Abstract
MurF is an essential enzyme of bacterial cell wall biosynthesis. Few MurF inhibitors have been reported, and none have displayed measurable antibacterial activity. Through the use of a MurF binding assay, a series of 8-hydroxyquinolines that bound to the Escherichia coli enzyme and inhibited its activity was identified. To derive additional chemotypes lacking 8-hydroxyquinoline, a known chelating moiety, a pharmacophore model was constructed from the series and used to select compounds for testing in the MurF binding and enzymatic inhibition assays. Whereas the original diverse library yielded 0.01% positive compounds in the binding assay, of which 6% inhibited MurF enzymatic activity, the pharmacophore-selected set yielded 14% positive compounds, of which 37% inhibited the enzyme, suggesting that the model enriched for compounds with affinity to MurF. A 4-phenylpiperidine (4-PP) derivative identified by this process displayed antibacterial activity (MIC of 8 μg/ml against permeable E. coli) including cell lysis and a 5-log10-unit decrease in CFU. Importantly, treatment of E. coli with 4-PP resulted in a 15-fold increase in the amount of the MurF UDP-MurNAc-tripeptide substrate, and a 50% reduction in the amount of the MurF UDP-MurNAc-pentapeptide product, consistent with inhibition of the MurF enzyme within bacterial cells. Thus, 4-PP is the first reported inhibitor of the MurF enzyme that may contribute to antibacterial activity by interfering with cell wall biosynthesis.
Bacterial cell wall biosynthesis has proven to be a rich source of targets for antibacterial agents, including β-lactams and glycopeptides (10). Synthesis of the cell wall pentapeptide precursor UDP-MurNAc-l-Ala-γ-d-Glu-meso-diaminopimelate-d-Ala-d-Ala (UDP-MurNAc-pentapeptide) in gram-negative bacteria (with l-Lys instead of meso-diaminopimelate in gram-positive bacteria) commences with the activity of MurA, which is the target of the drug fosfomycin (12). The last cytoplasmic step of UDP-MurNAc-pentapeptide synthesis is carried out by the MurF enzyme which catalyzes the ligation of d-Ala-d-Ala to UDP-MurNAc-l-Ala-γ-d-Glu-meso-diaminopimelate (UDP-MurNAc-tripeptide), with hydrolysis of ATP (1, 6). These three substrates bind to MurF in an ordered fashion, with ATP binding first and effecting a conformational change in the enzyme, followed by UDP-MurNAc-tripeptide and finally by d-Ala-d-Ala (1, 25, 30).
Unlike the MurA and MurB substrates, UDP-MurNAc-tripeptide is not commercially available, which has hampered efforts to assay MurF activity directly. Several methods have been employed to produce this MurF substrate, using MurA and MurB substrates and the cloned enzymes MurA, MurB, MurC, MurD, and MurE, either sequentially (19) or in a coupled reaction (7, 29). We have shown recently that muropeptide ligase can be used to synthesize UDP-MurNAc-tripeptide from synthetic tripeptide and the MurB product, UDP-MurNAc (2). However, it was not practical to synthesize sufficient quantities of UDP-MurNAc-tripeptide to enable high-throughput screening (HTS) of compounds for the identification of inhibitors of the MurF enzyme. Instead, we decided to employ as a primary screen a thermal stability assay (4, 18), which assesses the binding of putative inhibitors to the protein of interest by monitoring changes in protein melting temperature (Tm).
Few inhibitors of MurF have been reported (8, 23, 24). These include a nonhydrolyzable ATP analog (1) and phosphinate transition state analogs (16). Gu et al. have reported on a series of sulfonamides (11), and Longenecker et al. have cocrystallized an inhibitor with Streptococcus pneumoniae MurF (14). In addition, we have reported on a series of thiazolylaminopyrimidines that inhibit Escherichia coli MurF (2). All of these compounds appear to lack measurable antibacterial activity, possibly due to low permeability, lack of long-term enzyme inhibition, or to other, unidentified reasons. Although the possibility that MurF was inhibited in these studies but did not affect bacterial growth cannot be excluded, the existence of conditional lethal mutants of MurF in E. coli (15) and more recently, in Staphylococcus aureus (26), argues that inhibition of MurF within bacteria should be deleterious.
In the absence of a control compound that specifically inhibits the MurF enzyme within bacteria, it is instructive to examine the characteristics of conditional lethal mutants of MurF. In E. coli, a temperature-sensitive variant of MurF was employed, so that the enzyme was nonfunctional at the nonpermissive temperature (15); in S. aureus, production of MurF was dependent on expression from an inducible promoter (26). In both species, lack of functional MurF resulted in cell lysis, accumulation of UDP-MurNAc-tripeptide, and a decrease in the amount of UDP-MurNAc-pentapeptide. Thus, treatment of bacteria with a MurF inhibitor might also be expected to result in these changes.
