Abstract
We describe an unusual pathway of human immunodeficiency virus type 1 reverse transcriptase resistance during therapy with tenofovir-emtricitabine, characterized initially by the mutations K70E and M184V and later by K70G and M184V, with the two mutations coexisting on the same viral genome. Phenotypic resistance to lamivudine, emtricitabine, abacavir, didanosine, and tenofovir was observed, whereas susceptibility to zidovudine and stavudine was preserved.
A 32-year-old black African man was diagnosed with human immunodeficiency virus type 1 (HIV-1) infection (subtype G) in March 1997. He started antiretroviral therapy 6 months later and over the years received the following: zidovudine and lamivudine; zidovudine, lamivudine, and nevirapine; didanosine, stavudine and nevirapine; tenofovir, ritonavir-boosted lopinavir and saquinavir; tenofovir and ritonavir-boosted lopinavir; and ritonavir-boosted lopinavir and atazanavir. In April 2004, treatment was changed to a once-daily tablet of fixed-dose tenofovir-emtricitabine to facilitate adherence and at least partial virological control.
Genotypic resistance testing by population sequencing (Viroseq HIV-1 genotyping system; Celera Diagnostics) was performed on three occasions prior to the commencement of tenofovir-emtricitabine. In July 2001 (didanosine, stavudine, nevirapine), it showed the reverse transcriptase (RT) resistance mutations D67G and Q151M for the nucleos(t)ide RT inhibitors (NRTIs) and K103N and V106A and Y181C for the nonnucleoside RT inhibitors. In August 2003 (tenofovir, ritonavir-boosted lopinavir, saquinavir) and in April 2004 (ritonavir-boosted lopinavir, atazanavir) there were no detectable RT mutations, whereas the protease showed the major resistance mutations G48V, I54V, and V82T. In March 2005, 1 year after the start of tenofovir-emtricitabine, the genotype showed the RT mutations K70E and M184V (Table 1). The major protease resistance mutations were no longer detectable. Identical results were obtained in June 2005. In November 2005 the genotype showed the RT mutations K70G and M184V. Identical results were obtained 3 weeks later. The switch from K70E to K70G coincided with a CD4+-cell count drop from 90 to 56 cells/mm3 and a rise in plasma HIV-1 RNA load from 4.5 to 5.1 log10 copies/ml, possibly suggesting improved fitness of the K70G M184V mutant.
TABLE 1.
Timetable of HIV-1 plasma RNA load and genotypic resistance testing during dual antiretroviral therapy with fixed-dose tenofovir-emtricitabinea
| Date | No. of HIV-1 RNA copies/ml | Resistance mutation(s) |
|---|---|---|
| March 2004 | 18,500 | None |
| June 2004 | 7,700 | ND |
| March 2005 | 86,700 | K70E M184V |
| June 2005 | 32,800 | K70E M184V |
| November 2005 | 138,401 | K70G M184V |
| November 2005 | ND | K70G M184V |
ND, not determined.
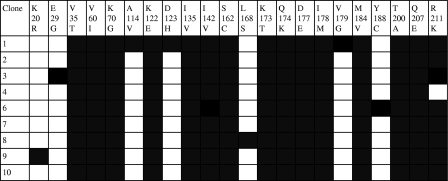
To determine whether K70G and M184V coexisted on the same viral genome, RT codons 1 to 243 were amplified by nested RT-PCR, and the PCR product was cloned into pCR2.1TOPO and expressed in Escherichia coli DH5α. Nine clones generated from three independent PCRs all contained K70G and M184V in combination and in the absence of other NRTI resistance mutations (Fig. 1).
FIG. 1.
Sequences of nine clones (RT codons 1 to 243) derived from a plasma sample showing the mutations K70G and M184V by population sequencing (reference HXB2). All mutations detected in RT are shown as shaded boxes.
Phenotypic testing (Antivirogram; Virco) was carried out on two samples (Table 2). In March 2005 (genotype K70E M184V), according to the assay biological cutoffs (BCOs), there was resistance to lamivudine and emtricitabine but preserved susceptibility to other NRTIs. In November 2005 (genotype K70G M184V), there was resistance to lamivudine, emtricitabine, didanosine, and abacavir but preserved susceptibility to tenofovir, zidovudine, and stavudine. As BCOs may fail to reflect the clinical significance of small changes in phenotypic susceptibility, the data were also analyzed using the Vircotype HIV-1 clinical cutoffs (CCOs). The Vircotype HIV-1 system provides a phenotype prediction based on linear modeling of Antivirogram data. Extrapolation of the proposed Vircotype HIV-1 lower and upper clinical CCOs indicated that K70E and M184V would virtually abrogate responses to lamivudine and reduce responses to didanosine, abacavir, and tenofovir. The K70G-M184V combination would virtually abrogate responses to lamivudine, didanosine, and abacavir and reduce responses to tenofovir. In both cases, preserved susceptibility to zidovudine and stavudine was predicted.
