Abstract
In Pseudomonas aeruginosa, azithromycin has been shown to reduce virulence factor production, to retard biofilm formation, and to exhibit bactericidal effects on stationary-phase cells. In this study we analyzed whether these azithromycin-mediated effects require interaction with the ribosome. We blocked the access of azithromycin to the ribosome in P. aeruginosa PAO1 by expressing the 23S rRNA methylase ErmBP from Clostridium perfringens. Ribosome protection prevented the azithromycin-mediated reduction of elastase and rhamnolipid production, as well as the inhibition of swarming motility. Ribosome protection also prevented the killing of stationary-phase cells, suggesting that the cell-killing effect of azithromycin does not result solely from membrane destabilization. We further show that rhamnolipids are involved in cell killing, probably by increasing the uptake of the hydrophobic azithromycin molecule. These results have important implications for the treatment with azithromycin of patients chronically colonized by P. aeruginosa and might explain the variability in the efficacy of azithromycin treatments.
Pseudomonas aeruginosa frequently colonizes the lungs of patients with cystic fibrosis (CF) or diffuse panbronchiolitis (DPB). Once colonization is established, eradication is unsuccessful, despite repeated antimicrobial therapies. Hence, alternative strategies to classical antimicrobial therapies are needed. The 14- and 15-C macrolides (erythromycin, azithromycin [AZM], and clarithromycin) are not bactericidal against P. aeruginosa but have been found to inhibit virulence factor production in vitro (17, 30), as well as in a murine model (36), and to interfere with biofilm formation (8, 11). Furthermore, macrolides and, in particular, AZM were shown to have immunomodulatory activities on host cells, resulting in a decreased inflammatory response to bacterial stimulations (25). Several clinical studies have now demonstrated the improvement of lung function in CF or DPB patients treated with AZM (5, 24, 35). However, none of those studies has precisely addressed whether this beneficial effect is due primarily to an anti-Pseudomonas effect or an immunomodulatory activity, or both.
While several studies have shown the effect of AZM on quorum sensing (QS)-dependent virulence factor production (30, 31), biofilm formation (8, 11), and cell killing in stationary phase (12), it remains uncertain how these anti-Pseudomonas activities are mediated. Although macrolides inhibit protein synthesis by interfering with the exit of peptides from the ribosomal channel, a recent microarray analysis showed that at subinhibitory concentrations AZM is able to both activate and repress different subsets of genes (20). It is therefore possible that macrolides might have so far uncharacterized nonribosomal targets which could explain their effect on transcription. We therefore used ribosomal protection to analyze the effect of AZM on QS-dependent virulence factor production and cell killing. Our results show that both effects require the interaction of AZM with the ribosome. We further found that stationary-phase killing of AZM is enhanced by the production of rhamnolipids, which probably facilitate the uptake of macrolides.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. All strains were derived from antibiotic-susceptible wild-type strain PT5. Unless otherwise stated, the strains were grown in Luria-Bertani (LB) medium at 37°C with agitation (250 rpm).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype | Reference or source |
|---|---|---|
| Strains | ||
| PT5 (PAO1) | Wild type | Laboratory collection |
| PT1300 | PT5(pEX1.8) Cbr | This study, 22 |
| PT1308 | PT5(pEXERM) Cbr | This study |
| 11B | PAO1 mexY::Tn501 Hgr | 23 |
| 12B | PAO1 mexZ::bla Cbr | P. Plésiat |
| PT712 | PT5 rhlA::Gm Gmr | 19 |
| PT1323 | PT712(pAKRHL) Cbr | This study |
| PT1332 | PT712(pAK1900) Cbr | 14 |
| Plasmids | ||
| pMLS001 | pMMB206 carrying ermBP, Cmr Eryr | 15, K. Poole |
| pEX1.8 | Cloning vector, Cbr | 22 |
| pAK1900 | Cloning vector, Cbr | 14 |
| pEXERM | pEX1.8 carrying ermBP from pMLS001, Cbr | 15 |
| pAKRHL | pAK1900 carrying rhlABRI from pJPP6, Cbr | 4, 14 |
Plasmid pMLS001, which contains the ermBP gene from Clostridium perfringens and which codes for a 23S rRNA methylase, was kindly provided by Keith Poole (Kingston, Ontario, Canada). The ermBP gene was amplified on a 737-bp fragment with primers ermBP-F (5′-GGATCCGGATCCAGAAGGAGTGATTACAGAAC-3′) and ermBP-R (5′-AAGCTTAAGCTTTAGAATTATTTCCTCCCGTTA-3′) (15) under the following PCR conditions: denaturation at 95°C for 1 min, followed by 25 cycles of 95°C for 30 s, 47°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min. The PCR product was digested with BamHI and HindIII and cloned into BamHI- and HindIII-restricted pEX1.8, yielding plasmid pEXERM. The rhlAB genes of P. aeruginosa were cloned into HindIII-SphI-restricted plasmid pAK1900, after the digestion of plasmid pJPP6 with HindIII and SphI, to yield plasmid pAKRHL. The plasmids were transferred into P. aeruginosa by electroporation.