In this study, we report on a series of 8-hydroxyquinoline derivatives that were initially identified by the ability to bind to purified E. coli MurF. Members of the series also inhibited MurF enzymatic activity, with 50% inhibitory concentration (IC50) values as low as 330 nM (4). Due to the well-known chelating ability of 8-hydroxyquinolines (9, 21), it was of interest to use this series as a starting point to search for analogs lacking this functionality. Accordingly, a pharmacophore model based on the 8-hydroxyquinoline series was constructed and used to select additional compounds for testing in the binding assay (5). From this process, a 4-phenylpiperidine (4-PP) derivative was identified that was shown to inhibit the MurF enzyme. The characterization of the compound is presented in this report.
MATERIALS AND METHODS
MurF binding assay.
Cloning and purification of E. coli MurF have been described previously (2). After buffer optimization experiments, thermal stability studies using ThermoFluor technology (18) were performed with 20 μg/ml (0.4 μM) MurF, 1 mM ATP, 25 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 7.2, 150 mM NaCl, 2 mM MgCl2, 1 mM reduced glutathione, 0.002% Tween 20, and 40 μM 1-anilino-8-naphthalenesulfonic acid unless stated otherwise. HTS was performed using 100 μM test compound. Dissociation constants (Kds) were determined from the protein Tm measured as a function of ligand concentration (13).
MurF enzymatic assay.
The enzymatic assay of E. coli MurF, using high-performance liquid chromatography (HPLC) to detect the appearance of the UDP-MurNAc-pentapeptide product, has been described previously (2). MurF enzyme (20 ng; final concentration of 4 nM) in 40 μl of 100 mM Tris-Cl, pH 8.5, 5 mM ATP, and 300 mM NaCl, was preincubated with compound or dimethyl sulfoxide (2 μl) in a 96-well plate for 10 min at room temperature, and then 10 μl of 1 mM d-Ala-d-Ala and 50 μl of a completed Mpl reaction containing UDP-MurNAc-l-Ala-γ-d-Glu-meso-diaminopimelate (UDP-MurNAc-tripeptide) were added (2). After incubation (15 min at 37°C), the reaction was terminated by the addition of 5 μl of 10% trifluoroacetic acid. Peaks corresponding to UDP-MurNAc-pentapeptide and UDP-MurNAc-tripeptide were detected by HPLC, and MurF IC50 values were determined as described previously (2).
Microbiology studies.
MIC assays were performed by the CLSI broth microdilution method (17) in cation-adjusted Mueller-Hinton broth. Bacterial strains were obtained from the American Type Culture Collection (Manassas, VA) or from the strain collection of Johnson & Johnson Pharmaceutical Research & Development, L.L.C. (Raritan, NJ); the E. coli NovaBlue strain was purchased from EMD Biosciences (La Jolla, CA). For bacterial growth curves, 100-μl samples taken from flasks from UDP-precursor pool studies were added to replicate wells of a Bioscreen C 100-well plate immediately after the addition of compound. The plate was incubated in the Bioscreen C microbiology reader (Oy Growth Curves AB Ltd., Helsinki, Finland) at 37°C with continuous shaking, with an absorbance reading taken every 15 min. For CFU determinations, a Whitley automatic spiral plater and a Synbiosis ProtoCOL colony counter (Microbiology International, Frederick, MD) were used, on serial dilutions in phosphate-buffered saline of an aliquot of cells removed from replicate wells of the Bioscreen C plate.
Analysis of UDP-linked precursor pool.
Muropeptides were purified as described previously (22) with modifications. Cultures (125 ml) of E. coli OC2530 were grown with aeration to mid-log phase (A600 of 0.3). Compounds (or 250 μl dimethyl sulfoxide) were added to a concentration of 2× MIC with continued incubation. After 30 min, cells were chilled in an ice-ethanol bath and harvested by centrifugation. Cells were rinsed in cold 0.9% NaCl, centrifuged, and lysed in cold 5% trichloroacetic acid on ice for 30 min, mixing occasionally. Lysates were centrifuged (10,000 × g for 10 min), and the supernatants were extracted six times with an equal volume of diethyl ether to remove trichloroacetic acid. The supernatants were lyophilized, suspended in 1 ml of H2O, and adjusted to pH 7 with NaOH. The extracts eluted with water from a Hi Prep 26/10 Sephadex column (GE Healthcare, Piscataway, NJ) at a flow rate of 2 ml/min, with detection of muropeptide peaks at A254. Muropeptide fractions were pooled, lyophilized, and suspended in 125 μl of H2O. Muropeptides were quantitated by HPLC as described above.
Pharmacophore modeling.
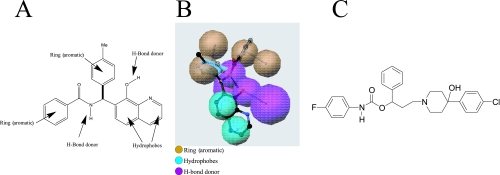
A pharmacophore model of the 8-hydroxyquinoline series (Fig. 1A and B) was constructed and used to search a compound library for structures that aligned to the pharmacophore, using the Common Features Module of the Catalyst program (3).
FIG. 1.
(A) Pharmacophore model of the 8-hydroxyquinoline series derived from the Common Features algorithm of the Catalyst program. Me, methyl. (B) Catalyst depiction of the pharmacophore model including directionality of the aromatic rings and H-bond donors. (C) Structure of the MurF inhibitor, 4-PP.
