Abstract
Rupintrivir (formerly AG7088) is an irreversible inhibitor of the human rhinovirus (HRV) 3C protease that has been demonstrated to have in vitro activity against all HRVs tested, consistent with its interaction with a strictly conserved subset of amino acids in the 3C protease. The potential for resistance was studied following in vitro serial passage of HRV serotypes 14, 2, 39, and Hanks in the presence of increasing rupintrivir concentrations. HRV variants with reduced susceptibilities to rupintrivir (sevenfold for HRV 14) or with no significant reductions in susceptibility but genotypic changes (HRV 2, 39, and Hanks) were initially isolated following 14 to 40 cumulative days in culture (three to six passages). Sequence analysis of the 3C protease identified one to three substitutions in diverse patterns but with common features (T129T/A, T131T/A, and T143P/S in HRV 14; N165T in HRV 2; N130N/K and L136L/F in HRV 39; T130A in HRV Hanks). Notably, three of the four HRV variants contained a substitution at residue 130 (residue 129 in HRV 14). Continued selection in the presence of escalating concentrations of rupintrivir (40 to 72 days) resulted in the accumulation of additional mutations (A121A/V and Y139Y/H in HRV 14, E3E/G and A103A/V in HRV 2, S105T in HRV 39), with only minimal further reductions in susceptibility (up to fivefold). The ability of specific substitutions to confer resistance was examined by susceptibility testing of HRV 14 variants constructed to contain 3C protease mutations. In summary, the slow accumulation of multiple amino acid substitutions with only minimal to moderate reductions in susceptibility highlight the advantages of 3C protease as an antiviral target.
Human rhinoviruses (HRVs), which are members of the family Picornaviridae, comprise over 100 different serotypes and are the predominant cause of the common cold. Although HRV infections are generally mild and self-limiting, they can also be associated with more serious illnesses, specifically, exacerbation of disease in individuals with underlying respiratory disorders (1, 2, 3, 5, 6, 18, 26, 29). Although no effective antiviral therapies have been approved for the treatment of HRV infections, chemotherapeutic approaches that utilize antiviral agents directed toward viral attachment, capsid uncoating, and 3C protease have been well studied (reviewed in reference 22). Our research efforts have focused on the inhibition of the picornavirus 3C protease, an enzyme absolutely required for viral replication (15, 21).
Rupintrivir (formerly AG7088) is a novel, irreversible inhibitor of 3C protease that was discovered by using structure-based drug design methodologies and was formulated for intranasal delivery in human clinical trials (8-11, 13, 19). Parallel efforts to identify a 3C protease inhibitor that was orally bioavailable led to the discovery of compound 1 (7, 24). Similar to rupintrivir, compound 1 is an irreversible inhibitor that incorporates a Michael acceptor moiety that forms a covalent bond with the 3C protease active-site cysteine (7). In vitro, both compounds have demonstrated potent and broad-spectrum activities against all HRV serotypes, HRV clinical isolates, and enteroviruses tested (4, 14, 23, 24). Clinical studies subsequently showed that rupintrivir treatment was able to moderate the severity of illness and reduce the viral load in a human experimental HRV challenge trial (13), thus providing a proof of concept for the mechanism of 3C protease inhibition. In subsequent natural infection studies, rupintrivir was not able to significantly affect virus replication or disease severity, and the further clinical development of rupintrivir, along with that of compound 1, was terminated (unpublished data).
Previous crystallographic studies of rupintrivir bound to the HRV serotype 2 3C protease identified an extensive set of interactions, including close binding between the inhibitor and side chains of 14 amino acids of 3C protease (19). Recently, sequence analyses of 3C proteases from 38 HRV serotypes and clinical isolates demonstrated the complete conservation of 13 of these 14 amino acids (4, 19, 20). This finding is consistent with the broad-spectrum activity demonstrated by rupintrivir against all laboratory strains of HRV (48 of 48) and clinical HRV isolates (23 of 23) tested to date (14, 23).
As an extension of these studies, we investigated the potential for the development of resistance by performing in vitro serial passage of HRV serotypes 14, 2, 39, and Hanks in the presence of increasing concentrations of rupintrivir. HRV variants emerged slowly during serial passage, contained multiple mutations in diverse patterns but with common features, and in general were associated with only minimal to moderate reductions in susceptibility. Our data, which describe a high genetic barrier to resistance for rupintrivir, are discussed in the context of known rupintrivir binding interactions with 3C protease.
MATERIALS AND METHODS
Compound.