Phenotypic assays.
The elastase activity in the supernatants of cultures grown for 7 h in Pseudomonas broth medium (6) was determined by the elastin Congo red (ECR) assay (32). Briefly, 50 μl of supernatant was added to 0.95 ml of 0.1 M Tris (pH 7.4)-1 mM CaCl2 with an excess of ECR (reaction volume, 4 mg/ml). After 18 h of incubation at 37°C, the samples were centrifuged and the degradation of ECR was measured by determination of the absorbance at 495 nm in the supernatant. Elastase activity is expressed as the ratio of the optical density at 495 nm (OD495)/OD600 of the culture.
The production of rhamnolipid was assayed on SW plates (27). SW is based on M9 minimal medium, in which NH4Cl is replaced by 0.1% glutamate and which is supplemented with 0.2% glucose and MgSO4 (final concentration, 1 mM). The inhibition of rhamnolipid production was estimated on SW plates containing an AZM concentration gradient from 0 to 50 μg/ml. Three-microliter droplets containing ca. 106 CFU were placed at equal distances from each other on the dried plates, which were incubated for 24 h at 37°C and then for 24 h at 25°C.
Swarming motility was tested on 0.5% agar plates, which is based on M9 medium supplemented with 0.2% glucose and 1 mM MgSO4 and in which 0.05% glutamate instead of ammonium chloride is used as the nitrogen source. Three microliters of an overnight LB culture was deposited on the plates, which were incubated for 18 h at 37°C.
Killing assays.
The strains were grown overnight in 2 ml LB medium supplemented, when required, with carbenicillin (100 μg/ml) or gentamicin (15 μg/ml). On the next morning, the strains were inoculated to an OD600 of 0.05 into 25 ml LB medium without antibiotics and then incubated at 37°C. Killing assays were performed with stationary-phase cultures (OD600, 3.0 to 3.5) grown in LB medium. At comparable cell densities, AZM was added to 2-ml aliquots at the concentrations indicated in the text. Incubation was continued for 20 to 22 h at 37°C, and the viable counts were determined by plating serial dilutions on LB agar plates.
RESULTS AND DISCUSSION
AZM, like other macrolides, inhibits protein synthesis by blocking the peptide exit channel of the 50S ribosomal subunit through interaction with the 23S rRNA. We tested whether the observed effects of AZM on virulence factor production require interaction with the ribosome. To address this question, we introduced plasmid pEXERM into our P. aeruginosa wild-type strain, strain PT5. Plasmid pEXERM expresses the 23S rRNA methylase ErmBP from Clostridium perfringens (15). This enzyme methylates the adenine moiety of ribonucleotide 2048 (Escherichia coli numbering) in the 23S rRNA, thereby blocking the access of macrolide antibiotics to the peptide exit channel. The presence of plasmid pEXERM (in strain PT1308) conferred resistance to erythromycin (data not shown) and AZM, permitting growth at concentrations above 1,000 μg/ml (Fig. 1A). As expected, the susceptibility of PT5 to nonmacrolide antimicrobials (ciprofloxacin, aztreonam, amikacin) was not affected by the plasmid (data not shown).
