Abstract
The excision of the staphylococcal chromosomal cassette mec (SCCmec) from methicillin-resistant Staphylococcus aureus (MRSA) strains results in methicillin-susceptible S. aureus (MSSA) strains. In order to determine the proportion and diversity of multidrug-resistant MSSA (MR-MSSA) strains derived from MRSA strains, 247 mecA-negative isolates recovered in 60 French hospitals between 2002 and 2004 were characterized. The spa types of all strains were determined, and a subset of the strains (n = 30) was further genotyped by multilocus sequence typing. The IDI-MRSA assay was used to test the isolates for the presence of the SCCmec element, which was detected in 68% of all isolates analyzed. Molecular analysis of the samples suggested that 92% of the MR-MSSA isolates were derived from MRSA clones of diverse genetic backgrounds, of which the clone of sequence type 8 and SCCmec type IVA accounted for most of the samples. High variations in incidence data and differences in the molecular characteristics of the isolates from one hospital to another indicate that the emergence of MR-MSSA resulted from independent SCCmec excisions from epidemic MRSA isolates, as well as the diffusion of methicillin-susceptible strains after the loss of SCCmec. MR-MSSA could constitute a useful model for the study of the respective genetic and environmental factors involved in the dissemination of S. aureus in hospitals.
In contrast to the vast amounts of data available on the epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) strains, our knowledge of methicillin-susceptible S. aureus (MSSA) strains recovered from hospitalized patients is limited. Nonetheless, interest in MSSA has increased recently since phylogenomic studies have shown that methicillin-resistant staphylococci emerge from susceptible backgrounds after the acquisition of a staphylococcal chromosomal cassette harboring the mecA gene (2, 26, 27, 36, 40). So far, five types of staphylococcal chromosomal cassette mec (SCCmec) have been described among S. aureus isolates, and these types differ by their epidemiological characteristics: strains with cassette types I, II, and III are limited to isolates from hospital-acquired infections, while type V has been reported in some community-acquired strains, and type IV MRSA strains are present in both categories (25, 33, 34).
MRSA is a worldwide problem associated with increased morbidity and mortality among hospitalized patients; therefore, the identification of the factors influencing its diffusion in hospitals is of great concern for the prevention of nosocomial infections (7). Nevertheless, the causes of MRSA propagation are still controversial, and contradictory conclusions have been drawn from different studies (35). On the one hand, pandemic clones belong to a few genetic lineages, and this argues strongly for the existence of intrinsic epidemicity properties encoded by genetic determinants (1, 32, 33, 36). On the other hand, some studies have focused on the acquisition of antibiotic resistance by methicillin-resistant strains along with antibiotic consumption in hospitals to explain the ability of these strains to spread (31, 39). An important challenge is to determine whether genetic elements could be involved in the dramatic spread of epidemic clones or, conversely, if environmental factors, e.g., antibiotic use, are mainly responsible for the propagation of MRSA (30, 41).
Staphylococcal chromosomal cassettes (SCCs) are mobile elements which are able to insert into and to be excised from the chromosome. We have recently shown that SCCmec excisions occurred in an epidemic caused by MRSA clones in French hospitals, resulting in MSSA isolates (11, 13). Deciphering the molecular basis of the MRSA strains and the MRSA-derived MSSA strains would be of great interest for comprehension of the epidemiology of S. aureus.
Excision of the SCC element can be complete or partial, in which some elements are left behind at the attB site, i.e., the integration site of SCCmec in the S. aureus chromosome. Since most hospital-acquired MRSA strains are resistant to multiple drugs and contain determinants of resistance other than mecA, isolates from which SCCmec is excised retain resistance to antibiotics other than β-lactams (11, 13). Therefore, an unusual resistance phenotype of MSSA strains would be a good marker for the detection of such strains. In this study, 247 MSSA isolates found to be resistant to two antibiotic families have been characterized by molecular methods in order to determine the diversity and estimate the proportion of MSSA strains derived from MRSA strains. In order to determine whether these isolates are able to spread, we have also compared the incidence of multidrug-resistant MSSA (MR-MSSA) strains along with their molecular characteristics in different hospitals and their geographical distribution in continental regions of France.
MATERIALS AND METHODS
Bacterial strains.
The antibiotic susceptibility profiles of S. aureus strains in French hospitals were obtained from the French National Observatory for Antibiotic Resistance Epidemiology (ONERBA), which coordinates the surveillance of antibiotic resistance in hospitals and community settings (ONERBA, 2002 [http://www.onerba.org/bin/res]). Since the data from ONERBA indicate that most hospital-acquired MRSA strains are resistant to fluoroquinolones, macrolides, and aminoglycosides, in this study we have defined MR-MSSA strains as isolates resistant to at least two antibiotics belonging to two of the three different antibiotic families other than β-lactams: aminoglycosides (kanamycin, gentamicin, tobramycin), macrolides-lincosamides (erythromycin and lincomycin), and fluoroquinolones. Susceptibility to methicillin was defined according to the recommendations of the Antibiogram Committee of the French Society for Microbiology; i.e., the diameter of the inhibition zone by the diffusion method was ≥27 mm or ≥20 mm around a disk containing 30 μg of cefoxitin or 5 μg of oxacillin, respectively, or the MIC was ≤2 μg/ml for oxacillin, as determined by another method (4).
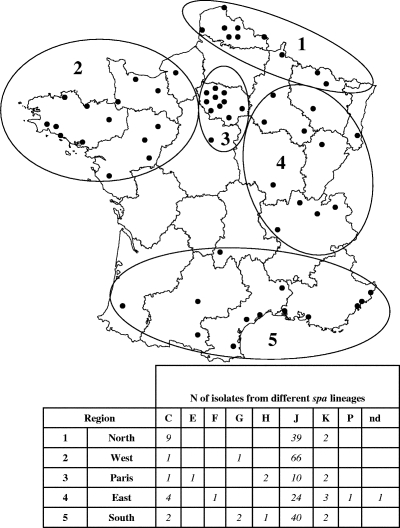
Sixty-two laboratory members of the Collège de Bactériologie-Virologie-Hygiène des Hôpitaux de France network (see Fig. 2) collected S. aureus isolates which were susceptible to methicillin and resistant to the other antibiotics, as defined above. Isolates were forwarded in primary conservation tubes (Bio-Rad, Marnes-la-Coquette, France) to the coordinating laboratory, where the samples were streaked on Trypticase soy agar and stocked in cryobank vials at −80°C. Duplicate conservation tubes were sent to the two other participating laboratories for the independent confirmation of the antibiograms by the diffusion method on Mueller-Hinton agar with Rosco tablets (Rosco Diagnostica, Taastrup, Denmark) (4).