Rupintrivir was synthesized at Pfizer Global Research and Development, San Diego, CA (8).
Cells and virus strains.
HRV serotypes 2, 14, and 39 were purchased from the American Type Culture Collection (ATCC; Manassas, VA). HRV serotype Hanks was kindly provided Ronald B. Turner (Department of Pediatrics, University of Virginia School of Medicine, Charlottesville). All HRV strains were propagated in and antiviral assays were performed with H1-HeLa cells (ATCC) incubated at 34°C. The cells were grown in minimal essential medium (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (HyClone, Logan, UT).
Isolation of HRV variants in the presence of increasing compound concentrations.
H1-HeLa cells were initially infected with HRV 14, 2, 39, or Hanks at a multiplicity of infection (MOI) of 0.1 in the presence of up to 3.5 times the 50% effective concentration (EC50) of rupintrivir. Supernatants were collected when the cytopathic effect (CPE) reached at least 50%. Subsequent infections were conducted by infecting fresh cells with 0.2 to 0.5 ml of supernatant from infected cell cultures in one- to threefold higher concentrations of rupintrivir and were monitored for the progression of the CPE. All supernatants were cleared of cellular debris by low-speed centrifugation and were stored at −80°C for subsequent analysis of HRV RNA and antiviral susceptibility.
DNA sequence analysis of 3C protease.
RNA was purified from cell-free HRV lysates by silica-based extraction by the RNeasy method (Qiagen, Valencia, CA), and then cDNA was synthesized from the viral RNA by using a First Strand synthesis kit (Amersham Pharmacia Biotech, Inc., Piscataway, NJ) and random nanomers. For each strain, the target region (corresponding to bases 5116 to 5827 in HRV 89) was amplified by PCR with Taq 2000 polymerase (Stratagene, San Diego, CA). The 3C protease nucleotide sequences from HRV 14, 2, 39, Hanks, and their respective variants were determined by sequencing of the PCR products. Clonal analysis was performed with late-stage passages of each virus. The PCR amplicons for clonal analysis were subcloned into pGEM-T Easy vectors (Promega, Madison, WI) and sequenced by using primers T7 and SP6. The PCR primers used have been described previously (4) and were specific for the corresponding HRV type. The DNA sequences were analyzed, the relevant regions were translated, and alignments were generated by using DNASTAR analysis software (DNASTAR Inc., Madison, WI). The reference peptide sequences for HRV 2 wild-type (WT) (X02316 [30]) and HRV 14 WT (K02121 [31]) were from GenBank, and the peptide sequences for HRV 39 WT and HRV Hanks WT were reported previously (4).
Construction of recombinant HRV variants.
A series of transcription vectors encoding the full-length HRV 14 cDNA were created by site-directed mutagenesis by using the Quick Change mutagenesis kit (Stratagene, San Diego, CA), according to the manufacturer's directions, and complementary primers. The transcription vector template, pET24/HRV 14, encodes the full-length HRV 14 cDNA and is driven by a T7 promoter. The following primers and their exact complements were used: for variant T129A, primer 5′-GACTTATTAATTTGAGTAGCGCCCCCACTAACAGAATGATTC-3′; for variant T131A, primer 5′-GAGTAGCACCCCCGCTAACAGAATGATTCGTTATG-3′; for variant T143P, primer 5′-CGTTATGATTATGCAACAAAACCTGGGCAGTGTGGAGGTGTG-3′; for variant Y139H, primer 5′-CGTTATGATCATGCAACAAAAACTGGGCAGTGTGG-3′; for variant T129A/T131A/T143P, primer 5′-GACTTATTAATTTGAGTAGCACCCCCACTAACAGAAT GATTC-3′ with variant T143P used as the template; and for variant T129A/T131A/Y139H/T143P, primer 5′-CGTTATGATCATGCAACAAAACCTGGGCAGTGTGG-3′, with variant T129A/T131A/T143P used as the template.
The coordinates for the primer locations in reference to the sequence of HRV 14 for the HRV resistance-associated mutations are 5604 to 5645 (T129A), 5617 to 5651 (T131A), 5645 to 5686 (T143P), 5645 to 5679 (Y139H), 5604 to 5645 (T129A/T131A/T143P), and 5645 to 5679 (T129A/T131A/Y139H/T143P). Viral RNA was subsequently transcribed by using a MEGAscript transcription kit (Ambion, Austin, TX) and was transfected into subconfluent H1-HeLa cells by using lipofectamine (Invitrogen, Carlsbad, CA), and the supernatants were harvested after a visible CPE was observed.