RESULTS
The three MurF substrates are known to exhibit ordered binding, with ATP binding first, effecting a conformational change in the enzyme which facilitates the binding of UDP-MurNAc-tripeptide, followed by d-Ala-d-Ala (1, 25, 30). We proposed previously that performing HTS in the presence of ATP may aid in identifying inhibitors that do not compete with ATP and that bind at the latter two substrate sites (2). To search for compounds able to bind to MurF, the method used is ThermoFluor technology, a miniaturized fluorescence-based thermal stability assay in which binding of a ligand to the protein of interest is monitored by an increase in protein Tm (18). Under our experimental conditions, the apparent Kd of ATP from MurF was determined to be 400 μM, comparable to the Km value of approximately 100 to 200 μM obtained by standard kinetic methodology (1, 6). The presence of 1 mM ATP increased the reference MurF Tm from 54.5°C to 56°C (Fig. 2).
FIG. 2.
ThermoFluor traces of normalized fluorescence intensity as a function of temperature for MurF. The vertical dashed lines indicate the Tms of MurF without ATP (triangles) and with 1 mM ATP (squares). The change in Tm is shown.
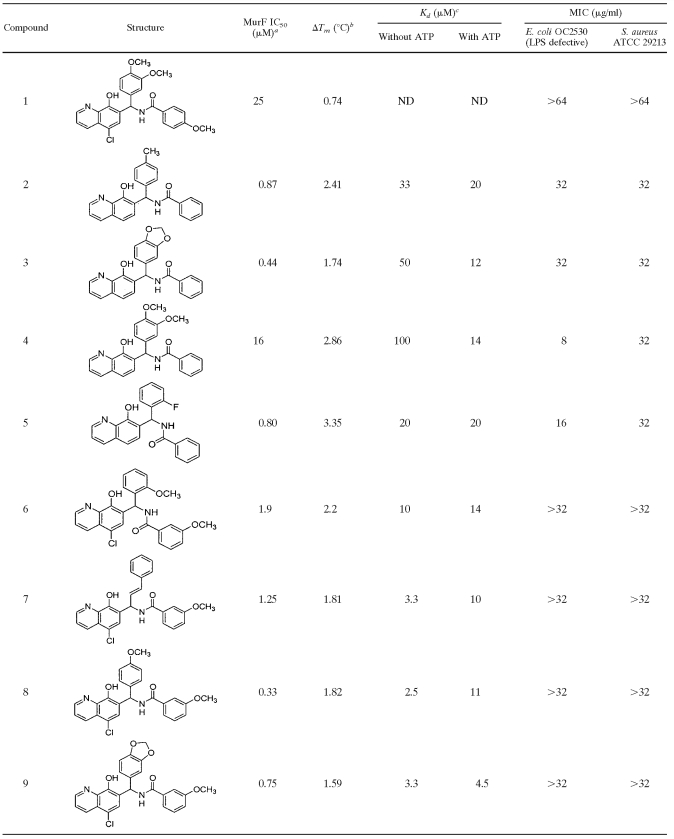
A chemical library of approximately 200,000 compounds was screened for binding to MurF, and 17 compounds that increased MurF Tm by ≥0.7°C were identified. These compounds were then tested for the ability to inhibit the enzymatic activity of MurF. Only 1 of the 17 compounds, the 8-hydroxyquinoline compound 1, inhibited MurF enzymatic activity, displaying an IC50 value of 25 μM (Table 1).
TABLE 1.
8-Hydroxyquinoline MurF inhibitors
MurF IC50 values were determined in the enzymatic inhibition assay.
ΔTm, shift in the melting temperature of MurF in the presence of 1 mM ATP as determined by Thermofluor analysis.
Kd values were determined in the absence and presence of 1 mM ATP. ND, not determined.
With the aim of improving potency, compound 1 was used to select a collection of approximately 4,000 closely related analogs. Testing of these compounds by the ThermoFluor methodology identified eight analogs with a change in the Tm (ΔTm) of ≥1.6°C (Table 1). Binding of the compounds to MurF in the absence and presence of ATP was investigated by determining Kd values (Table 1). Of the eight analogs, only compounds 7 and 8 exhibited a significant increase in Kd in the presence of ATP, suggesting that these two compounds may compete with ATP. In contrast, the other six compounds either exhibited similar Kd values with and without ATP, or in the case of compounds 3 and 4, appeared to bind more tightly to MurF in the presence of ATP, as demonstrated by a four- to sevenfold decrease in Kd. All eight compounds had lower IC50 values than that of compound 1 in the enzymatic assay, as low as 330 nM, but no correlation between IC50 values and either ΔTm or Kd was discernible.