FIG. 2.
Delineation of the regions, the localization of the hospitals, and the distributions of the spa lineages for 215 MR-MSSA isolates isolated in continental France in 2004.
Real-time PCR for detection of mecA gene.
Detection of the mecA gene was performed for 309 S. aureus strains by a real-time PCR mecA MRSA assay with a LightCycler apparatus (Roche Diagnostic, Meylan, France) (3), according to the manufacturer's recommendations. Extraction of DNA from the isolates was performed with an Instagen kit (Bio-Rad). For each run an MSSA strain (strain ATCC 25923) and a clinical MRSA strain (strain 856985) were used as negative and positive controls, respectively (13).
Detection of SCCmec elements.
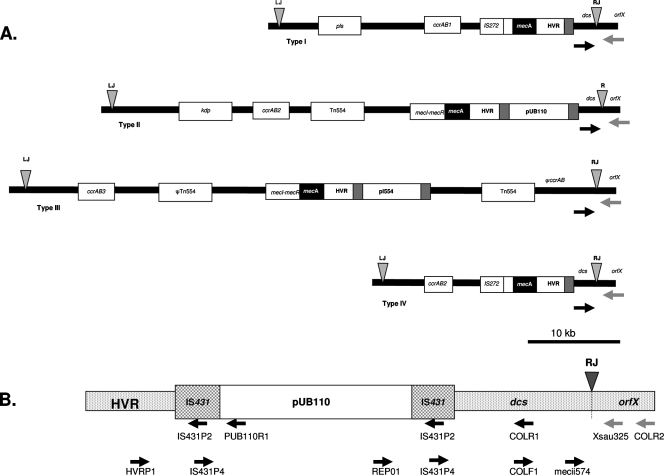
Detection of SCCmec elements inserted in the S. aureus chromosome at the attB site was performed by the IDI-MRSA assay method (Infectio Diagnostic Inc., Sainte-Foy, Quebec, Canada). The IDI-MRSA assay is a qualitative real-time PCR method for the detection of MRSA directly from nasal swabs (22, 23). This test uses five primers specific for the different SCCmec sequences located downstream of the mecA gene combined with primer Xsau325, a primer specific for the S. aureus chromosomal orfX gene, a highly conserved open reading frame flanking the SCCmec integration site, attB. As shown in Fig. 1A, the SCCmec primers have no direct relationship to structural genes, and only a small part of the amplified fragments (40 to 122 nucleotides long) corresponds to SCCmec. The MRSA-specific amplicons are detected by three molecular beacon probes. We have used this test to detect remnant SCCmec elements following partial excision, even when only short DNA fragments were retained.
FIG. 1.
(A) Schematic representation of SCCmec types I to IV (adapted from reference 33 with permission) with the positions of the primers used in the IDI-MRSA assay (black arrows) and primer Xsau325 in orfX (gray arrows) (for more details, see reference 22). (B) Representation of the structure HVR::IS431::pUB110::IS431::dcs linked to orfX in SCCmec types IA, II, and IVA, with the relative positions of the primers used for the characterization of the SCCmec elements in two J-8 MR-MSSA isolates shown.
In this study, the IDI-MRSA method was used directly with the S. aureus colonies to detect SCCmec elements. Briefly, S. aureus isolates were cultured overnight on blood Columbia agar (bioMérieux, Marcy l'Étoile, France), suspended in distilled water, and adjusted to the turbidity of a 1 McFarland standard. The suspensions were heated at 55°C for 15 min in a solution of achromopeptidase (0.05 U/μl; Sigma) and centrifuged (12,000 × g for 3 min). The supernatants were collected and the pellets were discarded. Extracts were stored at 4°C less than 1 h prior to analysis by the IDI-MRSA assay. The PCR assay was processed according to the manufacturer's instructions. The lyophilized master mix was reconstituted with diluent, and the DNA templates were added to the PCR tubes. The PCR was performed with a SmartCycler instrument (Cepheid, Sunnyvale, CA) for 45 min. An internal control was included in each reaction mixture, as described previously (22). Likewise, external positive and negative controls were also included.
Detection of ccr genes coding for recombinases A, B, and C.
PCR was used to detect ccr genes that code for different types of recombinases which act in the integration and excision of SCCmec in the staphylococcal chromosome. The primers used for identification (ccrAB, ccrC) and allotyping for ccrAB (ccrAB1, ccrAB2, ccrAB3) have been described previously (24, 25). DNA extracts from MRSA strains of SCCmec types I (strain COL), II (strain PM64), III (strain HT2004068), and V (strain LIM) (16) were used as controls for ccrAB and ccrAB1, ccrAB2, ccrAB3, and ccrC, respectively. MRSA strains HT2004068, PM64, and LIM were kind gifts from M. Bès and G. Lina (Centre National de Référence des Staphylocoques, Lyon, France), J. A. Lindsay (St George's University, London, United Kingdom), and F. Garnier (CHU, Limoges, France), respectively.
Characterization of SCCmec elements found in MR-MSSA strains.
SCCmec elements from two MR-MSSA isolates positive by the IDI-MRSA assay were characterized by PCR. We first performed five PCRs separately, each of them with one of the five IDI-MRSA assay primers specific for SCCmec: primers mecii574, meciii519, meciv511, mecv492, and mecvii512 (22). Then, a step-by-step amplification method was applied as reported previously (13) in order to identify structures found at the right extremities of SCCmec types I, IA, II, IV, and IVA (Fig. 1B).
Molecular typing.
