-H-E-K-A-L-K-Q-L-E-E-L-F-K-K-W-NH2; Mp-H2, Ac-N-L-E-E-F-L-K-K-F-Q-E-A-L-E-K-A-Q-K-L-L-K(Mp)-NH2; Mp-H3(StBu), Mp-G-N-A-L-E-L-H-E-K-A-L-K-Q-L-E-C(StBu)-L-L-K-Q-L-NH2; Mp-H4(Acm), Mp-G-N-A-L-E-L-H-E-K-A-L-K-C(Acm)-L-E-Q-L-L-K-Q-L-NH2; T(StBu) (Trt)2(Acm), cyclo[C(Acm)-A-C(Trt)-P-G-C(StBu)-A-C(Trt)-P-G-]; MOP2, T[Sp-H1][Sp-H2]2[Sp-H3]; MOP3, T[Sp-H1][Sp-H2]2[Sp-H4].
The helical peptides were synthesized by solid-phase peptide synthesis using standard 9-fluorenylmethoxycarbonyl/t-butyl protection strategy with the exception of Cys-16 of H3, Cys-13 of H4, and Lys-21 of H2, which were protected by t-butylthio (StBu), acetamidomethyl (Acm), and allyloxycarbonyl, respectively, and the use of PAL-polyethylene glycol-polystyrene as described (19). 3-Maleimidopropionic acid was coupled to the N terminus of the resin-bound peptides H1, H3, and H4 by using a 3-fold molar excess of the preformed symmetrical anhydride synthesized as described (20) except for the use of diisopropylcarbodiimide. H2 was acetylated by stirring the resin in 5% (vol/vol) acetic anhydride in dimethylformamide for 30 min. The allyloxycarbonyl group at the ɛ-amino group of Lys-21(H2) was removed by palladium-catalyzed hydrostannolysis (19) and 3-maleimidopropionic acid coupled as described above. The peptides were cleaved from the resin and simultaneously deprotected by using 23:1:1 trifluoroacetic acid (TFA)/anisole/DTT except for the Acm and StBu protecting groups. Crude peptides were precipitated with cold diethylether and purified to homogeneity by preparative reversed-phase HPLC on a Waters model 4000 system equipped with a Waters model 486 tunable absorbance detector and a Waters DeltaPak C18 PrepPak column (40 × 300 mm, 15 μm) at a flow rate of 50 ml/min by using aqueous-acetonitrile gradients containing 0.1% (vol/vol) TFA.
The cyclic template peptide was synthesized manually as described for a similar sequence (19). The StBu protecting group was removed by reduction with tri-n-butylphosphine: 0.07 mmol of the cyclic peptide were dissolved in 100 ml 3:2 (vol/vol) 1-propanol/0.1 M ammonium acetate buffer, pH 7.8. The solution was degassed by bubbling with argon for 10 min. After adding 12 ml of 0.6 M tri-n-butylphosphine in 1-propanol (7.2 mmol) the solution was stirred for 30 min under argon and evaporated to dryness, and the peptide was purified by reversed-phase HPLC.
Assembly of the Modular Peptides MOP2 and MOP3.
The chemoselective ligation of the unprotected helical peptide Mp-H1 with the StBu deprotected cyclic template T(Acm)trityl (Trt)2 was carried out by combining 65 mg (i.e., 19.1 μmol with six TFA counteranions per peptide molecule) of Mp-H1 with 27 mg (19 μmol) of T(Acm)(Trt)2 in 1 ml of 3:2 (vol/vol) 0.15 M sodium phosphate buffer, pH 7/acetonitrile previously deoxygenated by bubbling with argon. The addition of the SH-group of cysteine to the Mp group converts it to a thioether-linked 3-succinimidopropionyl (Sp) group. The reaction was terminated after stirring the solution under argon for 30 min at room temperature by adding 10 μl of mercaptoethanol to deactivate the surplus Mp groups. After 10 min the peptide was purified by preparative reversed-phase HPLC and lyophilized. The Trt protecting groups of T(Acm)(Trt)2[Sp-H1] were removed by adding 5 ml of 19:1 (vol/wt) TFA/DTT to the peptide and stirring for 30 min. The solution was concentrated to approximately 1 ml, and the peptide T(Acm)[Sp-H1] was precipitated with cold diethyl ether, purified by preparative reversed-phase HPLC, and lyophilized.
Sixty-nine milligrams (i.e., 16 μmol with six TFA counteranions) T(Acm)[Sp-H1] were ligated with 105 mg of Mp-H2 (i.e., 33 μmol with four TFA counteranions) in 2.5 ml of buffer as described above. After 30 min 10 μl of mercaptoethanol were added, and after an additional 10 min the product was lyophilized. The Acm protecting group of T(Acm)[Sp-H1][Sp-H2]2 was removed with mercuric acetate at pH 4 as described previously for the synthesis of MOP1 (19). After reversed-phase HPLC and lyophilization the helical peptides Mp-H3(StBu) and Mp-H4(Acm) were ligated with T[Sp-H1][Sp-H2]2 as described, and the StBu and Acm protecting groups of Cys-16(Mp-H3) and Cys-13(Mp-H4) were removed to yield MOP2 and MOP3, respectively. All crude and purified peptides were analyzed by electrospray mass spectrometry on a Finnigan model TSQ 700 tandem quadrupole mass spectrometer.