Although the parent 8-hydroxyquinoline, compound 1, lacked measurable antibacterial activity (MIC > 64 μg/ml for E. coli OC2530 and S. aureus ATCC 29213 [Table 1]), four members of the series did exhibit modest antibacterial activity (MICs of 8 to 32 μg/ml [Table 1]). To determine whether antibacterial activity was due to inhibition of MurF within bacterial cells, E. coli cultures were treated with either cycloserine, compound 2, or compound 4 as described in Materials and Methods; MurF UDP-MurNAc-tripeptide substrate and UDP-MurNAc-pentapeptide product were then purified and quantitated (Table 2). Untreated control cells contained relatively low levels of UDP-MurNAc-tripeptide compared to the levels of UDP-MurNAc-pentapeptide, as has been reported previously (15, 26). Treatment of the culture with cycloserine, which blocks production of the MurF substrate d-Ala-d-Ala by inhibiting both d-Ala racemase and d-Ala-d-Ala ligase (20), resulted in an accumulation of UDP-MurNAc-tripeptide and a decrease in the amount of UDP-MurNAc-pentapeptide. In contrast, treatment of cells with either compound 2 or compound 4 had no effect on the ratio of UDP-MurNAc-tripeptide to UDP-MurNAc-pentapeptide, suggesting that the observed antibacterial activity of these compounds was not associated with inhibition of MurF; the well-known chelation ability of 8-hydroxyquinolines (9, 21) may contribute to antibacterial activity.
TABLE 2.
Quantitation of UDP-MurNAc-tripeptide and UDP-MurNAc-pentapeptide from E. coli OC2530
| E. coli treatment | Peak area (mean ± SD)a
|
Tripeptide/pentapeptide ratio | |
|---|---|---|---|
| UDP-MurNAc-tripeptide | UDP-MurNAc-pentapeptide | ||
| None (control)b | 18 ± 3 | 543 ± 10 | 0.03 |
| Cycloserine | 545 ± 0 | 18 ± 2 | 30.3 |
| Compound 2 | 28 ± 2 | 1,106 ± 57 | 0.02 |
| Compound 4 | 37 ± 1 | 915 ± 10 | 0.04 |
Relative amounts of each muropeptide were determined by integration of the area of the corresponding HPLC peaks at A260.
No 8-hydroxyquinoline compound or cycloserine was added to the control.
To search for additional chemotypes with improved potency and antibacterial activity, a pharmacophore model (Fig. 1A and B) based on the 8-hydroxyquinoline series was constructed using the Common Features Module of the program Catalyst. The model consists of two ring aromatics, two hydrophobes, and two H-bond donors and was used to search a compound library to provide a list of compounds that aligned to the pharmacophore. Approximately 1,300 compounds selected by this method were tested for MurF binding, yielding 182 compounds which were then tested for inhibition of MurF enzymatic activity. Of the 182 compounds identified by the binding assay, 68 also inhibited the MurF enzyme. Among these compounds was the 4-PP derivative (Fig. 1C), which inhibited the MurF enzyme with an IC50 value of 26 ± 2 μM.
The antibacterial activity of 4-PP was assessed against a panel of gram-positive and gram-negative bacteria (Table 3). The compound displayed MICs of 8 to 16 μg/ml against a permeable, lipopolysaccharide (LPS)-defective E. coli strain, and against S. aureus (methicillin-susceptible and -resistant strains), Enterococcus faecalis, and Enterococcus faecium. However, against wild-type E. coli and Pseudomonas aeruginosa, 4-PP displayed MICs of >32 μg/ml.
TABLE 3.
MICs of 4-PP and cycloserine
| Bacterial strain | MIC (μg/ml)
|
|
|---|---|---|
| 4-PP | Cycloserine | |
| E. coli strains | ||
| OC2530 (LPS defective) | 8 | 64 |
| OC2605 | >32 | 16 |
| OC9040 | >32 | 32 |
| ATCC 25922 | >32 | 32 |
| P. aeruginosa ATCC 27853 | >32 | >128 |
| E. faecalis ATCC 29212 | 16 | 128 |
| E. faecium OC3312 | 16 | 64 |
| S. aureus strains | ||
| ATCC 29213 | 16 | 32 |
| COL OC3726a | 16 | 32 |
| OC2878a | 16 | 64 |
| OC4172 | 8 | 32 |
Methicillin-resistant S. aureus strains.
To examine whether impermeability of 4-PP was possibly responsible for the lack of measurable MICs for wild-type (non-LPS-defective) E. coli strains, polymyxin B nonapeptide (PMBN) was used to permeabilize the cells (27). The MICs of PMBN for wild-type E. coli strains ATCC 25922 and NovaBlue were 50 and 25 μg/ml, respectively, compared to 6.25 μg/ml for the LPS-defective strain OC2530 (Table 4). Accordingly, 4-PP was tested against these strains without and with 1 μg/ml PMBN, a concentration that is 6- to 50-fold below the PMBN MIC. The MICs of 4-PP against the two wild-type E. coli strains, without and with 1 μg/ml PMBN, were >64 and 16 μg/ml, respectively. Thus, it appears that the lack of antibacterial activity of 4-PP against wild-type E. coli may be due to lack of compound penetration. PMBN had little effect on the MIC of 4-PP for the permeable, LPS-defective strain of E. coli (8 and 4 μg/ml without and with PMBN, respectively).
TABLE 4.