The spa types of all isolates were determined. Typing was performed as previously described by Shopsin and coworkers (38) with primers spa1113F (5′-TAAAGACGATCCTTCGGTGAGC-3′) and spa1496R (5′-TTTGCTTTTGCAATGTCATTTACTG-3′). The sequences of both strands were determined with the BigDye (version 3.1) cycle sequencing kit and an 3730 XL DNA analyzer (Applera, Courtaboeuf, France), and sequence analysis was carried out with Sequencing Analysis software (version 5.1; Applera, Courtaboeuf, France). The spa types were determined and assigned through the spa-type database (http://tools.egenomics.com and http://www.ridom.de/spaserver).
Multilocus sequence typing (MLST) was performed for a subset of 30 strains selected according to their spa types and IDI-MRSA assay results. MLST was performed as previously described by Enright and coworkers (14) (http://saureus.mlst.net). The sequences of both strands were determined with the BigDye cycle sequencing kit and an ABI 7000 sequencer (Applera). Sequence analysis was carried out with the Sequence Analysis software package (Applera). MLST sequence types (STs) were assigned through the MLST database (www.mlst.net).
Comparison of pairs of MRSA and MR-MSSA strains that originated from the same patients.
Pulsed-field gel electrophoresis (PFGE) has been used to compare pairs of MRSA and MR-MSSA strains recovered from the same patients. Lysis and DNA purification of agarose-embedded bacteria were performed as described by Carles-Nurit and coworkers (5). After DNA restriction by SmaI, a CHEF-II system (Bio-Rad) was used for PFGE, which was performed for 22 h at 6 V/cm, with the switch times ranging from 5 s to 40 s. Pairs of related isolates, i.e., isolates with PFGE restriction profiles that differed by less than four bands, were then compared after Southern blotting and hybridization with a digoxigenin-labeled mecA probe.
Analysis of incidence data.
Data on the incidence of MRSA and MR-MSSA in 21 hospitals over a 9-month period (January to September 2004) were obtained. For each category, the rates were calculated as the number of isolates (one per patient) for 100 patient admissions over the period of the study. To compare hospitals with very different MRSA incidence rates, we used a ratio (PS/R), defined as the number of MR-MSSA isolates per 100 MRSA isolates over the same study period [PS/R = (number of MR-MSSA isolates/number of MRSA isolates) × 100]. For comparison, we postulated that if MR-MSSA isolates emerge randomly and independently from MRSA isolates, the numbers of isolates of each category recovered in a clinical microbiology laboratory would be proportional, with few variations in PS/R values expected from one hospital to another. Conversely, high variations would suggest that outbreaks of MR-MSSA have occurred in some health care facilities.
Study of geographical distribution of MR-MSSA strains.
In order to determine differences in the geographical distribution of MR-MSSA strains, the number of isolates clustered into different spa lineages throughout five regions of France (north, west, Paris, east, and south) were compared for 2004 (Fig. 2).
Statistical analysis.
The chi-square test was used to test the statistical significance of the association of the characteristics of the isolates.
RESULTS
Selection of strains by real-time PCR for mecA and antibiotic resistance patterns.
Three hundred nine strains were forwarded to the coordinating laboratory as MR-MSSA, and of these, 273 isolates were collected prospectively between January and August 2004. In addition, 36 isolates recovered during 2002 and 2003 in seven hospitals were also included in the study. Fifteen pairs of isolates, each pair of which comprised one MR-MSSA strain and one MRSA strain, originating from the same patient were also collected for comparative analysis.
Seventeen isolates determined to be positive for the mecA element by real-time PCR were excluded from further analysis. On the basis of the susceptibility results, 45 isolates which did not fulfill the criteria defined above were also excluded from the study: 11 of them were fully susceptible, while the remaining 34 were resistant to a single antibiotic family.
Finally, 247 MR-MSSA isolates recovered in 60 hospitals (29 in 2002 and 2003, 218 in 2004) were included for further analysis. Ninety-four percent of the isolates were resistant to macrolides, 88% to fluoroquinolones, and 30% to aminoglycosides. Among the MR-MSSA strains, 19 different resistance patterns were found, with 4 of them accounting for 85% of the isolates: 63% were resistant to erythromycin, lincomycin, and fluoroquinolones (ELF pattern); 11% to kanamycin, tobramycin, erythromycin, lincomycin, and fluoroquinolones (KTELF pattern), 6% to kanamycin and erythromycin (KE pattern), and 5% to erythromycin and fluoroquinolones (EF pattern). Eight other resistance patterns comprised only one isolate each; two included two isolates each; two included three isolates each; and each of the last three included four, six, and eight isolates, respectively.
spa typing and MLST results.
All MR-MSSA strains were spa typed by determination of short sequence repeat units. Since these elements are unstable, related spa types were classified into lineages with respect to the short sequence repeat composition and organization, as described previously (2, 37). Eleven lineages were found, of which five comprised at least three isolates each: lineages C, G, H, J, and K (Table 1). The most quantitatively significant lineage consisted of lineage J, which comprised 206 isolates with the spa type pattern YHGFMBQBLO (spa type 1 [according to http://tools.egenomics.com; B. Kreiswirth] or type t008 Ridom [http://www.ridom.de/spaserver]) or a related type. Seventeen lineage J isolates were further typed by MLST: most of them were ST8 (n = 12) or single-locus variants of ST8 (ST770, ST836), and three were ST247 or a single-locus variant of ST247 (ST247SLV). According to eBURST software, all these STs belong to MLST clonal complex 8.
TABLE 1.