Susceptibility assays.
Reductions in susceptibility to rupintrivir were determined in standard CPE inhibition assays by using the 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide (XTT) dye reduction method (28, 32) or in modified CPE inhibition assays (14, 33). In the standard CPE assay, H1-HeLa cells were initially suspended at a final concentration of 2 × 105 cells per ml and infected with virus at MOIs of 0.08, 0.1, 0.1, and 0.2 for HRV 14, HRV 2, HRV 39, and HRV Hanks, respectively. Three days (HRV 14, HRV 2, and HRV 39) or 4 days (HRV Hanks) later, XTT and phenazine methosulfate (PMS) were added to the test plates and the amount of formazan produced was quantified spectrophotometrically at 450 nm, with adjustment for the background at 650 nm. Data are expressed as the percentage of formazan produced in compound-treated cells compared to the amount of formazan produced in wells of uninfected, compound-free cells. The EC50 was calculated as the concentration of compound that increased the percentage of formazan production in infected, compound-treated cells to 50% of that produced by uninfected, compound-free cells. In the modified CPE assay, H1-HeLa cells were incubated with six serial 10-fold dilutions of HRV in quadruplicate to determine a 50% tissue culture infectious dose (TCID50) and in parallel at three 10-fold virus dilutions in the presence of compound. The EC50 values were determined by utilizing the inoculum dilution for which the calculated TCID50/ml ranged between 32 and 320. XTT and PMS were added to the test plates, and the EC50 values were calculated as described above.
Statistical analysis.
Antiviral susceptibility data were assessed for significant differences between HRV variants and WT HRV by determining the probability associated with the Student's t test for two-tailed, equal-variance analyses. P values were deemed significant for data sets with P values of ≤0.05.
RESULTS
In vitro selection of HRV variants with reduced susceptibilities to rupintrivir.
HRV variants were selecting by serial passaging WT HRV 14, 2, 39, and Hanks in the presence of increasing concentrations of rupintrivir (e.g., up to 3- to 90-fold above the WT EC50, which consisted of rupintrivir concentrations of 60 to 1,800 nM, respectively). HRV 2 and HRV 14 are representative of genetic subgroups A and B, respectively (4), and HRV 39 and Hanks are virus serotypes commonly used in experimental HRV infection models (13). The HRV variants were subsequently evaluated for phenotypic and genotypic changes (Tables 1 to 5; Fig. 1.) HRV 2 and HRV 14 were also passaged in parallel in the absence of compound as controls.
TABLE 1.
In vitro serial passage of HRV 14 in the presence of rupintrivir
| Passage historya
|
Phenotypeb as EC50 (nM [fold change]) | Genotypec as amino acid substitutions | ||
|---|---|---|---|---|
| Passage no. | No. of days (cumulative) | >Fold EC50 (concn [nM]) | ||
| WT | 0 | 0 | 21 ± 11 | — |
| 1 | 6 | 3.5 (70) | ND | — |
| 2 | 11 | 3.5 (70) | ND | — |
| 3 | 14 | 5 (100) | 152 ± 108 (7)d | T129T/A, T131T/A, T143P/S |
| 4 | 17 | 10 (200 | 227 ± 124 (11)d | T129A, T131T/A, T143P |
| 5 | 22 | 20 (400) | 96 ± 16 (5)d | T129A, T131T/A, T143P |
| 6 | 27 | 20 (400) | ND | T129A, T131T/A, T143P |
| 7 | 33 | 30 (600) | ND | T129A, T131T/A, T143P |
| 8 | 40 | 40 (800) | ND | T129A, T131T/A, T143P |
| 9 | 48 | 60 (1,200) | ND | T129A, T131A, T143P, Y139Y/H |
| 10 | 55 | 90 (1,800) | 324 ± 170 (15)d | T129A, T131A, T143P, Y139Y/H |
| 11 | 65 | 90 (1,800) | 335 ± 60 (16)d | A121A/V, T129A, T131A, T143P, Y139Y/H |
| 12 | 72 | 90 (1,800) | 414 ± 92 (20)d | A121A/V, T129A, T131A, N132S, Y139Y/H, T143P |
HRV 14 was serially passaged for the indicated number of times (passage number) or days in the presence of increasing concentrations of rupintrivir, as indicated.
The results represent the means ± the standard deviations from three or more experiments, as determined by CPE assays as described in Materials and Methods. ND, not determined. Values in parentheses (fold change) represent the ratio of the EC50 value obtained for mutant HRV compared to the EC50 value obtained for WT HRV 14.