Effects of PMBN on the MICs of 4-PP and cycloserine for LPS-defective and wild-type E. coli strains
| E. coli strain | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|
| 4-PP | 4-PP (+ 1 μg/ml PMBN) | Cycloserine | Cycloserine (+ 1 μg/ml PMBN) | PMBN | |
| OC2530 (LPS defective) | 8 | 4 | 64 | 64 | 6.2 |
| ATCC 25922 | >64 | 16 | 32 | 32 | 50 |
| NovaBlue | >64 | 16 | 64 | 64 | 25 |
The cell wall synthesis inhibitor, cycloserine, is an analog of d-Ala and uses d-Ala and Gly transport systems for uptake into bacteria (28). The MIC of cycloserine is therefore not expected to be enhanced either by the use of a permeable (LPS-defective) strain of E. coli or by the presence of PMBN, consistent with the results shown in Tables 3 and 4. This lack of an enhancement of cycloserine activity supports the idea that the reduction of the MICs for 4-PP through the use of PMBN or LPS-defective E. coli is due to increased permeability of the compound and that lack of activity of 4-PP in wild-type E. coli may be due to poor penetration.
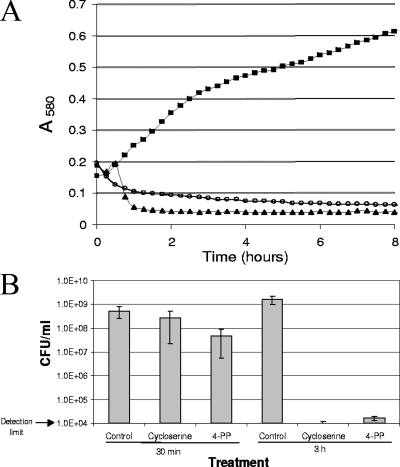
To determine the effect of 4-PP on bacterial growth, mid-log cultures of the LPS-defective E. coli strain were treated with either 4-PP or cycloserine at 2× MIC, and the growth of the cultures was monitored by both optical density (Fig. 3A) and by determination of CFU (Fig. 3B). The decrease in optical density for both the cycloserine- and 4-PP-treated samples (Fig. 3A) suggested that cell lysis occurred, as confirmed by microscopic examination of the cultures.
FIG. 3.
(A) Growth curves of E. coli treated with MurF inhibitor 4-PP or cycloserine. Strain OC2530 was grown to mid-log phase as described in Materials and Methods, and compounds were added to a final concentration of 2× MIC. Aliquots of cultures were transferred to the Bioscreen C plate, and culture growth was monitored by A580. A representative well of 10 wells for each culture (control, squares; cycloserine, triangles; 4-PP, open circles) is shown. (B) Quantitation of CFU after treatment with MurF inhibitor 4-PP or cycloserine. Aliquots of cultures shown in panel A were removed from the Bioscreen C plate at the indicated times for determination of CFU. The averages ± standard deviations (error bars) for two independent experiments are shown.
Viable bacteria from this experiment were counted 30 min and 3 h after the addition of compound (Fig. 3B). After 30 min of 4-PP exposure (at 2× MIC), a reduction in CFU of approximately 1 log10 unit relative to the control value was observed. By 3 h, 4-PP reduced CFU by about 5 log10 units relative to the control value, indicating a bactericidal mode of action. Cycloserine demonstrated similar efficacy, with reductions in CFU of approximately 0.5 log10 unit at 30 min and >5 log10 units at 3 h.
To determine whether bacterial killing could be attributed to inhibition of MurF in E. coli, the effect of 4-PP on the soluble muropeptide composition of cells treated with the compound was examined. From previous studies on MurF mutants (15, 26), it would be expected that inhibition of MurF activity within bacteria would alter muropeptide profiles, resulting in the accumulation of MurF substrate UDP-MurNAc-tripeptide and in a decrease in the amount of MurF product UDP-MurNAc-pentapeptide.
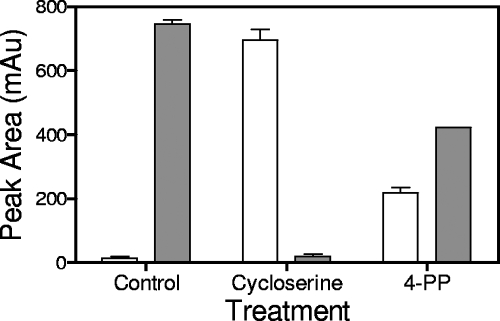
In the control culture of E. coli (Fig. 4), UDP-MurNAc-tripeptide was present in very small amounts compared to the amounts of UDP-MurNAc-pentapeptide, consistent with previous studies (15, 26). As expected, cycloserine treatment, which blocks production of d-Ala-d-Ala by inhibiting d-Ala racemase and d-Ala-d-Ala ligase (20), led to a pronounced reduction in the amount of UDP-MurNAc-pentapeptide with a concomitant accumulation of UDP-MurNAc-tripeptide. Treatment with 4-PP also altered the cellular muropeptide profile, albeit not as drastically as cycloserine treatment. In the 4-PP-treated culture, a 15-fold increase in MurF substrate UDP-MurNAc-tripeptide and a 50% decrease in MurF product UDP-MurNAc-pentapeptide were observed. Thus, it appears that 4-PP inhibited MurF enzymatic activity within bacterial cells.