Ru(bpy)2Cl2 (with bpy being 2,2′-bipyridine) was synthesized as described by Hitchcock et al. (21). The synthesis of 4-bromomethyl-4′-methyl-2,2′-bipyridine (Br-dmbpy) and the assembly of the ruthenium complex [Ru(Br-dmbpy)(bpy)2](PF6)2 was performed as described (22). The ruthenium complex was chemoselectively coupled to the deprotected cysteine residues of MOP2 or MOP3 by adding 13 mg (13.4 μmol) of [Ru(Br-dmbpy)(bpy)2](PF6)2 to a degassed solution of 15 mg (1.3 μmol) of MOP2 or MOP3 in 500 μl of 3:2 0.15 M sodium phosphate, pH 7.5/acetonitrile and stirring the solution under argon at room temperature for 14 h. The ruthenium-modified peptides were purified by reversed-phase HPLC. Size exclusion chromatography, the incorporation of heme, and the determination of the heme extinction coefficients by the pyridine-hemochrome method were performed as described (19). Peptide concentrations were estimated from the absorbance of tryptophan at 280 nm or of Ru(dmbpy)(bpy)2 at 452 nm by using an extinction coefficient of 5,700 or 13,500 M−1⋅cm−1 (22), respectively. For comparison the ruthenium complex [Ru(Br-dmbpy)(bpy)2](PF6)2 also was coupled to Cys-102 of yeast cytochrome c (22).
The midpoint potentials of the heme group in heme-Ru-MOP2 and heme-Ru-MOP3 at 5 μM in 50 mM sodium phosphate buffer, pH 7, 100 mM NaCl were determined by redox potentiometry (19, 23). CD spectra of the peptides were recorded at 20°C on a Mark V dichrograph from ISA Jobin-Yvon (Longjumeau, France) with an optical path length of 1 mm. Three spectra were averaged.
Kinetic Measurements.
The samples were excited at 532 nm with a Nd:YAG laser (Spektrum, Berlin) producing pulses of 115 mJ and 5-ns duration. The measuring light source with a 150 W Xenon arc lamp (Osram, Berlin) was boosted for 5 ms to about 500 W. The light beam was passed through a Calflex C filter and a 440-nm interference filter with 60 nm full width at half maximum and limited to 10 ms by an optical shutter (Vincent Associates, Rochester, NY). The light was focused through the sample on the entrance slit of a f/2 monochromator (Jobin-Yvon) set to 5 nm width and detected by a Silicon PIN photodiode (Hamamatsu, Middlesex, NJ). Phosphorescence was measured at 630 nm with the measuring light turned off. The rise time (10/90) of the system was 15 ns. The signals were digitized into 1,000 data points by a 1-GHz digital oscilloscope (Tektronix). Data acquisition was controlled by a Hewlett Packard 9000 series 700 VXI UNIX system. Trigger electronics and software were of local design. Fifty signals were averaged with a repetition rate of 5 Hz. The samples containing 10 μM peptide in 100 mM phosphate buffer, pH 7, were kept under argon at 25°C during the measurements. The kinetics were analyzed with the program gnuplot (Gnu software) based on an exponential decay (24) of the metal-to-ligand and charge transfer (MLCT) triplet state RuII* formed immediately after excitation (25). The MLCT triplet state decays by the parallel processes of phosphorescence, radiationless deactivations, and ET to the oxidized heme, RuII*-heme(FeIII) → RuIII-heme(FeII). The heme is reoxidized by the backreaction, RuIII-heme(FeII) → RuII-heme(FeIII). For analysis the following extinction coefficients of the Ru(bpy)3 complex have been used: ɛ452 (RuII) = 15.7 mM−1⋅cm−1, ɛ452 (RuII*) = 2.1 mM−1⋅cm−1 (26), and ɛ420 (RuIII) = 3.3 mM−1⋅cm−1 (27). ET pathway analysis was performed by using the algorithm of Betts et al. (28) with the parameter set of Beratan et al. (29), which defines factors approximating the ability to couple ET via covalent and hydrogen bonds and through-space jumps at van der Waals interactions. Hydrogen bonds were identified by the program insight ii (Molecular Simulations, Waltham, MA).
RESULTS AND DISCUSSION
Protein Design.