All amino acid substitutions in the 3C protease-coding region relative to an HRV 14 reference sequence are reported. —, no change relative to the reference sequence.
P ≤ 0.05 by comparison of antiviral susceptibility data between the indicated HRV variants and WT HRV.
TABLE 5.
In vitro serial passage of HRV Hanks in the presence of rupintrivir
| Passage historya
|
Phenotypeb as EC50 (nM [fold change]) | Genotypec as amino acid substitution | ||
|---|---|---|---|---|
| Passage no. | No. of days (cumulative) | >Fold EC50 (concn [nM]) | ||
| WT | 0 | 0 | 19 ± 9 | — |
| 1 | 7 | 1 (26) | ND | — |
| 2 | 17 | 2 (40) | ND | — |
| 3 | 35 | 2 (40) | ND | — |
| 6 | 40 | 3 (60) | 51 ± 27 (3) | T130A |
| 9 | 59 | 3 (60) | ND | T130A |
HRV Hanks was serially passaged for the indicated number of times (passage number) or days in the presence of increasing concentrations of rupintrivir, as indicated.
The results represent the means ± the standard deviations from three or more experiments or individual values from independent experiments. ND, not determined. EC50 and EC90 values were determined by modified CPE assays, as described in Materials and Methods, or by plaque reduction assay. Values in parentheses (fold change) represent the ratio of the EC50 or the EC90 of passaged virus to the EC50 or the EC90 of WT HRV.
All amino acid substitutions in the 3C protease-coding region relative to an HRV Hanks reference sequence are reported. —, no change relative to the reference sequence.
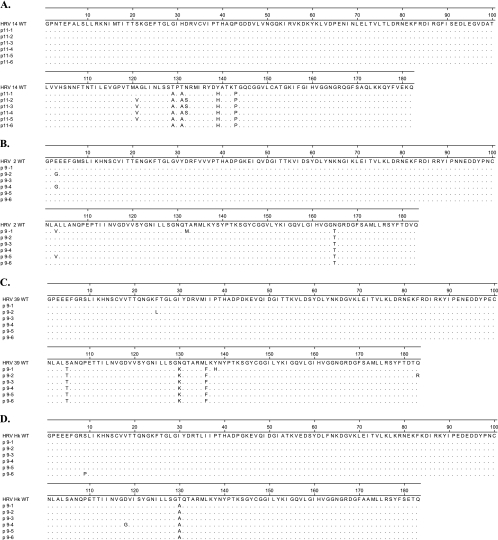
FIG. 1.
Deduced amino acid sequence of 3C proteases from clones of HRV 14 (A), HRV 2 (B), HRV 39 (C), and HRV Hanks (D) isolated following 11, 9, 9, and 9 passages, respectively. Dots indicate no change from the sequence of the HRV WT.
An HRV 14 variant with reduced susceptibility to rupintrivir was initially identified at passage 3 (p3) following 14 cumulative days in culture (Table 1). Genotypic analysis of HRV 14 at p3 demonstrated three amino acid substitutions of T129T/A, T131T/A, and T143P/S. Phenotypic analysis demonstrated a moderate reduction in susceptibility (sevenfold) to rupintrivir (Table 1) with no significant reduction in susceptibility to pleconaril, an inhibitor of capsid binding/uncoating (data not shown). Subsequent passage in the presence of increasing concentrations of rupintrivir for periods up to 72 days led to the selection of HRV 14 variants characterized by the sequential accumulation of three additional substitutions of Y139Y/H, A121A/V, and N132S, but with only a 3-fold greater reduction in susceptibility compared to that of HRV 14 at p3 (20- and 7-fold changes for the HRV 14 variant at p12 and p3, respectively). In parallel experiments, no amino acid substitutions developed in an HRV 14 WT strain passaged in the absence of compound for 12 passages over 48 days (data not shown).
To relate the specific amino acid changes observed to rupintrivir resistance, we performed susceptibility assays with HRV 14 variants constructed to contain HRV 14-specific amino acid substitutions in the 3C protease (Table 2). No reduction in susceptibility was observed for any HRV 14 variant containing only a single amino acid substitution (T129A, T131A, T143P, or Y139H). In contrast, 7- to 16-fold reductions were observed for an HRV 14 variant containing all four mutations. No reductions in susceptibility to the control compound, pleconaril, were observed for any HRV 14 variant tested (data not shown). Although variants were not constructed to contain specific combinations of double and triple mutations, on the basis of the results of the linkage experiments described below, it can be presumed that the reductions in susceptibilities described for isolated HRV variants in Table 1 should approximate the levels of reductions for the HRV variants constructed.