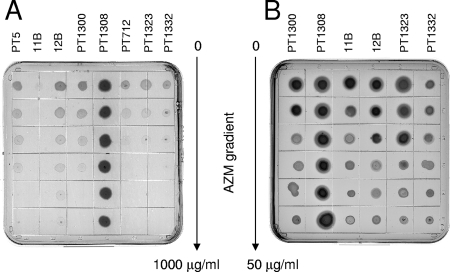
FIG. 1.
Strain susceptibilities to AZM. (A) The strains listed in Table 1 were streaked onto an LB agar plate containing an AZM gradient with a maximal concentration of 1 mg/ml. The plates were incubated at 37°C for 18 h. Note the hypersusceptibility of mexX mutant 11B and the resistance of methylase-carrying strain PT1308. (B) The effect of AZM on rhamnolipid production, visible as halos around the colonies, was compared on SW blue plates containing an AZM gradient with a maximal concentration of 50 μg/ml.
Effect of ribosomal protection on AZM modulation of virulence factor production.
AZM at 2 μg/ml was previously shown to reduce the production of QS-dependent virulence factors, including elastase and rhamnolipids, due to the reduced expression of the autoinducer synthetase genes lasI and rhlI (30, 31). We analyzed rhamnolipid production on SW plates in the presence of an AZM gradient (maximal concentration, 50 μg/ml). While AZM inhibited rhamnolipid production in strain PT1300 at one-third of the gradient plate, strain PT1308 carrying the methylase gene produced rhamnolipids even at AZM concentrations of 50 μg/ml (Fig. 1B). Therefore, AZM has to interact with the ribosome in order to decrease rhamnolipid production. Expression of the MexXY efflux pump is induced by antimicrobials that interfere with the ribosome (chloramphenicol, tetracycline, macrolides) (15). We tested whether the reduction of rhamnolipid production by AZM was affected by the expression level of this efflux pump. As shown in Fig. 1B, rhamnolipid production was inhibited at lower AZM concentrations in AZM-hypersusceptible mexX deletion mutant 11B than in strain 12B, which overexpresses mexXY due to inactivation of the repressor gene mexZ. This observation is of relevance for AZM-treated CF patients, since P. aeruginosa isolates overexpressing the MexXY efflux pump have been reported recently (13, 29, 33). Furthermore, the mexZ gene was one of the major mutational targets in isolates from 29 CF patients (28). Such mutants are expected to require higher AZM concentrations to achieve a reduction of QS-dependent gene expression. This observation could explain the weak effect of AZM in vitro on isolates from chronically infected CF patients (34).
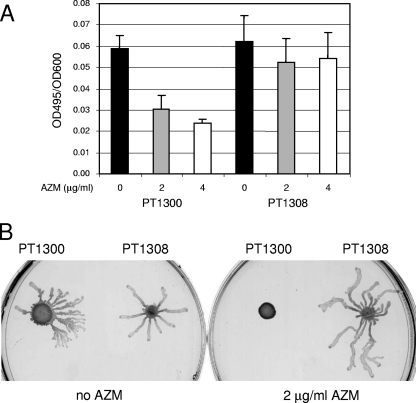
We further tested the effect of ribosome protection on elastase production. While the elastase activity in the supernatants of vector-carrying strain PT1300 showed a dose-dependent decrease in the presence of AZM, the elastase activity in the supernatants of the methylase-carrying strain (PT1308) was not significantly affected by AZM (Fig. 2A).
FIG. 2.
Determination of elastase activity and swarming motility. (A) Strains were grown for 7 h in Pseudomonas broth medium, and the elastase activity in the culture supernatants was determined by the ECR assay. The results are expressed as the OD495/OD600 of culture after 7 h growth. The results represent the means ± standard deviations of five independent experiments. (B) Swarming was assessed on M9-based medium, as described in Materials and Methods. The plates were incubated for 18 h at 37°C.