Classification and distribution of 247 MR-MSSA isolates by spa lineage with IDI-MRSA results, antibiotic resistance patterns, and MLST results
| No. of strains | spa lineagea |
spa type
|
No. of isolates with the following IDI-MRSA assay result:
|
Antibiotic resistance patternf
|
Corresponding ST or CC (n)g | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PHRIb | Ridomc | N1d | N2e | Positive | Negative | ELF | KTELF | KE | EF | Others | |||
| 1 | B | ZZ2PNGKBKGOLB | t166 | 1 | 1 | 0 | 1 | 1 | NDh | ||||
| 18 | C | TJMBMDMGMK | t002 | 11 | 7 | 5 | 6 | 1 | 6 | 4 | ST5 (3) | ||
| TJJMBMDMGMK | t071 | 2 | 2 | 1 | 1 | 2 | ST125 (1) | ||||||
| TJMBMDMGMKK | t214 | 1 | 1 | 1 | 1 | ST146 (1) | |||||||
| TJMBMDMMMK | 1 | 1 | 1 | 1 | ST5SLVs (2) | ||||||||
| TJMGMK | t062 | 1 | 1 | 1 | 1 | ||||||||
| TM | t2379 | 1 | 1 | 1 | 1 | CC5 (7) | |||||||
| TO2MBMDMBMDMGMK | t041 | 1 | 1 | 1 | 1 | ||||||||
| 1 | E | UJGFGMDMGGM | t148 | 1 | 1 | 1 | 1 | ND | |||||
| 1 | F | I2Z2EGMJH2M | t645 | 1 | 1 | 1 | 1 | ST837 (1) | |||||
| 4 | G | UJFKBPE | t127 | 1 | 1 | 1 | 1 | ST1 (2) | |||||
| UJFK | t2279 | 2 | 2 | 2 | 2 | ||||||||
| UJFMGJAGJ | t796 | 1 | 1 | 1 | 1 | CC1 (2) | |||||||
| 3 | H | A2MBKE | 2 | 1 | 2 | 2 | ST45 (1) | ||||||
| XKAX2BMBMB | t1768 | 1 | 1 | 1 | 1 | CC45 (1) | |||||||
| 206 | J | YHGFMBQBLO | t008 | 151 | 49 | 104 | 47 | 116 | 20 | 2 | 4 | 9 | |
| YHFGFMBQBLO | t051 | 3 | 2 | 3 | 1 | 2 | |||||||
| YFGH3MBQBLO | 1 | 1 | 1 | 1 | ST8 (12) | ||||||||
| YFMBQBLO | t648 | 1 | 1 | 1 | 1 | ST770 (1) | |||||||
| YHGFMBQLO | t1617 | 1 | 1 | 1 | 1 | ST836 (1) | |||||||
| YGFMBOBLO | 2 | 1 | 2 | 2 | |||||||||
| YGFMBQBLO | t024 | 12 | 11 | 7 | 5 | 8 | 3 | 1 | |||||
| YHGCMBQBLO | t064 | 1 | 1 | 1 | 1 | ||||||||
| YHFMBQBLO | t121 | 8 | 7 | 7 | 1 | 5 | 2 | 1 | |||||
| YHGFMBBLO | t400 | 1 | 1 | 1 | 1 | ||||||||
| YHGFMBLO | t622 | 1 | 1 | 1 | 1 | ST247 (2) | |||||||
| YHGFMBOBLO | t2054 | 5 | 4 | 2 | 3 | 4 | 1 | ST247SLV (1) | |||||
| YHGFMBQBM | t1578 | 3 | 3 | 1 | 2 | 3 | |||||||
| YHGFMBQBMO | t801 | 2 | 2 | 1 | 1 | 1 | 1 | ||||||
| YHGFMBQBQBLO | t967 | 1 | 1 | 1 | 1 | ||||||||
| YHGGBQBLO | t574 | 1 | 1 | 1 | 1 | ||||||||
| YHHGGFMBQBLO | 2 | 1 | 1 | 1 | 2 | CC8 (17) | |||||||
| YHHGFMBQBLO | t068 | 1 | 1 | 1 | 1 | ||||||||
| YHBLO | t1259 | 1 | 1 | 1 | 1 | ||||||||
| YMBQBLO | t190 | 1 | 1 | 1 | 1 | ||||||||
| YHBQLO | 1 | 1 | 1 | 1 | |||||||||
| YC2BQBLO | t197 | 1 | 1 | 1 | 1 | ||||||||
| YC2FMBQBLO | t304 | 4 | 3 | 4 | 4 | ||||||||
| ZHGFMBQBLO | 1 | 1 | 1 | 1 | |||||||||
| 9 | K | UKBBGGJAGJ | t760 | 1 | 1 | 1 | 1 | ||||||
| TJGBBGGJAGJ | t491 | 1 | 1 | 1 | 1 | ST15 (1) | |||||||
| UJGGBGGJAGJ | t499 | 7 | 6 | 6 | 1 | 1 | 6 | ||||||
| 2 | P | UJGFMB | t189 | 2 | 2 | 2 | 2 | ST188 (1) | |||||
| 1 | ND | ZDMOB | t441 | 1 | 1 | 1 | 1 | ND | |||||
| 1 | ND | I2GJAABB | t587 | 1 | 1 | 1 | 1 | ND | |||||
| Total | 168 | 79 | 158 | 28 | 13 | 12 | 36 | ||||||
The spa lineage nomenclature has been described previously (2).
spa types according to the Public Health Research Institute nomenclature (PHRI, Newark, NJ).
spa types were obtained from the Ridom spa server (SpaServer.ridom.de).
N1, numbers of isolates of each spa type.
N2, numbers of hospitals where isolates of each spa type have been recovered.
E, erythromycin; L, lincomycin; K, kanamycin; T, tobramycin; F, fluoroquinolones.
Number of isolates of each ST and clonal complex (CC) in the subset of 30 isolates used for ST determination.
ND, not determined.
Lineage C included 18 isolates with the spa type pattern TJMBMDMGMK (spa type 2 Kreiswirth or type t002 Ridom) or a related type (Table 1). Seven isolates of lineage C were typed by MLST to be ST5 (n = 3) or single-locus variants of ST5: ST125, ST146, and two not yet identified STs (ST5SLV1 and ST5SLV2). According to the eBURST database, all these STs belong to MLST clonal complex 5.
The spa-type patterns found that lineages G, H, and K corresponded to MLST ST1, ST45, and ST15, respectively (Table 1). Each cluster was designated by combining the spa lineage and the MLST clonal complex (or the ST when the clonal complex was not defined), e.g., J-8 and K-15.
Antibiotic resistance patterns of different types of MR-MSSA isolates.