TABLE 2.
In vitro susceptibilities of HRV 14 variants to rupintrivira
| HRV 14 variant | Phenotype
|
|
|---|---|---|
| EC50 (nM)b | Fold changec | |
| WT | 52, 17 | |
| T129A | 46, 14 | 1, 1 |
| T131A | 32, 14 | 1, 1 |
| Y139H | 14, 18 | 1, 1 |
| T143P | 36, 15 | 1, 1 |
| T129A/T131A/Y139H/T143P | 359, 268 | 7, 16 |
HRV 14 WT cDNA was subjected to site-directed mutagenesis to generate a panel of HRV 14 variants with rupintrivir resistance-associated amino acid substitutions, as described in Material and Methods.
The results represent individual values from two independent assays. EC50 values were determined by CPE assays, as described in Materials and Methods.
The fold change represents the ratio of the EC50 value obtained for mutant HRV compared to the EC50 value obtained for WT HRV 14.
HRV 14 at p11 was further evaluated by clonal analysis to identify the linkage of specific amino acids on single RNA genomes (Fig. 1). The results indicated that mutations appeared on the same RNA molecule. Specifically, six of six clones possessed the T129A, T131A, Y139H, and T143P substitutions; four of six of the clones also had the A121V substitution, and three of those four clones had the N132S substitution that was detected in HRV 14 at p12 (Table 1).
HRV 2 variants with genotypic changes in the 3C protease but with no significant reduction in susceptibility to rupintrivir were initially identified at p4 (24 cumulative days in culture) (Table 3). Genotype analysis revealed the presence of one amino acid substitution, N165T. Similar to HRV 14, subsequent passage in the presence of increasing concentrations of rupintrivir for periods of up to 66 days led to the selection of HRV 2 variants with two additional substitutions of E3E/G and A103A/V but only a fivefold (HRV 2 p9) reduction in susceptibility to rupintrivir compared to that of the WT (Table 3). Clonal analysis of HRV 2 at p9 showed that six of six clones contained the N165T substitution and that two of six clones had either A103V or E3G but not both (Fig. 1). One of two clones with A103V also contained a change not detected by population sequencing, e.g., T132M. As with HRV 14, no amino acid substitutions were detected in an HRV 2 WT strain passaged in parallel in the absence of compound for 12 passages over 53 days (data not shown).
TABLE 3.
In vitro serial passage of HRV 2 in the presence of rupintrivir
| Passage historya
|
Phenotypeb as EC50 (nM [fold change]) | Genotypec as amino acid substitution(s) | ||
|---|---|---|---|---|
| Passage no. | No. of days (cumulative) | >Fold EC50 (concn [nM]) | ||
| WT | 0 | 0 | 30 ± 9 | — |
| 1 | 6 | 1 (30) | ND | — |
| 2 | 10 | 1 (30) | ND | — |
| 3 | 17 | 3 (100) | 50, 34 (2, 1) | — |
| 4 | 24 | 5 (150) | 44, 46 (2, 2) | N165T |
| 5 | 30 | 5 (150) | 57 ± 24 (2) | N165T |
| 6 | 37 | 5 (150) | 67 (2) | N165T |
| 7 | 45 | 5 (150) | ND | A103A/V, N165T |
| 8 | 52 | 15 (450) | ND | A103A/V, N165T |
| 9 | 62 | 15 (450) | 140 ± 63 (5)d | E3E/G, A103A/V, N165T |
| 10 | 66 | 15 (450) | ND | E3E/G, A103A/V, N165T |
HRV 2 was serially passaged for the indicated number of times (passage number) or days in the presence of increasing concentrations of rupintrivir, as indicated.
The results represent the means ± the standard deviations from three or more experiments or individual values from independent experiments, as determined by CPE assays as described in Materials and Methods. ND, not determined. Values in parentheses (fold change) represent the ratio of the EC50 value obtained for mutant HRV compared to the EC50 value obtained for WT HRV 2.
All amino acid substitutions in the 3C protease-coding region relative to an HRV 2 reference sequence are reported. —, no change relative to the reference sequence.
P ≤ 0.05 by comparison of antiviral susceptibility data between the indicated HRV variant and WT HRV.