FIG. 4.
Altered muropeptide profile in 4-PP-treated E. coli. Cells were treated with 4-PP or cycloserine (2× MIC for 30 min) as described in Materials and Methods, and muropeptides were extracted and quantitated. White bars, UDP-MurNAc-tripeptide; gray bars, UDP-MurNAc-pentapeptide. The results of one representative experiment of two independent experiments are shown. mAu, milli absorbance units.
DISCUSSION
In these studies, we report on a MurF inhibitor, an 8-hydroxyquinoline, which was initially identified using a binding assay. The compound itself lacked antibacterial activity but was used to construct a pharmacophore model which led to the identification of a 4-PP derivative whose antibacterial activity was accompanied by an alteration in the E. coli muropeptide profile, with both the accumulation of MurF substrate (UDP-MurNAc-tripeptide) and a decrease in MurF product (UDP-MurNAc-pentapeptide). The altered muropeptide profile is consistent with the expected behavior of inhibition of MurF within bacterial cells from studies on conditional lethal MurF mutants (15, 26). Thus, 4-PP would appear to be the first reported inhibitor of the MurF enzyme displaying an effect on the pool of cell wall precursors.
Although several MurF inhibitors with IC50 values as low as 22 nM against purified enzyme have been reported, no antibacterial activity was observed, despite efforts to render cells permeable with EDTA or nisin, and the use of efflux-defective E. coli, leading the authors to speculate that MurF might not catalyze a rate-limiting step of cell wall biosynthesis (11). In contrast, our results suggest that it is feasible to inhibit MurF within E. coli, as demonstrated by the altered muropeptide profile. Similar to cycloserine-treated cells and to a MurF deletion mutant of E. coli (15), 4-PP treatment resulted in cell lysis and a 5-log10-unit reduction in CFU. An important question is whether inhibition of MurF is actually responsible for the antibacterial activity of 4-PP. The effect of 4-PP on muropeptide profiles, with a 50% decrease in the amount of UDP-MurNAc-pentapeptide, was less pronounced than the action of cycloserine, which resulted in the near absence of detectable pentapeptide and a concomitant increase in tripeptide. However, for a conditional MurF deletion mutant of S. aureus (26), even a two- to threefold reduction in the level of UDP-MurNAc-pentapeptide was sufficient to interfere with cell growth. We cannot exclude the possibility that the antibacterial activity of 4-PP could be due to multiple mechanisms of action, one of which may be inhibition of MurF.
Our data suggest that permeability of 4-PP into E. coli was a problem which could be circumvented (for proof of the principle of the utility of MurF inhibitors) either by the use of an LPS-defective strain or by use of the permeabilizing agent PMBN. In fact, with PMBN, the efficacy of 4-PP against wild-type E. coli strains was superior to that of the cell wall synthesis inhibitor cycloserine, with 4-PP displaying MICs two- to fourfold lower than cycloserine. In the absence of PMBN, 4-PP also exhibited eightfold-lower MICs compared to the MIC for cycloserine for LPS-defective E. coli and for E. faecalis and two- to fourfold-lower MICs against S. aureus (both methicillin-susceptible and -resistant strains) and E. faecium. Possible contributions of efflux mechanisms to high MICs (>32 μg/ml) against gram-negative bacteria were not examined.
The observation that in the primary HTS assay, only about 0.01% (17/200,000) of compounds bound to MurF but that in the pharmacophore-selected set of compounds, 14% (182/1,300) bound suggests that pharmacophore modeling was an effective means to enrich for compounds with affinity for MurF. However, it is evident that binding to MurF does not necessarily cause inhibition of MurF enzymatic activity. From the primary HTS, only 6% of compounds that bound to MurF (1/17) also inhibited MurF enzymatic activity. This low frequency of binders which are also enzyme inhibitory is probably not a limitation of the ThermoFluor technique, as another binding detection method, capillary electrophoresis (2; data not shown) yielded similar results. The pharmacophore-selected set of compounds showed a stronger correlation between MurF binding and enzyme inhibition compared to the general set of diverse compounds screened in the primary HTS, with 37% (68/182) of the binders also inhibiting the enzyme, again suggesting that pharmacophore modeling enriched for compounds with MurF affinity.
The finding that MurF binding does not necessarily result in MurF inhibition may suggest that the compound binds to the protein in such a way that does not affect enzymatic activity, e.g., outside of the active site and may explain the lack of correlation between ΔTm and MurF IC50 or Kd values (Table 1). The crystal structures of MurF from E. coli (30) and S. pneumoniae (14) indicate that these are monomeric enzymes with multiple domains; it is possible that some of the binding compounds alter domain interactions, stabilizing the protein without inhibiting the enzyme. Since the ThermoFluor assay identified compounds that produced an increase in MurF Tm, presumably by binding to and stabilizing the protein, it is unlikely that the compounds that inhibited MurF activity act by causing protein denaturation, since denaturation should decrease the MurF melting temperature.