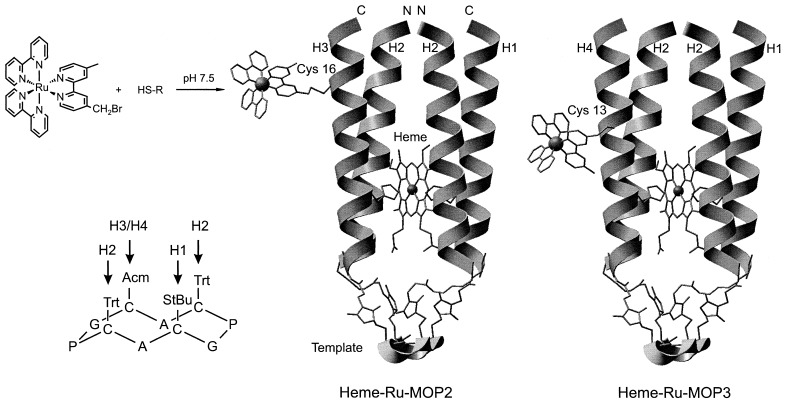
The complex de novo design of the two antiparallel four-helix bundle proteins takes advantage of assembling different helices on a template. For simplicity these modular proteins are termed MOP2 and MOP3. Our previous design of a bis-heme binding cytochrome b (19) based on a template with pairs of two diagonally located coupling sites, termed MOP1, is extended to three different coupling sites that can be selectively addressed. The single heme binding pocket in the hydrophobic interior adopts some of the features of one of the two hemes in cytochrome b. A length of the helices of 21 amino acids has been chosen to ensure a sufficient stabilization by association of the hydrophobic core of the four-helix bundle. It is also long enough to allow a variability of the attachment site for the ruthenium complex introduced at the hydrophilic side of helices H3 and H4. His-7 at the hydrophobic face of one of these helices and at that of H1 serve as the ligand for the iron of the heme group. The two helices H2 are oriented antiparallel to H1 and H3/H4 and shield the binding pocket from the solvent. The modular proteins MOP2 and MOP3 are assembled in the last step by binding Mp-H3 and Mp-H4, respectively, to the same template with three attached helices T[Sp-H1][Sp-H2]2. The Arg residue in H1 provides a single positive charge near the intended position of the propionate groups of the heme. The protected Cys-16 and Cys-13 in the hydrophilic face of H3 and H4, respectively, are positioned at different distances relative to His-7 and the heme. After deprotection they serve as attachment sites for the bromomethyl group of the ruthenium complex by selective thioether formation. Features to stabilize the helical conformation (30) and association of the helix bundle are (i) association of the hydrophobic faces of the four amphiphilic helices, (ii) antiparallel orientation of the helices, (iii) N termini of shielding helices acetylated, C termini amidated, (iv) salt bridges between Glu and Lys spaced by four positions at the hydrophilic outer surface of the helices, (v) Asn at the N-terminal end of a helix as N-cap, i.e., forming H-bonds with the NH of the backbone, (vi) branched structure of the synthetic four-helix bundle protein fixed to the template, (vii) cyclic peptide template, and (viii) binding of the heme group. A schematic representation of this design is shown in Fig. 1.
Figure 1.
Structural models of the de novo proteins heme-Ru-MOP2 (Middle) and heme-Ru-MOP3 (Right), coupling of the bromomethylated Ru-complex and protection chemistry of the cyclic template (Left). The backbone of the cyclic template and of the helices is shown as ribbon. Amino acid side chains are shown to illustrate the thioether linkage of the four Cys of the template with the Sp moiety formed by addition of SH to the Mp group at the N-terminal Gly of H1, H3, and H4 and the C-terminal Lys of H2. The His residues and the bound heme group are shown as sticks. N and C indicate the N and C terminus, respectively, of the helices. The program insight ii was used.
Synthesis.
The template-assembled synthetic proteins with an antiparallel four-helix bundle, Ru-MOP2 and Ru-MOP3, were synthesized according to the following principles.
Synthesis of the building blocks (modules).
(i) Synthesis of the helical peptides was achieved by automated solid-phase peptide synthesis and modification with 3-maleimidopropionic acid for chemoselective coupling of the purified and deprotected peptides with cysteine thiol groups. Antiparallel orientation of the helices was achieved by coupling the Mp group either to the N terminus or the ɛ-amino group of a C-terminal lysine. (ii) The cyclic decapeptide template was manually synthesized with four cysteins with three different protecting groups, which allow to address specific cysteine residues. (iii) The bromomethylated ruthenium complex was synthesized for chemoselective coupling to cysteine.
Assembly of the four-helix bundle.
The four-helix bundle was assembled by consecutive deprotection of the cysteine residues of the template and almost quantitative chemoselective coupling with the helices in solution. The protecting groups StBu, Trt, and Acm are cleavable with high selectivity in the given sequence. The template with the three constant helices was synthesized in stock. With the fourth helix a further cysteine residue was introduced as coupling site for the ruthenium complex. The protection of this cysteine is necessary during the synthesis of the protein and its characterization without the ruthenium complex to avoid disulfide formation.
Introduction of the cofactors.
After deprotection of the helical cysteine the ruthenium complex was coupled to form Ru-MOP2 and Ru-MOP3. Finally, the heme group was incorporated. Alternatively, the heme group was bound to the proteins with protected cysteine as reference. We have used both the StBu and the Acm groups to protect the cysteine inserted for binding the ruthenium complex to helices H3 and H4, respectively. The advantage of the StBu group is its easy removal by reduction of the S-S bond but may interfere with the redox titration. The very stable Acm group is more hydrophilic than the StBu group and is of advantage at the hydrophilic face of the helix for studies in the absence of the ruthenium complex.