TABLE 2.
Phenotypic resistancea of HIV-1 clinical isolates with the RT mutations K70E M184V and K70G M184V
| Drug | Change (fold) in IC50
|
Antivirogram BCO | Vircotype CCOb
|
||
|---|---|---|---|---|---|
| K70E+M184V | K70G+M184V | Lower | Upper | ||
| Lamivudine | >29.3 | >84.5 | 2.4 | 1.0 | 3.4 |
| Emtricitabine | >40.8 | >63.4 | 3.5 | NA | NA |
| Zidovudine | <0.3 | <0.8 | 2.7 | 1.2 | 9.6 |
| Stavudine | 0.4 | 0.9 | 2.3 | 0.9 | 2.0 |
| Didanosine | 1.6 | 5.3 | 2.2 | 0.9 | 2.6 |
| Abacavir | 1.3 | 3.2 | 2.2 | 0.8 | 1.9 |
| Tenofovir | 1.7 | 1.5 | 2.1 | 0.9 | 2.1 |
Phenotypic resistance measured by the Antivirogram assay.
NA, not available.
A search of the Virco database, containing 190,485 sequences from routine clinical samples, showed a low frequency (<0.5%) of both K70E and K70G (Fig. 2). The mutations showed a small but significant increase in frequency over time (P < 0.01). In the same database, the prevalence of the thymidine analogue mutation K70R declined from 18.3% in 1999 to 10.4% in 2005 (data not shown). Among seven clinical isolates in the database showing K70G and M184V in the absence of other NRTI resistance mutations, the median (range) change (n-fold) in 50% inhibitory concentration (IC50) was 0.8 (0.2 to 1.5) for zidovudine, 1.0 (0.4 to 2.2) for stavudine, 1.8 (0.7 to 3.3) for didanosine, 3.1 (1.4 to 7.0) for abacavir, and 1.1 (0.7 to 1.3) for tenofovir. These data were consistent with the findings from the case. One clinical isolate with K70G alone (in the absence of M184V and other NRTI resistance mutations) showed a change (n-fold) in IC50 of 4.0 for lamivudine, 1.0 for zidovudine, 1.3 for stavudine, 2.1 for didanosine, 2.1 for abacavir, and 1.9 for tenofovir. These data indicate that the phenotypic effects of K70G on NRTI susceptibility are similar to those observed with K70E and K65R and suggest that M184V may have a resensitizing effect for tenofovir in the presence of K70G.
FIG. 2.
Prevalence of the RT mutations K70E and K70G in routine clinical samples submitted for resistance testing between 1999 and 2005 (Virco database, n = 190,485).
The K70E mutation was first described after serial passage of HIV-1IIIb in increasing concentrations of adefovir (1) and more recently after serial passage of HIV-1LAI in increasing concentrations of the cytidine analogue D-d4FC (3). Unlike the thymidine analogue mutation K70R, K70E is uncommon in treatment-experienced persons, although an increased prevalence has been observed since the introduction of tenofovir in 2001 (4). We similarly observed a trend towards an increased prevalence of K70E over time, although the overall frequency remained <0.5%. Previous reports have described the occurrence of K70E among persons receiving the following: tenofovir, abacavir, and lamivudine with or without didanosine (9, 2); tenofovir, lamivudine, and efavirenz (9, 5); adefovir either alone or in combination with zidovudine and lamivudine (7); and abacavir, didanosine and stavudine (8). These observations indicate that the selective pressure on K70E is similar to that observed for K65R. At the genomic level K70E has not been found to be coselected with K65R, although it has been proposed that emergence of K70E may precede that of K65R (9).
In earlier studies, site-directed recombinant virus carrying K70E showed reduced susceptibility to adefovir and lamivudine (1, 6). More recently, Sluis-Cremer et al. demonstrated that K70E allows the RT enzyme to discriminate between the natural substrate and the NRTI triphosphate (10). Compared to the wild-type enzyme, K70E RT showed 2.1-, 2.3-, and 3.5-fold resistance toward tenofovir diphosphate, carbovir triphosphate, and lamivudine triphosphate, respectively.