The binding assay was performed in the presence of ATP. As noted previously (2), ATP was included since it binds to MurF first, inducing a conformational change that facilitates the binding of the other substrates, UDP-MurNAc-tripeptide and d-Ala-d-Ala (1, 25, 30). The inclusion of ATP might allow compounds to bind to these latter two sites and might also serve as a filter to exclude nonspecific compounds that bind to the ATP site of MurF and potentially of other ATP-utilizing enzymes. The validity of this approach is supported by the determination of apparent Kd values in the presence and absence of ATP. A compound that is competitive with ATP is expected to have a higher apparent Kd in the presence of ATP. Most (six/eight) of the 8-hydroxyquinolines tested in this study did not appear to compete with ATP. In fact, compounds 3 and 4 seemed to bind more tightly to MurF in the presence of ATP. We note that performing the binding assay in the presence of ATP could introduce a complication in interpreting the results, especially in the primary HTS: ATP causes an increase in MurF Tm of 1.5°C. If a compound binds to MurF and displaces ATP, the net ΔTm would therefore be decreased by 1.5°C, possibly masking the effect of the compound.
In summary, the 4-PP derivative that appeared to inhibit MurF within whole cells of E. coli has been identified. Although permeability into E. coli apparently restricted the compound's activity to an LPS-defective strain and to wild-type E. coli strains rendered permeable by sub-MIC levels of PMBN, 4-PP provides proof of the principle that a MurF inhibitor might serve as an antibacterial agent. Efforts to identify additional inhibitors of MurF with greater potency and a broader spectrum of activity could provide compounds suitable for future consideration as antibiotics.
Acknowledgments
We thank Ellyn Wira, Yan Kong, and Yan Wang for performing microbiology or ThermoFluor studies. We thank Darren Abbanat for useful comments on the manuscript.
Footnotes
Published ahead of print on 1 October 2007.
REFERENCES
- 1.Anderson, M. S., S. S. Eveland, H. R. Onishi, and D. L. Pompliano. 1996. Kinetic mechanism of the Escherichia coli UDPMurNAc-tripeptide d-alanyl-d-alanine-adding enzyme: use of a glutathione S-transferase fusion. Biochemistry 35:16264-16269. [DOI] [PubMed] [Google Scholar]
- 2.Baum, E. Z., S. M. Crespo-Carbone, D. Abbanat, B. Foleno, A. Maden, R. Goldschmidt, and K. Bush. 2006. Utility of muropeptide ligase for identification of inhibitors of the cell wall biosynthesis enzyme MurF. Antimicrob. Agents Chemother. 50:230-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clement, O. O., and A. T. Mehl. 1999. HipHop: pharmacophores based on multiple common-feature alignments, p. 69-84. In O. F. Guner (ed.), Pharmacophore perception, development, and use in drug design. International University Line, La Jolla, CA.
- 4.Crespo-Carbone, S. M., A. Klinger, Y. Wang, B. Foleno, E. Wira, K. Bush, and E. Z. Baum. 2005. ThermoFluor-based identification of inhibitors of MurF, an essential bacterial cell wall synthesis enzyme. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-1849.
- 5.Crespo-Carbone, S. M., I. Turchi, B. Foleno, Y. Kong, E. Wira, M. Macielag, K. Bush, and E. Z. Baum. 2006. A MurF inhibitor which disrupts biosynthesis of the bacterial cell wall. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F2-1171.
- 6.Duncan, K., J. Van Heijenoort, and C. T. Walsh. 1990. Purification and characterization of the d-alanyl-d-alanine-adding enzyme from Escherichia coli. Biochemistry 29:2379-2386. [DOI] [PubMed] [Google Scholar]
- 7.El Zoeiby, A., F. Sanschagrin, P. C. Havugimana, A. Garnier, and R. C. Levesque. 2001. In vitro reconstruction of the biosynthetic pathway of peptidoglycan cytoplasmic precursor in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 201:229-235. [DOI] [PubMed] [Google Scholar]
- 8.El Zoeiby, A., F. Sanschagrin, and R. C. Levesque. 2003. Structure and function of the Mur enzymes: development of novel inhibitors. Mol. Microbiol. 47:1-12. [DOI] [PubMed] [Google Scholar]
- 9.Fraser, R. S., and J. Creanor. 1975. The mechanism of inhibition of ribonucleic acid synthesis by 8-hydroxyquinoline and the antibiotic lomofungin. Biochem. J. 147:401-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green, D. W. 2002. The bacterial cell wall as a source of antibacterial targets. Exp. Opin. Ther. Targets 6:1-19. [DOI] [PubMed] [Google Scholar]