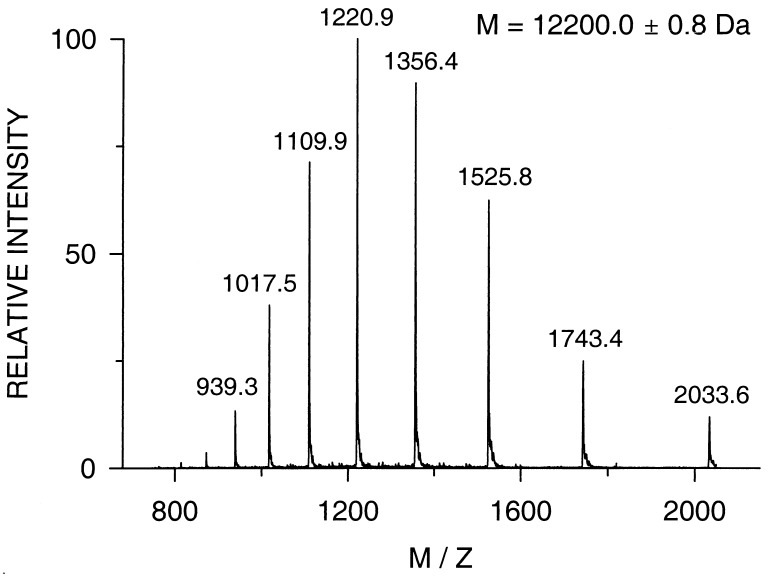
The masses of the intermediate products determined during the synthesis are compared with the calculated values in Table 1. The mass spectrum of Ru-MOP2 is shown in Fig. 2. The detected mass of 12,200.0 ± 0.8 Da verifies the correct primary structure, and the spectrum shows the high purity of the protein.
Table 1.
Molecular masses of intermediate and final products determined during synthesis and assembly of the proteins
| Peptide | Calculated average mass | Experimental mass |
|---|---|---|
| T(StBu)(Trt)2(Acm) | 1,507.0 | 1,505.7 |
| T(Trt)2(Acm) | 1,418.8 | 1,417.8 |
| Mp-H1 | 2,718.1 | 2,717.9 ± 0.2 |
| Mp-H2 | 2,740.3 | 2,739.7 ± 0.1 |
| Mp-H3(StBu) | 2,630.2 | 2,629.9 ± 1.0 |
| Mp-H4(Acm) | 2,613.1 | 2,612.9 ± 0.1 |
| T(Trt)2(Acm)[Sp-H1] | 4,136.9 | 4,136.5 ± 0.3 |
| T(Acm)[Sp-H1] | 3,652.3 | 3,651.8 ± 0.3 |
| T(Acm)[Sp-H1][Sp-H2]2 | 9,132.9 | 9,131.7 ± 0.5 |
| T[Sp-H1][Sp-H2]2 | 9,061.8 | 9,062 ± 0.6 |
| T[Sp-H1][Sp-H2]2[Sp-H3(StBu)] | 11,692.0 | 11,691.2 ± 0.5 |
| T[Sp-H1][Sp-H2]2[Sp-H4(Acm)] | 11,674.9 | 11,674.6 ± 1.0 |
| T[Sp-H1][Sp-H2]2[Sp-H3] = MOP2 | 11,603.8 | 11,602 ± 1.1 |
| T[Sp-H1][Sp-H2]2[Sp-H4] = MOP3 | 11,602.5 ± 0.9 | |
| T[Sp-H1][Sp-H2]2[Sp-H3-Ru] = Ru-MOP2 | 12,199.5 | 12,200.0 ± 0.8 |
| T[Sp-H1][Sp-H2]2[Sp-H4-Ru] = Ru-MOP3 | 12,201.2 ± 1.0 | |
| Ru-cytochrome c | 13,306.2 | 13,304.5 ± 1.4 |
Figure 2.
Electrospray ionization mass spectrum of the purified modular protein Ru-MOP2. The spectrum reveals the high purity and that the protein with a calculated mass of 12,199.5 Da possesses the correct primary structure.
Cofactor Binding.
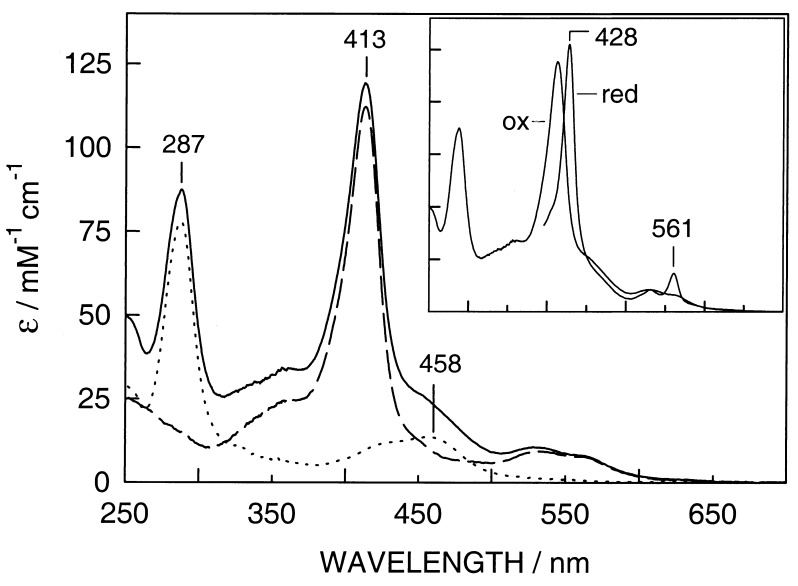
Heme in dimethyl sulfoxide added to an aqueous solution of the peptides is spontaneously incorporated. Fig. 3 shows the UV-visible spectra of Ru-MOP2 without heme, heme-MOP2, and heme-Ru-MOP2 with oxidized heme. The normalized spectrum of heme-Ru-MOP2 is approximately the sum of the spectra of Ru-MOP2 and heme-MOP2, which indicates that there is no major effect of the ruthenium complex on the spectrum of the bound heme group. The oxidized heme complexes display a characteristic Soret band at 413 nm and a broad band around 532 nm. Reduction of heme-Ru-MOP2 leads to a characteristic red shift of the Soret or γ-band to 428 nm and to distinct α and β bands at 530 and 561 nm, respectively (Fig. 3, Inset), which are typical for bis-histidine ligated b-type cytochromes. Very similar spectra were observed with heme-Ru-MOP3 (not shown).