A strong association between spa lineage J and resistance patterns ELF and KTELF was found. ELF-KTELF isolates (99%) belonged to spa lineage J, and reciprocally, 90% of spa lineage J isolates had a resistance pattern of ELF or KTELF. Among the 17 lineage J isolates for which the MLST types were determined, all ST8 or single-locus variant isolates (n = 14) had the ELF or the KTELF pattern. In contrast, the three isolates with ST247 or a single-locus variant had a gentamicin-resistant background and resistance pattern KTGELF.
Other spa lineages contained MR-MSSA isolates with 1 of the 15 minor resistance phenotypes; the exceptions were C-5 isolates, which had primarily the KE or the EF resistance pattern.
Detection and characterization of SCCmec elements.
Detection of the SCCmec elements by the IDI-MRSA method, performed with 247 MR-MSSA isolates, identified 169 positive samples (68%). No difference in the rates of positivity was observed between spa lineage J isolates (141/206) and isolates of other spa lineages (28/41). For each one of the five main groups (C-5, G-1, H-45, J-8, and K-15), the test proved positive for at least half of the isolates, including four of four and eight of nine isolates in groups G-1 and K-15, respectively (Table 1).
SCCmec elements were characterized in two representative MR-MSSA isolates (ST8, spa type Ridom t008 or t024) with the respective resistance patterns ELF and KTELF. When PCR was performed with the IDI-MRSA assay primers separately, only primer mecii574 combined with either primer Xsau325 or primer COLR2 allowed amplification in both isolates (Fig. 1B). Since primer mecii574 has been described to identify the sequence at the right extremity of SCCmec in MRSA strains of SCCmec type I, II, or IV (22), we screened the isolates for structures linked to orfX in these types, i.e., HVR::IS431::dcs in types I and IV and HVR::IS431::pUB110::IS431::dcs in types IA, II, and IVA. Characterization by PCR indicated that isolates with the KTELF resistance pattern contained the structure HVR::IS431::pUB110::IS431::dcs, as described previously (13). In contrast, in isolates with the ELF resistance pattern, only the dcs element was detected by using either primer mecii574 or primer COLF1.
PCR for detection of ccr genes coding for recombinases A, B, and C.
SCCs are not restricted to SCCmec: other mobile elements (e.g., SCC476) which do not harbor the mecA gene are able to insert and be excised from the chromosome by the action of the same recombinase enzymes (20). The ccr genes coding for one of the three types of recombinases (A, B, or C) are lost during the partial excision of the SCCmec elements. In order to verify the hypothesis that MR-MSSA strains originate from MRSA strains by the excision of SCCmec and do not harbor another cassette at the attB site, a subset of 14 representative isolates selected from the results of molecular typing and containing SCCmec elements (6 isolates from the J-8 cluster; 4 from the C-5 cluster; and 1 each from clusters G-1, H-45, K-15, and P-188) were tested for the presence of ccr genes by PCR amplification. None was found to be positive for the ccrA, ccrB, or ccrC gene by PCR.
Comparison of PFGE profiles of methicillin-resistant and -susceptible isolates from the same patients.
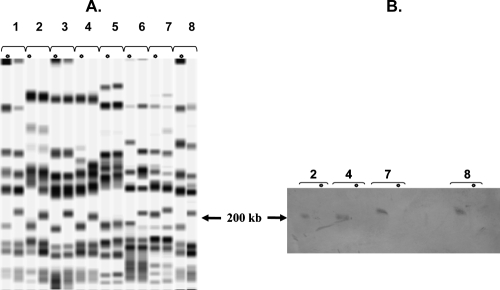
Fifteen pairs of MRSA and MSSA isolates not concomitantly recovered from 15 patients hospitalized in 10 different hospitals were compared by PFGE after restriction by SmaI. All 15 MSSA isolates belonged to the J-8 group. For four pairs, the restriction profiles were different by four or more bands; hence, the susceptible isolate was considered not to be related to the resistant one recovered from the same patient (data not shown). For 11 pairs, the restriction profiles of the MRSA and MSSA isolates were similar and differed by less than four bands, and for 10 of them comparison of the restriction profiles of the MSSA and MRSA isolates showed a loss of DNA fragments with molecular masses estimated to be from 10 kb to 40 kb on the 200-kb band (Fig. 3). Hybridization of the DNA from these 10 methicillin-resistant isolates with a mecA probe after Southern blotting confirmed that the 200-kb band harbors the mecA gene, and thus SCCmec, in J-8 MRSA strains (Fig. 3B).
FIG. 3.
(A) PFGE profiles for eight pairs of MRSA and MR-MSSA strains isolated from the same patients (lane pairs 1 to 8, respectively). Except for the samples from patient 5, the restriction profiles for both isolates originating from each patient are similar but not identical: the MSSA strain loses a DNA fragment at about the 200-kb band, which carries SCCmec in ST8. (B) Four pairs of related MR-MSSA and MRSA isolates after PFGE, Southern blotting, and hybridization with a digoxigenin-labeled mecA probe. The probe hybridizes with the 200-kb band in MRSA but not with bands of smaller sizes in MR-MSSA. Asterisks, lanes with MR-MSSA strains.
Incidence and comparison of molecular characteristics of MR-MSSA strains.