- 11.Gu, Y. G., A. S. Florjancic, R. F. Clark, T. Zhang, C. S. Cooper, D. D. Anderson, C. G. Lerner, J. O. McCall, Y. Cai, C. L. Black-Schaefer, G. F. Stamper, P. J. Hajduk, and B. A. Beutel. 2004. Structure-activity relationships of novel potent MurF inhibitors. Bioorg. Med. Chem. Lett. 14:267-270. [DOI] [PubMed] [Google Scholar]
- 12.Kahan, F. M., J. S. Kahan, P. J. Cassidy, and H. Kropp. 1974. Mechanism of action of fosfomycin (phosphonomycin). Ann. N. Y. Acad. Sci. 235:364-386. [DOI] [PubMed] [Google Scholar]
- 13.Klinger, A. L., D. F. McComsey, V. Smith-Swintosky, R. P. Shank, and B. E. Maryanoff. 2006. Inhibition of carbonic anhydrase-II by sulfamate and sulfamide groups: an investigation involving direct thermodynamic binding measurements. J. Med. Chem. 49:3496-3500. [DOI] [PubMed] [Google Scholar]
- 14.Longenecker, K. L., G. F. Stamper, P. J. Hajduk, E. H. Fry, C. G. Jakob, J. E. Harlan, R. Edalji, D. M. Bartley, K. A. Walter, L. R. Solomon, T. F. Holzman, Y. G. Gu, C. G. Lerner, B. A. Beutel, and V. S. Stoll. 2005. Structure of MurF from Streptococcus pneumoniae co-crystallized with a small molecule inhibitor exhibits interdomain closure. Protein Sci. 14:3039-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lugtenberg, E. J. J., and A. Van Schijndel-Van Dam. 1972. Temperature-sensitive mutants of Escherichia coli K-12 with low activities of the l-alanine adding enzyme and the d-alanyl-d-alanine adding enzyme. J. Bacteriol. 110:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, D. J., S. M. Hammond, D. Anderluzzi, and T. D. H. Bugg. 1998. Aminoalkylphosphinate inhibitors of d-Ala-d-Ala adding enzyme. J. Chem. Soc. Perkin Trans. 1:131-142. [Google Scholar]
- 17.NCCLS/CLSI. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M7-A6. NCCLS/CLSI, Wayne, PA.
- 18.Pantoliano, M. W., E. C. Petrella, J. D. Kwasnoski, V. S. Lobanov, J. Myslik, E. Graf, T. Carver, E. Asel, B. A. Springer, P. Lane, and F. R. Salemme. 2001. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screening 6:429-440. [DOI] [PubMed] [Google Scholar]
- 19.Reddy, S. G., S. T. Waddell, D. W. Kuo, K. K. Wong, and D. L. Pompliano. 1999. Preparative enzymatic synthesis and characterization of the cytoplasmic intermediates of murein biosynthesis. J. Am. Chem. Soc. 121:1175-1178. [Google Scholar]
- 20.Reitz, R. H., H. D. Slade, and F. C. Neuhaus. 1967. Biochemical mechanisms of resistance by streptococci to the antibiotics d-cycloserine and O-carbamyl-d-serine. Biochemistry 6:2561-2570. [DOI] [PubMed] [Google Scholar]
- 21.Rohde, W., B. Cordell, R. Webster, and W. Levinson. 1977. Inhibition of amino acyl tRNA synthetase activity by copper complexes of two metal binding ligands. N-methyl isatin beta-thiosemicarbazone and 8-hydroxyquinoline. Biochim. Biophys. Acta 477:102-111. [DOI] [PubMed] [Google Scholar]
- 22.Sieradzki, K., and A. Tomasz. 1997. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J. Bacteriol. 179:2557-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silver, L. L. 2006. Does the cell wall of bacteria remain a viable source of targets for novel antibiotics? Biochem. Pharmacol. 71:996-1005. [DOI] [PubMed] [Google Scholar]
- 24.Silver, L. L. 2003. Novel inhibitors of bacterial cell wall synthesis. Curr. Opin. Microbiol. 6:431-438. [DOI] [PubMed] [Google Scholar]
- 25.Smith, C. A. 2006. Structure, function and dynamics in the mur family of bacterial cell wall ligases. J. Mol. Biol. 362:640-655. [DOI] [PubMed] [Google Scholar]
- 26.Sobral, R. G., A. M. Ludovice, H. de Lencastre, and A. Tomasz. 2006. Role of murF in cell wall biosynthesis: isolation and characterization of a murF conditional mutant of Staphylococcus aureus. J. Bacteriol. 188:2543-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsubery, H., I. Ofek, S. Cohen, and M. Fridkin. 2000. Structure-function studies of polymyxin B nonapeptide: implications to sensitization of gram-negative bacteria. J. Med. Chem. 43:3085-3092. [DOI] [PubMed] [Google Scholar]
- 28.Wargel, R. J., C. A. Shadur, and F. C. Neuhaus. 1971. Mechanism of d-cycloserine action: transport mutants for d-alanine, d-cycloserine, and glycine. J. Bacteriol. 105:1028-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong, K. K., D. W. Kuo, R. M. Chabin, C. Fournier, L. D. Gegnas, S. T. Waddell, F. Marsilio, B. Leiting, and D. L. Pompliano. 1998. Engineering a cell-free murein biosynthetic pathway: combinatorial enzymology in drug discovery. J. Am. Chem. Soc. 120:13527-13528. [Google Scholar]
- 30.Yan, Y., S. Munshi, B. Leiting, M. S. Anderson, J. Chrzas, and Z. Chen. 2000. Crystal structure of Escherichia coli UDPMurNAc-tripeptide d-alanyl-d-alanine-adding enzyme (MurF) at 2.3 Å resolution. J. Mol. Biol. 304:435-445. [DOI] [PubMed] [Google Scholar]