Figure 3.
UV-visible spectra of Ru-MOP2 (dotted line), heme-MOP2 (broken line), and heme-Ru-MOP2 (solid line) with oxidized heme. (Inset) The spectrum of heme-Ru-MOP2 with reduced heme in addition to that with oxidized heme. The heme group was reduced with a minimum amount of solid sodium dithionite.
Size Exclusion Chromatography.
The apparent molecular mass in solution and a possible association of the modular peptides was analyzed by size exclusion chromatography. The elution times of MOP2 and heme-MOP2 indicate an apparent molecular mass of 13.8 and 14.2 kDa, respectively (data not shown); the respective values of MOP3 are 13.6 and 13.3 kDa. These proteins elute in highly symmetrical size exclusion chromatograms, which indicate homogeneous samples. Binding of the ruthenium complex causes a considerable delay of the elution and an asymmetric peak indicating interactions of the ruthenium complex with Sephadex. Aggregates were not observed in any of the peptide samples.
CD and Stability.
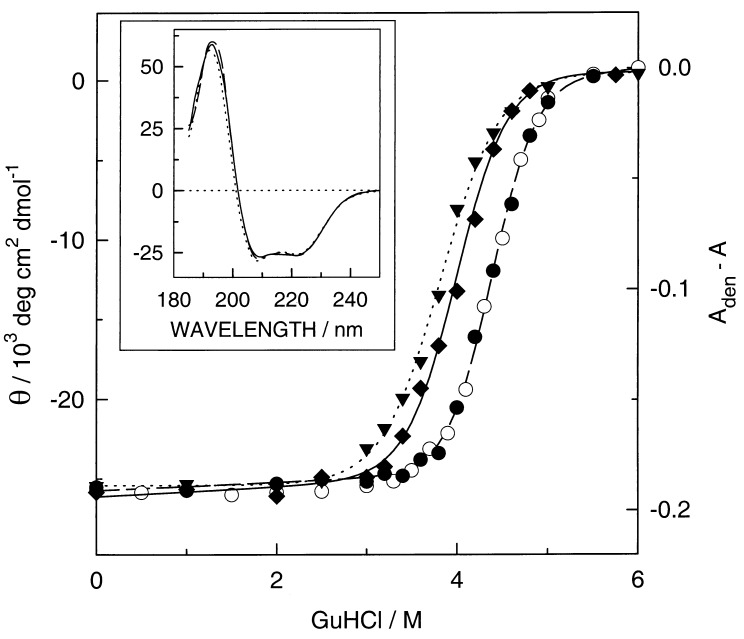
Fig. 4 (Inset) shows the CD spectra of MOP2, heme-MOP2, and heme-Ru-MOP2 with heme in the oxidized state. The spectra are almost identical with a mean residue molar ellipticity at 222 nm of −26,000 deg cm2⋅dmol−1. From this value a helicity of about 75% or 81% can be estimated whether 100% helicity corresponds to a value of −34,700 deg cm2⋅dmol−1 as calculated from the formula of Chen et al. (31) or of −32,200 deg cm2⋅dmol−1 as determined for tropomyosin (32), respectively. If all residues fold as modeled in Fig. 1 we would expect a helicity of 87%. The high α-helix content is not affected by the attachment of the rather large Ru(bpy)3 group.
Figure 4.
Guanidine hydrochloride (GuHCl) induced unfolding of the empty MOP2 (▾), heme-Ru-MOP2 (⧫), and heme-MOP2 (•) at 14 μM monitored by CD at 222 nm and of heme-MOP2 at 3.5 μM monitored by the absorbance at 413 nm (○) at 20°C with heme always in the oxidized state. The mean residue molar ellipticity Θ was calculated by dividing the measured ellipticity by the number of 94 residues. The lines are based on the parameters obtained by a nonlinear least-squares fit of the data points to the two-state transition model (33). For the visible region the absorbance difference at 413 nm of the fully denaturated sample minus that of the sample is plotted as a function of the GuHCl concentration. The denaturing experiment was performed as described (19). (Inset) The CD as a function of the wavelength of MOP2 (dotted line), heme-MOP2 (broken line), and heme-Ru-MOP2 (solid line) in 20 mM potassium phosphate, pH 7, at 0 M GuHCl.