The K70G mutation has been rarely observed in clinical studies, and the phenotypic susceptibility of virus with K70G has been reported only in the context of multiple major NRTI mutations, including Q151M and thymidine analogue mutations, which prevents an appreciation of the phenotypic effects of the mutation. We found that during therapy with tenofovir-emtricitabine, the mutation occurred with M184V on the same viral genome and in the absence of other NRTI resistance mutations. The phenotypic effects of K70G and M184V were similar to those observed with K70E and M184V and those reported for K65R and M184V, affecting multiple NRTIs but with preserved phenotypic susceptibility to zidovudine and stavudine. The wild-type lysine at RT position 70 is encoded by the triplet AAA or AAG, and the wild-type triplet was AAA in this patient. A switch to glutamic acid requires a change to GAA or GAG, and in this patient K70E was encoded by GAA. Glycine is encoded by GGT, GGC, GGA, or GGG, and in this patient K70G was encoded by GGA. Thus, K70G required a two-base change from the wild-type but only one base change from K70E. Taken together, these data indicate that K70G and M184V may evolve from K70E and M184V as an alternative pathway of resistance to tenofovir-emtricitabine. Further studies are required to determine the mechanisms of K70G-mediated resistance to the NRTIs, including the assessment of the phenotypic profile of K70G site-directed mutants. Meanwhile it is recommended that interpretation algorithms score the mutation pattern K70G M184V with an approach similar to that used for K70E and M184V.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in GenBank under accession numbers EU099970 (population sequence 03/05); EU099971 (population sequence 11/05); EU099972 (phenotyped sequence 03/05); EU099973 (phenotyped sequence 11/05); EU099974 (clone 1); EU099975 (clone 2); EU099976 (clone 3); EU099977 (clone 4); EU099978 (clone 6); EU099979 (clone 7); EU099980 (clone 8); EU099981 (clone 9); EU099982 (clone 10).
Footnotes
Published ahead of print on 17 September 2007.
REFERENCES
- 1.Cherrington, J. M., A. S. Mulato, M. D. Fuller, and M. S. Chen. 1996. Novel mutation (K70E) in human immunodeficiency virus type 1 reverse transcriptase confers decreased susceptibility to 9-[2-(phosphonomethoxy)ethyl]adenine in vitro. Antimicrob. Agents Chemother. 40:2212-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delaugerre, C., L. Roudiere, G. Peytavin, C. Rouzioux, J. P. Viard, and M. L. Chaix. 2005. Selection of a rare resistance profile in an HIV-1-infected patient exhibiting a failure to an antiretroviral regimen including tenofovir DF. J. Clin. Virol. 32:241-244. [DOI] [PubMed] [Google Scholar]
- 3.Hammond, J. L., U. M. Parikh, D. L. Koontz, S. Schlueter-Wirtz, C. K. Chu, H. Z. Bazmi, R. F. Schinazi, and J. W. Mellors. 2005. In vitro selection and analysis of human immunodeficiency virus type 1 resistant to derivatives of beta-2′,3′-didehydro-2′,3′-dideoxy-5-fluorocytidine. Antimicrob. Agents Chemother. 49:3930-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kagan, R., L. Ross, M. Winters, T. Merigan, P. Heseltine, and M. Lewinski. 2005. Adefovir-associated HIV-1 RT mutation K70E in the age of tenofovir. Antivir. Ther. 10:S103. [Google Scholar]
- 5.Margot, N. A., B. Lu, A. Cheng, and M. D. Miller. 2006. Resistance development over 144 weeks in treatment-naive patients receiving tenofovir disoproxil fumarate or stavudine with lamivudine and efavirenz in Study 903. HIV Med. 7:442-450. [DOI] [PubMed] [Google Scholar]
- 6.Miller, M. D., P. D. Lamy, M. D. Fuller, A. S. Mulato, N. A. Margot, T. Cihlar, and J. M. Cherrington. 1998. Human immunodeficiency virus type I reverse transcriptase expressing the K70E mutation exhibits a decrease in specific activity and processivity. Mol. Pharmacol. 54:291-297. [DOI] [PubMed] [Google Scholar]
- 7.Mulato, A. S., P. D. Lamy, M. D. Miller, W.-X. Li, K. E. Anton, N. S. Hellman, and J. M. Cherrington. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from AIDS patients after prolonged adefovir dipivoxil therapy. Antimicrob. Agents Chemother. 42:1620-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roge, B. T., T. L. Katzenstein, N. Obel, H. Nielse, O. Kirk, and J. Gerstoft. 2002. Genotypic and phenotypic changes in antiretroviral-naïve patients experiencing failure on randomised treatment with abacavir, didanosine and stavudine. Antivir. Ther. 7:S125. [Google Scholar]
- 9.Ross, L., P. Gerondelis, Q. Liao, B. Wine, M. Lim, M. Shaefer, A. Rodriguez, J. Gallant, K. Limoli, W. Huang, N. Parkin, and R. Lanier. 2005. Selection of the HIV-1 reverse transcriptase mutation K70E in antiretroviral-naïve subjects treated with tenofovir/abacavir/lamivudine therapy. Antivir. Ther. 10:S102. [Google Scholar]
- 10.Sluis-Cremer, N., C. W. Sheen, S. Zelina, P. S. Argoti Torres, U. M. Parikh, and J. W. Mellors. 2007. Molecular mechanism by which K70E in human immunodeficiency virus type 1 reverse transcriptase confers resistance to nucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 51:48-53. [DOI] [PMC free article] [PubMed] [Google Scholar]