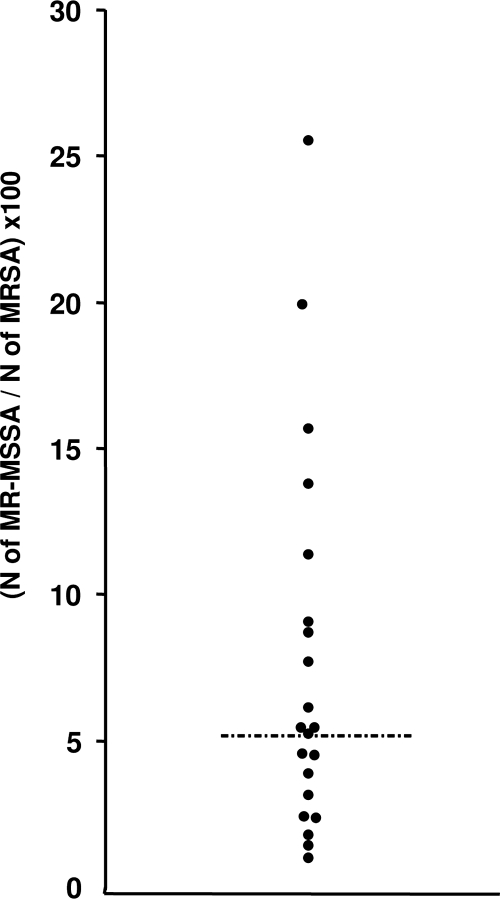
The incidence rates obtained from the 21 hospitals ranged from 0.16 to 1.22 for the MRSA strains and from 0.01 to 0.21 for the MR-MSSA strains. In order to compare hospitals with different MRSA and MSSA incidence rates, we have defined the PS/R ratio (see Materials and Methods), which ranged from 1.43 to 25.57, which had a median value of 5.48, and which did not have a Gaussian distribution. Two times the median value was used as a cutoff level to delimit two groups: 16 hospitals with PS/R values less than 11 and 5 hospitals with values of more than 11 (Fig. 4).
FIG. 4.
Distribution of PS/R ratio values for 21 hospitals. The dashed-dotted line corresponds to the median.
In hospitals with high PS/R values, ST8 isolates were more frequently spa type Ridom t008 than a variant type (P = 0.017), and these isolates more frequently had remnant SCCmec elements in their chromosomes than isolates from other hospitals (85% versus 63% IDI-MRSA assay-positive isolates; P = 0.015).
Geographical distribution of spa types.
There was a statistically significant difference in the distribution of the spa types throughout the five regions (P < 0.001), in part due to the higher than expected incidence of the lineage J isolates in the west and a greater diversity of isolates in regions 3 and 4 (Paris and eastern France, respectively) than in the other regions (Fig. 2).
At least half (n = 9) of the C-5 isolates were recovered from the north, including seven isolates with an EF resistance pattern. In contrast, C-5 isolates from the other regions did not exhibit such a resistance phenotype.
DISCUSSION
The loss of methicillin resistance by the excision of SCCmec in MRSA strains has been reported by many authors (11), but so far this phenomenon has not been investigated on a large scale. To undertake such a study, MRSA-derived isolates first had to be distinguished from other unrelated MSSA strains. In this work, we have selected MSSA strains resistant to at least two antibiotic families other than β-lactams as a selection method. The use of the IDI-MRSA assay to identify MR-MSSA isolates with SCCmec elements among the MSSA strains with uncommon resistance phenotypes led to a 15-fold increase in the proportion of positive isolates (68%). This is in sharp contrast to the 4.6% previously reported among 519 MSSA strains collected worldwide, whereby antibiotic resistance patterns were not taken into account (22). The absence of genes coding for the different recombinase types in representative MR-MSSA strains indicates that elements inserted in the attB site are not complete SCCs deprived of the mecA gene, as described elsewhere (6, 20). Instead, they constitute residual elements of SCCmec resulting from partial excision. According to these results and to our knowledge about the epidemiology of MRSA in France, MR-MSSA strains emerge from the MRSA background, mostly from an ST8-SCCmec type IVA clone, also called the “Lyon” clone (15), which has predominated in French hospitals since 1992 (11, 12, 15, 19, 28, 29). MSSA isolates derived from this MRSA clone may display either the ELF or the KTELF resistance phenotype (11, 13). Strains with the KTELF resistance phenotype result from the partial excision and retention of the HVR::IS431::pUB110::IS431::dcs structure and are positive by the IDI-MRSA method (27/28 tested). The ELF pattern results either from the total excision of SCCmec in 49% of isolates with this resistance phenotype or from a large, but incomplete, excision of SCCmec, including all elements except the downstream constant sequence (dcs). PFGE was a useful tool for comparison of the pairs of methicillin-susceptible and -resistant J-8 isolates, including MSSA isolates either positive or negative by the IDI-MRSA method, and for confirmation of the relationship between MRSA and MR-MSSA strains. For 11 pairs, the restriction profiles for both isolates originating from each patient were similar but not identical: the MR-MSSA isolates differed from the MRSA isolates by a DNA band corresponding to the loss of a fragment ranging in size from 10 to 40 kb on the approximately 200-kb band which carries SCCmec in ST8 MRSA strains, as recently reported by others (17). Consequently, a digoxigenin-labeled probe for the mecA gene hybridized with this fragment in MRSA isolates but not in MR-MSSA isolates (Fig. 3B). On the basis of these findings, 227 MR-MSSA isolates resulted from the loss of SCCmec in MRSA isolates: 168 MR-MSSA isolates (68%) resulted from partial excisions, and among the isolates without remnant SCCmec elements, 59 J-8 S. aureus isolates with the unusual ELF resistance pattern probably derived from MRSA isolates by total excision. It is noteworthy that 68% of the MR-MSSA isolates were IDI-MRSA assay positive and were found in spa lineages that included at least two isolates, suggesting that the partial excision of SCCmec is more common than complete excision and is independent of the genetic background of the MRSA.
The differences in the diversity of the spa types and the geographical distributions of the MR-MSSA isolates observed indicate a greater diversity of MRSA strains in Paris and the northern and eastern regions of France rather than in the western and southern regions. This is probably due to important population movements in Paris and across borders with Germany and Belgium. For instance, half of the C-5 isolates were recovered near the Belgian border, were associated to a particular resistance pattern (pattern EF), and were probably related to an ST5 MRSA clone with same resistance phenotype which is epidemic in Belgian hospitals (9). The transfer of patients between health care facilities in northern France and Belgium is frequent, especially between nursing homes. This transborder movement of strains has been highlighted in recent studies on the international spread of nosocomial pathogens (10).