To assess the stability of MOP2 and to determine the effects of the heme group and the ruthenium complex on the folding of the peptide, we have measured the molar ellipticity of MOP2, oxidized heme-MOP2, and heme-Ru-MOP2 at 222 nm as a function of guanidine hydrochloride concentration. In addition, the binding of the oxidized heme was monitored by measuring the absorbance at 413 nm. CD detects an integral value of the protein, whereas the latter indicates the ligation of the heme group. The superposition of both curves suggests a rather large cooperativity during the folding process. The free energies of folding in the absence of denaturant (ΔGH2O) (33) at 20°C are −29, −44, and −36 kJ/mol for MOP2, heme-MOP2, and heme-Ru-MOP2, respectively. Thus, binding of the heme stabilizes the structure of MOP2 by 15 kJ/mol, whereas the ruthenium complex destabilizes heme-MOP2 by 8 kJ/mol. For the respective values of m we found −7.6, −8.8, and −10 kJmol−1⋅M−1. The ligation of the heme increases the stability of the apoprotein by 15 kJ/mol as compared with 8.3 kJ/mol per heme in our previous bis-heme protein (19). The stability of heme-Ru-MOP3 was determined by measuring the absorbance at 413 nm as a function of guanidine hydrochloride (data not shown). For ΔGH2O a value of −35 kJ/mol (m = −9.2 kJ⋅mol−1⋅M−1) was determined, very similar to the values of heme-Ru-MOP2.
Redox Potential.
The electrochemical midpoint potentials of heme-Ru-MOP2 and heme-Ru-MOP3 were determined by monitoring the absorbance in the α-band at 561 nm as a function of the ambient redox potential (data not shown). The analysis of three titrations showed a midpoint potential of −170 mV vs. standard hydrogen electrode (SD ± 6 mV) for the heme group in heme-Ru-MOP2. For heme-Ru-MOP3 a midpoint potential of −164 mV was determined.
ET.
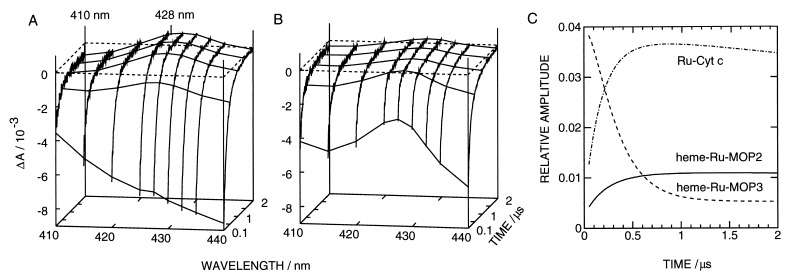
The absorbance changes of heme-Ru-MOP2, heme-Ru-MOP3, and Ru-Cyt c have been measured as a function of time at wavelengths between 410 and 440 nm from 50 ns to 8 μs after laser excitation. Fig. 5 A and B shows these kinetics for the two synthetic proteins from 0.1 to 2 μs. Transient spectra are plotted at several time points. At 2 μs the spectra of both proteins correspond to the difference spectrum of reduced minus oxidized heme (compare to Fig. 3) plus that of RuIII minus RuII (27), with extrema at 410 and 428 nm. These data show the occurrence of ET in both proteins. However, at 100 ns the spectra are significantly different. Although that of heme-Ru-MOP2 is nearly identical to the difference spectrum of the excited state, RuII* minus RuII (26), with heme-Ru-MOP3 the spectrum of reduced heme appears at a larger amplitude than at 2 μs. The ET in heme-Ru-MOP3 must be faster than in heme-Ru-MOP2, which is in agreement with the distances between heme and the ruthenium complex. The time course measured with Ru-Cyt c (not shown) was similar to the one of heme-Ru-MOP2. An analysis showed complex kinetics for all three proteins, which made a direct determination of rate constants difficult.
Figure 5.
Laser-induced absorbance changes of heme-Ru-MOP2 (A) and heme-Ru-MOP3 (B) at wavelengths between 410 and 440 nm. The transient absorbance spectra at 0.1, 0.5, 1, 1.5, and 2 μs are visualized by connecting lines through kinetics obtained by a global fit of the kinetics. The kinetics were obscured during the initial 50 ns by a laser-induced artifact. The extrema of the spectrum at 2 μs at 410 and 428 nm are labeled. (C) The fraction of reduced heme as a function of time relative to the amount of RuII* formed by laser excitation. The latter was determined from the amplitude by extrapolation to 0 s and the extinction coefficients. Heme-Ru-MOP2, solid line; heme-Ru-MOP3, broken line; Ru-Cyt c, dash-dotted line.