Similar results were obtained for the swarming of P. aeruginosa. Vector-carrying strain PT1300 was inhibited by 2 μg/ml AZM, while methylase-carrying strain PT1308 was able to swarm at this AZM concentration (Fig. 2B). Swarming inhibition very likely results from the decreased production of rhamnolipids, which are required as a surfactant for this type of motility (19). Since swarming was recently shown to be involved in biofilm formation (26), the retardation of biofilm formation by AZM (9) might thus result from the inhibition of swarming motility.
AZM killing requires interaction with the ribosome.
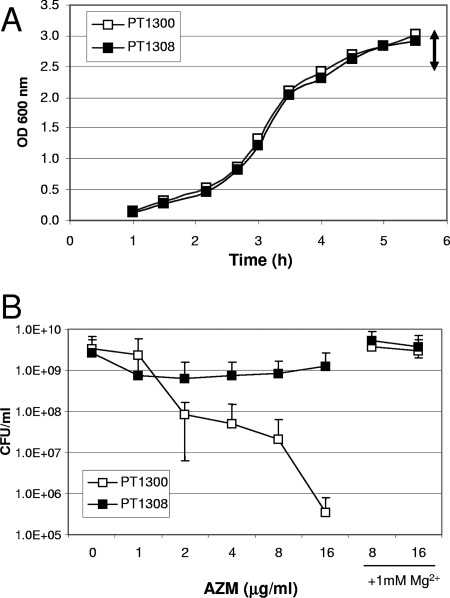
Recently, Imamura et al. (12) showed that AZM displays bactericidal effects against PAO1 in stationary phase but not in exponential growth phase. The authors suggested that AZM displaces Mg2+ ions from the phospholipid moiety of lipopolysaccharide, thereby destabilizing the outer membrane. However, it was not clear whether the observed bactericidal effect resulted solely from cell lysis. We therefore tested the effect of ribosome protection on the killing effect of AZM on stationary-phase cells. PT1300 and PT1308 were grown to stationary phase (OD600, 3.5 to 4) (Fig. 3A) and then exposed to various AZM concentrations. While vector-carrying strain PT1300 showed dose-dependent killing in the presence of AZM, the viability of methylase-expressing strain PT1308 was not significantly affected by AZM (Fig. 3B). Since both strains were cultured to stationary phase in the absence of AZM, the observed difference does not result from AZM-induced effects during prior growth. As shown by Imamura et al. (12), addition of 1 mM Mg2+ in combination with AZM prevented the killing of Pseudomonas cells (Fig. 3B). These observations suggest that AZM might first destabilize the outer membrane by displacing Mg2+ ions and thereby promote its own uptake, as reported previously for Escherichia coli (7) and for aminoglycosides in P. aeruginosa (10), thus increasing intracellular AZM concentrations. While Imamura et al. (12) concluded in their study that AZM has a bactericidal effect due to membrane disruption, our experiments suggest a two-step process in which AZM first permeabilizes the outer membrane and then causes cell death by inhibiting protein synthesis and/or ribosome assembly (3).
FIG. 3.
Effect of ribosome protection on AZM-mediated killing of stationary-phase cells. (A) Representative growth curves of strain PT5 harboring vector pEX1.8 (strain PT1300) or the ermBP-carrying plasmid pEXERM (strain PT1308) in LB medium. The double-headed arrow indicates the OD600 range for addition of AZM. (B) After addition of AZM or AZM plus MgSO4 at the indicated concentrations, the cultures were incubated at 37°C for 22 h. Viable counts were determined by serial dilutions and plating on LB agar plates. The results are expressed as the means ± standard deviations from three independent experiments.
Cell killing by AZM is enhanced by rhamnolipids.