In order to investigate the capacity of MR-MSSA strains to disseminate, we first compared the respective incidences of MR-MSSA and MRSA isolates and then the molecular characteristics of MR-MSSA isolates from hospitals with different incidences. On the basis of the variations in PS/R ratios, 21 hospitals could be divided into two groups (Fig. 4): a group of 16 hospitals for which PS/R values were less than 11, i.e., double the median value, where only the sporadic and independent emergence of MR-MSSA isolates from MRSA isolates would probably exist, and a second one of 5 hospitals with PS/R values ranging from 11.40 to 25.57 where an epidemic diffusion of MR-MSSA isolates could be suspected. Further investigations are needed to explain the differences from one hospital to another, and more particularly, the evolution of MR-MSSA isolates must be examined along with the consumption of antibiotics other than β-lactams. For instance, in the hospital displaying the highest PS/R value, most isolates of MR-MSSA were recovered from patients hospitalized in a long-term care facility where the level of consumption of fluoroquinolones was very high (A. Le Coustumier, personal communication). The nosocomial diffusion of S. aureus is more common for MRSA isolates than for MSSA isolates, since the latter are usually susceptible to most antibiotics and the widespread use of antibiotics promotes the survival of MRSA strains over MSSA strains in hospitalized patients (21). MR-MSSA isolates are susceptible to oxacillin, amoxicillin-clavulanate, and cephalosporins but are resistant to fluoroquinolones and macrolides, so they might theoretically have a selective advantage in wards where other antibiotics are used preferentially over β-lactams.
Most of the MR-MSSA isolates were derived from the predominant MRSA clone, a clone of ST8 and SCCmec type IVA, but there were significant differences in the molecular characteristics between isolates from hospitals with different PS/R values. The most frequently encountered MR-MRSA isolates with remnant SCCmec elements were of spa type Ridom t008 and were isolated from hospitals with high PS/R values. It is unclear whether this simply reflects the dissemination of MR-MSSA strains which result preferentially from partial excision or whether the insertion of elements at the attB site induces a greater capacity to spread, perhaps by activation of the genes involved in the mechanisms of colonization. Regardless, this argues strongly for the clonal expansion of ST8 MR-MSSA strains which have lost mecA. ST8 strains resistant to penicillin G, tetracyclines, and streptomycin were epidemic in Danish hospitals in the premethicillin era, and the significance of these clones in the evolution of both MSSA and MRSA has been highlighted in recent reports (8, 18, 32, 36). The ST8 lineage appears to be highly epidemic, and related clones are able to spread in hospitals under antibiotic selective pressure.
In conclusion, the emergence of MR-MSSA strains in French hospitals results from independent SCCmec excisions in epidemic MRSA isolates in low-incidence hospitals, as well as the clonal diffusion of methicillin-susceptible strains after the partial loss of SCCmec in high-incidence hospitals. To our knowledge, the epidemic diffusion of MR-MSSA strains in hospitals has not been described since the introduction of methicillin for therapeutic use. MR-MSSA could constitute a useful model for the better determination of the genetic and environmental factors involved in the spread of this major pathogen and the epidemiology of MRSA.
Acknowledgments
We are indebted to Duarte Oliveira and Herminia de Lencastre for the method of characterization of SCCmec elements by PCR. We thank Marie-France Travert, Philippe Gautier, and the staffs of the microbiology laboratories of Cahors and Rennes Hospitals for their excellent technical assistance.
Footnotes
Published ahead of print on 20 August 2007.
REFERENCES
- 1.Aires de Sousa, M., and H. de Lencastre. 2004. Bridges from hospitals to the laboratory: genetic portraits of methicillin-resistant Staphylococcus aureus clones. FEMS Immunol. Med. Microbiol. 40:101-111. [DOI] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., T. Conceicao, C. Simas, and H. De Lencastre. 2005. Comparison of genetic backgrounds of methicillin-resistant and -susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community. J. Clin. Microbiol. 43:5150-5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Haj-Hussein, B. T., M. A. Al-Shehri, E. A. Azhar, I. M. Ashankyty, and A. O. Osoba. 2005. Evaluation of two real-time PCR assays for the investigation of mecA gene in clinical isolates of MRSA in western Saudi Arabia. Saudi Med. J. 26:759-762. [PubMed] [Google Scholar]
- 4.Anonymous. 2006. Communiqué 2006. Comité de l'Antibiogramme de la Société Française de Microbiologie. http://www.sfm.asso.fr.
- 5.Carles-Nurit, M. J., B. Christophle, S. Broche, A. Gouby, N. Bouziges, and M. Ramuz. 1992. DNA polymorphisms in methicillin-susceptible and methicillin-resistant strains of Staphylococcus aureus. J. Clin. Microbiol. 30:2092-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corkill, J. E., J. J. Anson, P. Griffiths, and C. A. Hart. 2004. Detection of elements of the staphylococcal cassette chromosome (SCC) in a methicillin-susceptible (mecA gene negative) homologue of a fucidin-resistant MRSA. J. Antimicrob. Chemother. 54:229-231. [DOI] [PubMed] [Google Scholar]
- 7.Cosgrove, S. E., G. Sakoulas, E. N. Perencevich, M. J. Schwaber, A. W. Karchmer, and Y. Carmeli. 2003. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteraemia: a meta-analysis. Clin. Infect. Dis. 36:53-59. [DOI] [PubMed] [Google Scholar]
- 8.Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denis, O., A. Deplano, C. Nonhoff, R. De Ryck, R. de Mendonca, S. Rottiers, R. Vanhoof, and M. J. Struelens. 2004. National surveillance of methicillin-resistant Staphylococcus aureus in Belgian hospitals indicates rapid diversification of epidemic clones. Antimicrob. Agents Chemother. 48:3625-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deplano, A., W. Witte, W. J. Van Leeuwen, Y. Brun, and M. J. Struelens. 2000. Clonal dissemination of epidemic methicillin-resistant Staphylococcus aureus in Belgium and neighbouring countries. Clin. Microbiol. Infect. 6:239-245. [DOI] [PubMed] [Google Scholar]