The transient absorptions of the synthetic proteins and Ru-Cyt c were followed with a slow time base at 430 and 420 nm, respectively, to detect the slow recombination RuIII-heme(FeII) → RuII-heme(FeIII). A fit with a single exponential in the range from 5 to 80 μs yielded rate constants of 0.02, 0.07, and 0.06 μs−1 for heme-Ru-MOP2, heme-Ru-MOP3, and Ru-Cyt c, respectively. We also have measured the phosphorescence decay of all proteins at 630 nm to determine the quenching of the metal-to-ligand charge transfer triplet state of ruthenium (not shown). The kinetics could be detected at times ≥50 ns after laser excitation and showed a two-exponential decay in all samples with rate constants of 4.8 and 2.2, 9.5 and 1.9, and 6.2 and 2.7 μs−1 for heme-Ru-MOP2, heme-Ru-MOP3, and Ru-Cyt c, respectively. The respective ratios of the amplitudes were 1:0.22, 1:0.34, and 1:0.22. A different approach was followed to determine the amount of reduced heme as a function of time. To improve the signal-to-noise ratio we used a global fit between 50 ns and 4 μs to a sum of three exponentials with wavelength independent rate constants for heme-Ru-MOP2 and Ru-Cyt c. Two of the rate constants were those determined from the phosphorescence decay with their corresponding amplitude ratios, the third one that of the slow reoxidation. For heme-Ru-MOP3 a fourth exponential was needed with a rate constant of 3.6 μs−1. None of the residuals showed a deviation larger than noise. By using these smoothed data, the overall spectra could be deconvoluted at each point in time into the spectrum of RuII* minus RuII and that of FeII minus FeIII plus RuIII minus RuII. The amplitude of the second one as a function of time is shown in Fig. 5C for all proteins. Heme Ru-MOP2 and Ru-Cyt c show very similar reduction kinetics except for a three times higher amplitude of the latter. This factor between the amplitudes and the very similar phosphorescence decay in these proteins indicate that the rate constant of the ET ket is about three times higher in Ru-Cyt c than in heme-Ru-MOP2. This conclusion is drawn from the deactivation of an excited state by parallel processes with a rate constant representing the sum Σk over all rate constants of these processes including ket. The fraction of total RuII* oxidized by ET or the relative amount of reduced heme is given by the fraction ket/Σk.
For heme-Ru-MOP3 the global fit yielded a rather fast reoxidation of heme in Fig. 5C. This finding indicates that most of the heme has been reduced within the first 50 ns. It also implies that a fraction of the phosphorescence decay escaped detection. By comparing the extrapolated amplitudes of RuII* of the two synthetic proteins we conclude that the fast ET does not occur in a fraction larger than 15% of total heme-Ru-MOP3. This fraction is likely to contribute the fast portion of the reoxidation with the rate constant of 3.6 μs−1. This value is two orders of magnitude larger than those of the other proteins.
The Ru(bpy)3 complex bound to a cysteine should be able to rotate as rapidly as shown for cysteine-bound eosine (34). It is of note that we found no substantial difference in all kinetic measurements between Ru-Cyt c and heme-Ru-MOP2. For a qualitative comparison the ET through the helix carrying the ruthenium complex and the heme-binding histidine we have followed the procedure of Beratan et al. (35) to estimate possible ET pathways. From the Cα-atom of the Cys in MOP2 and MOP3 to the iron center we estimate tunneling lengths through the helix of 27 and 23 Å, respectively. Because the driving force and the reorganization energy should be very similar in both proteins, the difference of 4 Å indicates a factor of about 20 between the rate constants, if we assume a drop of the rate constant by an order of magnitude for every 3.15 Å increase in tunneling length (36). This finding is reasonably consistent with the measured ET rates. In the model of heme-Ru-MOP3, there is a conformation of the ruthenium complex with a short pathway to the heme after a through space jump from a bipyridine ligand. Such a conformation could be stable and give rise to fast ET in a fraction of the molecules. Future work will use a shorter linkage to the ruthenium complex to reduce the range of possible conformations in an attempt to simplify the ET kinetics.
CONCLUSIONS
De novo metalloproteins can be synthesized in high purity by well-defined assembly of peptide modules on a template. The variation of one helix in the complex structure of the antiparallel four-helix bundle demonstrates that modular assembly can be extended to a series of synthetic proteins. The introduction of an additional selectively addressable group with a helix, which made it possible to link a ruthenium complex, could be used to assemble even more complex structures. For studies of ET across different distances it is an advantage if both metal centers are bound to the same helix being stabilized within the template-based four-helix bundle. Extra redox groups also can be linked to the hydrophobic interior of the assembled proteins.
Acknowledgments
We thank Patric Hörth for the measurements of the mass spectra and Rosemarie Loyal for technical assistance. We also thank Hugo Scheer for help with the CD measurements. Support by the Volkswagen Stiftung and BMBF/BEO22/0311151 is gratefully acknowledged.