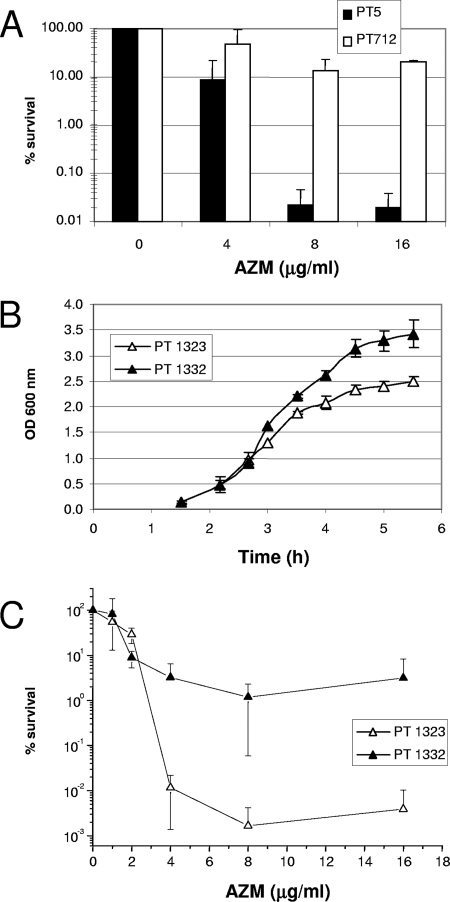
During the killing experiments we noticed that an rhlA mutant (strain PT712) deficient in rhamnolipid production was almost 1,000-fold less susceptible to the AZM-mediated killing than its parent strain, strain PT5 (Fig. 4A). To confirm these data, we repeated the killing experiments with the rhlA mutant carrying either the vector alone (strain PT1332) or complemented with plasmid pAKRHL (strain PT1323) expressing the rhamnosyltransferase genes rhlAB. The presence of plasmid pAKRHL restored rhamnolipid production in the rhlA mutant (Fig. 1B), without affecting susceptibility to AZM (Fig. 1A; compare the results for strains PT1323 and PT1332 with those for strain PT712). Interestingly, when rhlA mutant PT712 was complemented with rhlAB-expressing plasmid pAKRHL, susceptibility to AZM-mediated killing was restored, resulting in a reduction in the viable counts of 4 to 5 orders of magnitude (see the results for strain PT1323 in Fig. 4C). In comparison, the rhlA mutant harboring control vector pAK1900 showed a reduction in viable counts of only 1 to 2 orders of magnitude (see the results for strain PT1332 in Fig. 4C). Strain PT1323 showed reduced growth compared to that of the control, PT1332, which could result from (premature) overexpression of the plasmid-encoded rhlAB genes (Fig. 4B). This, however, did not affect the viabilities of the strains in the absence of AZM since both strains reached comparable numbers of CFU/ml after the 20-h incubation period (data not shown).
FIG. 4.
AZM-mediated killing of stationary-phase cells is enhanced by rhamnolipids. (A) Strains were grown to stationary phase in LB medium, as described in the legend to Fig. 3, and exposed to AZM at the indicated concentrations for 22 h. Viable counts were determined by serial dilutions and plating on LB agar plates. (B) Growth curves in LB medium of an rhlA mutant harboring the vector pAK1900 (strain PT1332) or rhlAB-encoding plasmid pAKRHL (strain PT1323). (C) The killing by AZM was assessed after 22 h incubation by determination of viable counts, as described above for panel A. The results are expressed as the means ± standard deviations of three independent experiments.
How can rhamnolipids promote AZM-mediated killing? Rhamnolipids have initially been identified as amphiphilic agents able to permit the growth of Pseudomonas isolates on aliphatic hydrocarbons, like hexadecane, which are not efficiently taken up by the cell (18, 21). Due to their detergent-like structure, rhamnolipids increase the uptake of these hydrophobic molecules by micelle formation. This fact was recently illustrated by the increased activity of the hydrophobic signaling molecule PQS in the presence of externally added rhamnolipids (2). Furthermore, rhamnolipids were shown to increase cell surface hydrophobicity by promoting the release of hydrophilic lipopolysaccharide from the outer membrane (1). It is therefore tempting to speculate that rhamnolipids promote the uptake of AZM. Since rhamnolipids are under the control of the QS system and are not expressed until late exponential phase, their effect is seen only under the conditions of the killing assay when AZM is added to stationary-phase cells. AZM susceptibility is not affected by rhamnolipids when cells grow exponentially, as in conventional MIC determinations or on the AZM gradient plates (Fig. 1A). Furthermore, rhamnolipids did not cause AZM-mediated cell killing during exponential phase when they were produced from rhlAB-encoding plasmid pAKRHL (data not shown). This suggests that changes in the outer membrane composition or the depletion of divalent cations that might occur in stationary phase are required for the permeability enhancement of AZM by rhamnolipids. Thus, our data provide an explanation of why in vitro AZM-mediated killing is observed only for cells in stationary phase and not cells in exponential growth phase (12). However, the relevance of AZM-mediated cell killing in the clinical situation remains to be established, since the growth conditions and the metabolic state of the bacteria in the patient largely remain unknown.