- 11.Donnio, P.-Y., L. Louvet, L. Preney, D. Nicolas, J. L. Avril, and L. Desbordes. 2002. Nine-year surveillance of methicillin-resistant Staphylococcus aureus in a hospital suggests instability of mecA DNA region in an epidemic strain. J. Clin. Microbiol. 40:1048-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnio, P.-Y., L. Preney, A.-L. Gautier-Lerestif, J.-L. Avril, and N. Lafforgue. 2004. Changes in staphylococcal cassette chromosome type and antibiotic resistance profile in methicillin-resistant Staphylococcus aureus isolates from a French hospital over an 11 year period. J. Antimicrob. Chemother. 53:808-813. [DOI] [PubMed] [Google Scholar]
- 13.Donnio, P.-Y., D. C. Oliveira, N. A. Faria, N. Wilhelm, A. Le Coustumier, and H. de Lencastre. 2005. Partial excision of the chromosomal cassette containing the methicillin resistance determinant results in methicillin-susceptible Staphylococcus aureus. J. Clin. Microbiol. 43:4191-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferry, T., M. Bes, O. Dauwalder, H. Meugnier, G. Lina, F. Forey, F. Vandenesch, and J. Etienne. 2006. Toxin gene content of the Lyon methicillin-resistant Staphylococcus aureus clone compared with that of other pandemic clones. J. Clin. Microbiol. 44:2642-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garnier, F., A. Tristan, B. Francois, J. Etienne, M. Delage-Corre, C. Martin, N. Liassine, W. Wannet, F. Denis, and M. C. Ploy. 2006. Pneumonia and new methicillin-resistant Staphylococcus aureus clone. Emerg. Infect. Dis. 12:498-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goering, R. V., L. K. McDougal, G. E. Fosheim, K. K. Bonnstetter, D. J. Wolter, and F. C. Tenover. 2007. Epidemiologic distribution of the arginine catabolic mobile element among selected methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates. J. Clin. Microbiol. 45:1981-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomes, A. R., H. Westh, and H. de Lencastre. 2006. Origins and evolution of methicillin-resistant Staphylococcus aureus clonal lineages. Antimicrob. Agents Chemother. 50:3237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gueudet, P., L. Gazagne, E. Lecaillon, A. Le Coustumier, R. Bismuth, and le Collège de Bactériologie, Virologie et Hygiène des Hôpitaux de France. 1997. Emergence of new gentamicin susceptible strains and important decrease in associated resistances in French methicillin-resistant Staphylococcus aureus, abstr. P-E122. Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 20.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooper, D. C. 2002. Fluoroquinolone resistance among gram-positive cocci. Lancet Infect. Dis. 2:530-538. [DOI] [PubMed] [Google Scholar]
- 22.Huletsky, A., R. Giroux, V. Rossbach, M. Gagnon, M. Vaillancourt, M. Bernier, F. Gagnon, K. Truchon, M. Bastien, F. J. Picard, A. van Belkum, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2004. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J. Clin. Microbiol. 42:1875-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huletsky, A., P. Lebel, F. J. Picard, M. Bernier, M. Gagnon, N. Boucher, and M. G. Bergeron. 2005. Identification of methicillin-resistant Staphylococcus aureus carriage in less than 1 hour during a hospital surveillance program. Clin. Infect. Dis. 40:976-981. [DOI] [PubMed] [Google Scholar]
- 24.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Layer, F., B. Ghebremedhin, W. König, and B. König. 2006. Heterogeneity of methicillin-susceptible Staphylococcus aureus strains at a German university hospital implicates the circulating-strain pool as a potential source of emerging methicillin-resistant S. aureus clones. J. Clin. Microbiol. 44:2179-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lelièvre, H., G. Lina, M. E. Jones, C. Olive, F. Forey, M. Roussel-Delvallez, M.-H. Nicolas-Chanoine, C. M. Bébéar, V. Jarlier, A. Andremont, F. Vandenesch, and J. Etienne. 1999. Emergence and spread in French hospitals of methicillin-resistant Staphylococcus aureus with increasing susceptibility to gentamicin and other antibiotics. J. Clin. Microbiol. 37:3452-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemaître, N., W. Sougakoff, A. Masmoudi, M.-H. Fievet, R. Bismuth, and V. Jarlier. 1998. Characterization of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus involved in nosocomial spread. J. Clin. Microbiol. 36:81-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindsay, J. A., and M. T. Holden. 2004. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 12:378-385. [DOI] [PubMed] [Google Scholar]
- 31.Monnet, D. L., F. M. MacKenzie, J. M. Lopez-Lozano, A. Beyaert, M. Camacho, R. Wilson, D. Stuart, and I. M. Gould. 2004. Antimicrobial drug use and methicillin-resistant Staphylococcus aureus, Aberdeen, 1996-2000. Emerg. Infect. Dis. 10:1432-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliveira, D. C., A. Tomasz, and H. De Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 33.Oliveira, D. C., A. Tomasz, and H. De Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 34.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riley, L. W. 2004. Hospital infections: Staphylococcus aureus, p. 249-280. In L. W. Riley (ed.), Molecular epidemiology of infectious diseases: principles and practices. ASM Press, Washington, DC.
- 36.Robinson, D. A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruppitsch, W., A. Indra, A. Stöger, B. Mayer, S. Stadlbauer, G. Wewalka, and F. Allerberger. 2006. Classifying spa types in complexes improves interpretation of typing results for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 44:2442-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shopsin, B., B. Mathema, X. Zhao, J. Martinez, J. Kornblum, and B. N. Kreiswirth. 2000. Resistance rather than virulence selects for the clonal spread of methicillin-resistant Staphylococcus aureus: implications for MRSA transmission. Microb. Drug Resist. 6:239-244. [DOI] [PubMed] [Google Scholar]
- 40.Vivoni, A. M., B. A. Diep, A. C. De Gouveia Magalhaes, K. R. N. Santos, L. W. Riley, G. F. Sensabaugh, and B. M. Moreira. 2006. Clonal composition of Staphylococcus aureus isolates at a Brazilian university hospital: identification of international circulating lineages. J. Clin. Microbiol. 44:1686-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woodford, N., and D. Livermore. 2001. Can we beat MRSA now we know its genome sequence? Lancet Infect. Dis. 1:9-10. [DOI] [PubMed] [Google Scholar]