ABBREVIATIONS
- Acm
acetamidomethyl
- bpy
2,2′-bipyridine
- Br-dmbpy
4-bromomethyl-4′-methyl-2,2′-bipyridine
- ET
electron transfer
- Mp
3-maleimidopropionyl
- Sp
3-succinimidopropionyl
- StBu
t-butylthio
- TFA
trifluoroacetic acid
- Trt
trityl
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. Dawson P E, Muir T W, Clark-Lewis I, Kent S B H. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 2.Ghadiri M R, Soares C, Choi C. J Am Chem Soc. 1992;114:825–831. [Google Scholar]
- 3.Sasaki T, Kaiser E T. J Am Chem Soc. 1989;111:380–381. [Google Scholar]
- 4.Åkerfeld K S, DeGrado W F. Tetrahedron. 1994;35:4489–4492. [Google Scholar]
- 5.Mutter M, Altmann E, Altmann K-H, Hersperger R, Koziej P, Nebel K, Tuchscherer G, Vuilleumier S, Gremlich H U, Müller K. Helv Chim Acta. 1988;71:835–847. [Google Scholar]
- 6.Dumy P, Eggleston I M, Cervigni S, Sila U, Sun X, Mutter M. Tetrahedron. 1995;36:1255–1258. [Google Scholar]
- 7.Gibney B R, Johansson J S, Rabanal F, Skalicky J L, Wand A J, Dutton P L. Biochemistry. 1997;36:2798–2806. doi: 10.1021/bi9618225. [DOI] [PubMed] [Google Scholar]
- 8.Mutter M, Vuilleumier S. Angew Chem Int Ed Engl. 1989;28:535–554. [Google Scholar]
- 9.Choma C T, Lear J D, Nelson M J, Dutton P L, Robertson D E, DeGrado W F. J Am Chem Soc. 1994;116:856–865. [Google Scholar]
- 10.Robertson D E, Farid R S, Moser C C, Urbauer J L, Mulholland S E, Pidikiti R, Lear J D, Wand A J, DeGrado W F, Dutton P L. Nature (London) 1994;368:425–432. doi: 10.1038/368425a0. [DOI] [PubMed] [Google Scholar]
- 11.McCafferty D G, Friesen D A, Danielson E, Wall C G, Saderholm M J, Erickson B W, Meyer T J. Proc Natl Acad Sci USA. 1996;93:8200–8204. doi: 10.1073/pnas.93.16.8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozlov G V, Ogawa M Y. J Am Chem Soc. 1997;119:8377–8378. [Google Scholar]
- 13.Mutz M W, McLendon G L, Wishart J F, Gaillard E R, Corin A F. Proc Natl Acad Sci USA. 1996;93:9521–9526. doi: 10.1073/pnas.93.18.9521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray H B, Winkler J R. Annu Rev Biochem. 1996;65:537–561. doi: 10.1146/annurev.bi.65.070196.002541. [DOI] [PubMed] [Google Scholar]
- 15.Marcus R A, Sutin N. Biochim Biophys Acta. 1985;811:265–322. [Google Scholar]
- 16.Moser C C, Dutton P L. Biochim Biophys Acta. 1992;1101:171–176. [PubMed] [Google Scholar]
- 17.Beratan D N, Betts J N, Onuchic J N. Science. 1991;252:1285–1288. doi: 10.1126/science.1656523. [DOI] [PubMed] [Google Scholar]
- 18.Rau H K, Haehnel W. Ber Bunsenges Phys Chem. 1996;100:2052–2056. [Google Scholar]
- 19.Rau H K, Haehnel W. J Am Chem Soc. 1998;120:468–476. [Google Scholar]
- 20.Atherton E, Sheppard R, Ward P. J Chem Soc Perkin Trans. 1985;I:2065–2073. [Google Scholar]
- 21.Hitchcock, P. B., Seddon, K. R., Turp, J. E., Yousif, Y. Z., Zora, J. A., Constable, E. C. & Wernberg, O. (1988) J. Chem. Soc. Dalton Trans. 1837–1842.
- 22.Geren L, Seung H, Durham B, Millet F. Biochemistry. 1991;30:9450–9457. doi: 10.1021/bi00103a009. [DOI] [PubMed] [Google Scholar]
- 23.Dutton P L. Methods Enzymol. 1978;54:411–435. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- 24.Durham B, Ping Pan L, Long J E, Millet F. Biochemistry. 1989;28:8659–8665. doi: 10.1021/bi00447a057. [DOI] [PubMed] [Google Scholar]
- 25.Damrauer N H, Cerullo G, Yeh A, Boussie T R, Shank C V, McCusker J K. Science. 1997;275:54–57. doi: 10.1126/science.275.5296.54. [DOI] [PubMed] [Google Scholar]
- 26.Lachish U. Chem Phys Lett. 1979;62:317–319. [Google Scholar]
- 27.Kalyanasundaram K. Coord Chem Rev. 1982;46:159–244. [Google Scholar]
- 28.Betts J N, Beratan D N, Onuchic J N. J Am Chem Soc. 1992;114:4043–4046. [Google Scholar]
- 29.Beratan D N, Betts J N, Onuchic J N. J Phys Chem. 1992;96:28522–2855. [Google Scholar]
- 30.Bryson J W, Betz S F, Lu H S, Suich D J, Zhou H X, O’Neil K T, DeGrado W F. Science. 1995;270:935–941. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y-H, Yang J T, Chau K H. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 32.Lau S Y M, Taneja A K, Hodges R S. J Biol Chem. 1984;259:13253–13261. [PubMed] [Google Scholar]
- 33.Santoro M M, Bolen D W. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 34.Sabbert D, Junge W. Proc Natl Acad Sci USA. 1997;94:2312–2317. doi: 10.1073/pnas.94.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beratan D N, Onuchic J N, Betts J N, Bowler B E, Gray H B. J Am Chem Soc. 1990;112:7915–7921. [Google Scholar]
- 36.Casimiro D R, Richards J H, Winkler J R, Gray H B. J Phys Chem. 1993;97:13073–13077. [Google Scholar]