In contrast, the AZM-mediated reduction of rhamnolipid production might be clinically relevant, as rhamnolipids were recently shown to promote the invasion of a reconstituted epithelium (38) and to kill polymorphonuclear leukocytes (16). The reduction of rhamnolipid production during AZM treatment might thereby inhibit one of the early steps in the progression from colonization toward infection, an effect that might be useful for preventive strategies.
Conclusions.
In this study, we show that the effect of AZM as a virulence attenuator can be moderately affected by the level of expression of the MexXY efflux pump and is seriously compromised by ribosome protection. Indeed, QS-dependent virulence factor production and swarming motility, as well as stationary-phase killing, were insensitive to the presence of AZM when the 23S rRNA of the 50S ribosome was protected by methylation. Ribosome protection in P. aeruginosa has been described so far only for the 16S rRNA, due to the plasmid-encoded rmtA gene that confers resistance to aminoglycosides (37). Interestingly, analysis against the PAO1 genome (www.pseudomonasv2.com) with the BLAST program revealed an open reading frame (in strain PA4067) that displayed 47% amino acid identity to ErmBP (data not shown). Whether the gene for this protein is involved in 23S rRNA methylation and could thus lead to clinically relevant resistance to the antivirulence effects of macrolides remains to be determined. We further show that AZM-mediated cell killing in stationary phase is enhanced by rhamnolipids, which could increase the uptake of this hydrophobic macrolide by P. aeruginosa. These observations might be of importance for CF patients receiving AZM and could explain the variabilities in treatment efficacies.
Acknowledgments
This work was supported by research grants from the Swiss National Science Foundation (NRP49 research grant 4049-063239 to T.K. and C.V.D. and research grant 32000-108106 to C.V.D.).
Footnotes
Published ahead of print on 17 September 2007.
REFERENCES
- 1.Al Tahhan, R. A., T. R. Sandrin, A. A. Bodour, and R. M. Maier. 2000. Rhamnolipid-induced removal of lipopolysaccharide from Pseudomonas aeruginosa: effect on cell surface properties and interaction with hydrophobic substrates. Appl. Environ. Microbiol. 66:3262-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calfee, M. W., J. G. Shelton, J. A. McCubrey, and E. C. Pesci. 2005. Solubility and bioactivity of the Pseudomonas quinolone signal are increased by a Pseudomonas aeruginosa-produced surfactant. Infect. Immun. 73:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Champney, W. S., and M. Miller. 2002. Inhibition of 50S ribosomal subunit assembly in Haemophilus influenzae cells by azithromycin and erythromycin. Curr. Microbiol. 44:418-424. [DOI] [PubMed] [Google Scholar]
- 4.De Kievit, T. R., Y. Kakai, J. K. Register, E. C. Pesci, and B. H. Iglewski. 2002. Role of the Pseudomonas aeruginosa las and rhl quorum-sensing systems in rhlI regulation. FEMS Microbiol. Lett. 212:101-106. [DOI] [PubMed] [Google Scholar]
- 5.Equi, A., I. M. Balfour-Lynn, A. Bush, and M. Rosenthal. 2002. Long term azithromycin in children with cystic fibrosis: a randomised, placebo-controlled crossover trial. Lancet 360:978-984. [DOI] [PubMed] [Google Scholar]
- 6.Essar, D. W., L. Eberly, and I. P. Crawfords. 1990. Identification and characterization of genes for a second anthranilate synthetase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farmer, S., Z. S. Li, and R. E. Hancock. 1992. Influence of outer membrane mutations on susceptibility of Escherichia coli to the dibasic macrolide azithromycin. J. Antimicrob. Chemother. 29:27-33. [DOI] [PubMed] [Google Scholar]
- 8.Favre-Bonté, S., T. Köhler, and C. Van Delden. 2003. Biofilm formation by Pseudomonas aeruginosa: role of the C4-HSL cell-to-cell signal and inhibition by azithromycin. J. Antimicrob. Chemother. 52:598-604. [DOI] [PubMed] [Google Scholar]
- 9.Gillis, R. J., and B. H. Iglewski. 2004. Azithromycin retards Pseudomonas aeruginosa biofilm formation. J. Clin. Microbiol. 42:5842-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock, R. E., and A. Bell. 1988. Antibiotic uptake into gram-negative bacteria. Eur. J. Clin. Microbiol. Infect. Dis. 7:713-720. [DOI] [PubMed] [Google Scholar]
- 11.Ichimiya, T., K. Takeoka, K. Hiramatsu, K. Hirai, T. Yamasaki, and M. Nasu. 1996. The influence of azithromycin on the biofilm formation of Pseudomonas aeruginosa in vitro. Chemotherapy 42:186-191. [DOI] [PubMed] [Google Scholar]
- 12.Imamura, Y., Y. Higashiyama, K. Tomono, K. Izumikawa, K. Yanagihara, H. Ohno, Y. Miyazaki, Y. Hirakata, Y. Mizuta, J. Kadota, B. H. Iglewski, and S. Kohno. 2005. Azithromycin exhibits bactericidal effects on Pseudomonas aeruginosa through interaction with the outer membrane. Antimicrob. Agents Chemother. 49:1377-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Islam, S., S. Jalal, and B. Wretlind. 2004. Expression of the MexXY efflux pump in amikacin-resistant isolates of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 10:877-883. [DOI] [PubMed] [Google Scholar]
- 14.Jansons, I., G. Touchie, R. Sharp, K. Almquist, M. A. Farinha, J. S. Lam, and A. M. Kropinski. 1994. Deletion and transposon mutagenesis and sequence analysis of the pRO1600 OriR region found in the broad-host-range plasmids of the pQF series. Plasmid 31:265-274. [DOI] [PubMed] [Google Scholar]
- 15.Jeannot, K., M. L. Sobel, F. El Garch, K. Poole, and P. Plesiat. 2005. Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J. Bacteriol. 187:5341-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen, P. O., T. Bjarnshold, R. Phipps, T. B. Rasmussen, H. Calum, L. Christoffersen, C. Moser, P. Williams, T. Pressler, M. Givskov, and N. Hoiby. 2007. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153:1329-1338. [DOI] [PubMed] [Google Scholar]
- 17.Kita, E., M. Sawaki, D. Oku, A. Hamuro, K. Mikasa, M. Konishi, M. Emoto, S. Takeuchi, N. Narita, and S. Kashiba. 1991. Suppression of virulence factors of Pseudomonas aeruginosa by erythromycin. J. Antimicrob. Chemother. 27:273-284. [DOI] [PubMed] [Google Scholar]
- 18.Koch, A. K., O. Kappeli, A. Fiechter, and J. Reiser. 1991. Hydrocarbon assimilation and biosurfactant production in Pseudomonas aeruginosa mutants. J. Bacteriol. 173:4212-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Köhler, T., L. Kocjancic-Curty, F. Barja, C. Van Delden, and J. C. Pechère. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nalca, Y., L. Jansch, F. Bredenbruch, R. Geffers, J. Buer, and S. Haussler. 2006. Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: a global approach. Antimicrob. Agents Chemother. 50:1680-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noordman, W. H., and D. B. Janssen. 2002. Rhamnolipid stimulates uptake of hydrophobic compounds by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 68:4502-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramos-Aires, J., T. Köhler, H. Nikaido, and P. Plésiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saiman, L., B. C. Marshall, N. Mayer-Hamblett, J. L. Burns, A. L. Quittner, D. A. Cibene, S. Coquillette, A. Y. Fieberg, F. J. Accurso, and P. W. Campbell. 2003. Azithromycin in patients with cystic fibrosis chronically infected with Pseudomonas aeruginosa: a randomized controlled trial. JAMA 290:1749-1756. [DOI] [PubMed] [Google Scholar]
- 25.Scaglione, F., and G. Rossoni. 1998. Comparative anti-inflammatory effects of roxithromycin, azithromycin and clarithromycin. J. Antimicrob. Chemother. 41(Suppl. B):47-50. [DOI] [PubMed] [Google Scholar]
- 26.Shrout, J. D., D. L. Chopp, C. L. Just, M. Hentzer, M. Givskov, and M. R. Parsek. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264-1277. [DOI] [PubMed] [Google Scholar]
- 27.Siegmund, I., and F. Wagner. 1991. New method for detecting rhamnolipids excreted by Pseudomonas species during growth in mineral agar. BioTechniques 5:265-268. [Google Scholar]
- 28.Smith, E. E., D. G. Buckley, Z. Wu, C. Saenphimmachak, L. R. Hoffman, D. A. D'Argenio, S. I. Miller, B. W. Ramsey, D. P. Speert, S. M. Moskowitz, J. L. Burns, R. Kaul, and M. V. Olson. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 103:8487-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sobel, M. L., G. A. McKay, and K. Poole. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 47:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tateda, K., R. Comte, J. C. Pechere, T. Köhler, K. Yamaguchi, and C. Van Delden. 2001. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45:1930-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tateda, K., Y. Ishii, T. Matsumoto, T. Kobayashi, S. Miyazaki, and K. Yamaguchi. 2000. Potential of macrolide antibiotics to inhibit protein synthesis of Pseudomonas aeruginosa: suppression of virulence factors and stress response. J. Infect. Chemother. 6:1-7. [DOI] [PubMed] [Google Scholar]
- 32.Van Delden, C., E. C. Pesci, J. P. Pearson, and B. H. Iglewski. 1998. Starvation selection restores elastase and rhamnolipid production in a Pseudomonas aeruginosa quorum-sensing mutant. Infect. Immun. 66:4499-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogne, C., J. R. Aires, C. Bailly, D. Hocquet, and P. Plesiat. 2004. Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 48:1676-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner, T., G. Soong, S. Sokol, L. Saiman, and A. Prince. 2005. Effects of azithromycin on clinical isolates of Pseudomonas aeruginosa from cystic fibrosis patients. Chest 128:912-919. [DOI] [PubMed] [Google Scholar]
- 35.Wolter, J., S. Seeney, S. Bell, S. Bowler, P. Masel, and J. McCormack. 2002. Effect of long term treatment with azithromycin on disease parameters in cystic fibrosis: a randomised trial. Thorax 57:212-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanagihara, K., K. Tomono, Y. Imamura, Y. Kaneko, M. Kuroki, T. Sawai, Y. Miyazaki, Y. Hirakata, H. Mukae, J. Kadota, and S. Kohno. 2002. Effect of clarithromycin on chronic respiratory infection caused by Pseudomonas aeruginosa with biofilm formation in an experimental murine model. J. Antimicrob. Chemother. 49:867-870. [DOI] [PubMed] [Google Scholar]
- 37.Yokoyama, K., Y. Doi, K. Yamane, H. Kurokawa, N. Shibata, K. Shibayama, T. Yagi, H. Kato, and Y. Arakawa. 2003. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 362:1888-1893. [DOI] [PubMed] [Google Scholar]
- 38.Zulianello, L., C. Canard, T. Kohler, D. Caille, J. S. Lacroix, and P. Meda. 2006. Rhamnolipids are virulence factors that promote early infiltration of primary human airway epithelia by Pseudomonas aeruginosa. Infect. Immun. 74:3134-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]