HRV 39 variants containing genotypic changes in the 3C protease were initially isolated at p4 following 24 cumulative days in culture (Table 4). Genotype analyses revealed two amino acid substitutions of N130N/K and L136L/F that conferred a threefold reduction in susceptibility to rupintrivir (HRV 39 at p5). Subsequent passage in the presence of increasing concentrations of rupintrivir for periods up to 50 days led to the selection of an HRV 39 variant with one additional substitution of S105T but with no further reduction in susceptibility to rupintrivir (fourfold for HRV 39 at p9). HRV 39 replicated poorly in the presence of increasing concentrations of rupintrivir, with the recovery of replication-competent virus possible only at concentrations of rupintrivir less than sixfold above the WT EC50 (150 nM) (Table 4). Clonal analysis of HRV 39 at p9 confirmed that six of six clones had S105T, N130K, and L136F (Fig. 1). Additional substitutions not detected by population-based sequencing included Y138H in one of six clones and F25L and Q183R in one of six clones.
TABLE 4.
In vitro serial passage of HRV 39 in the presence of rupintrivir
| Passage historya
|
Phenotypeb as EC50 (nM [fold change]) | Genotypec as amino acid substitutions | ||
|---|---|---|---|---|
| Passage no. | No. of days (cumulative) | >Fold EC50 (concn [nM]) | ||
| WT | 0 | 0 | 24 ± 5 | — |
| 1 | 7 | 3 (74) | ND | — |
| 2 | 12 | 4 (100) | ND | — |
| 3 | 14 | 6 (150) | ND | — |
| 4 | 24 | 6 (150) | ND | N130N/K, L136L/F |
| 5 | 29 | 6 (150) | 83 ± 18 (3)d | N130N/K, L136L/F |
| 6 | 33 | 6 (150) | ND | S105S/T, N130K, L136F |
| 9 | 50 | 6 (150) | 94 ± 13 (4)d | S105T, N130K, L136F |
HRV 39 was serially passaged for the indicated number of times (passage number) or days in the presence of increasing concentrations of rupintrivir, as indicated.
The results represent the means ± the standard deviations from three or more experiments, as determined by CPE assays as described in Materials and Methods. ND, not determined. Values in parentheses (fold change) represent the ratio of the EC50 value obtained for mutant HRV compared to the EC50 value obtained for WT HRV 39.
All amino acid substitutions in the 3C protease-coding region relative to an HRV 39 reference sequence are reported. —, no change relative to the reference sequence.
P ≤ 0.05 by comparison of antiviral susceptibility data between the indicated HRV variants and WT HRV.
HRV Hanks variants with genotypic changes (T130A) in the 3C protease but with no significant reduction in susceptibility to rupintrivir were initially identified at p6 (40 cumulative days in culture) (Table 5). Similar to HRV 39, HRV Hanks replicated poorly in the presence of escalating concentrations of compound; isolation of virus in the presence of concentrations of rupintrivir greater than threefold above the WT EC50 (60 nM) could not be achieved at up to nine passages during 59 days of continued culture. Clonal analysis of the HRV Hanks variant at p9 demonstrated a T130A substitution in six of six clones and a S9P or D118G substitution in one of six clones (Fig. 1).
Stability of genotypic changes and viral fitness.
To evaluate the stability of the genotypic changes associated with reduced susceptibility to rupintrivir, variants of HRV 14 at p11 and HRV 2 at p9 were propagated in the absence of compound for eight passages for 41 to 43 days. Sequence analysis of HRV 14 at p11 after eight passages without compound revealed no changes from the sequence of HRV 14 at p11 (genotype A121A/V, T129A, T131A, Y139Y/H, T143P; data not shown), suggesting that these mutations did not significantly affect the overall fitness of HRV 14. This finding is consistent with those of in vitro studies that evaluated the ability of an earlier HRV variant that contained three mutations (HRV 14 at p5) to produce a CPE. In these experiments, HRV 14 WT and HRV 14 at p5 exhibited the same percentage of cell killing (90.2 and 88.2%, respectively) 3 days following infection at an MOI of 0.02 (data not shown).
Sequence analysis of HRV 2 at p9 after eight passages without compound revealed two alterations compared to the sequence of HRV 2 at p9 (the genotype E3E/G, A103A/V, N165T became E3G, A103A, N165T; data not shown). The reversion of V/A103 to the WT (A103) suggests that A103 is the preferable amino acid and may be associated with enhanced fitness.
DISCUSSION
In this study, we performed in vitro serial passage studies with four different HRV serotypes to identify the phenotypic and genotypic changes that confer resistance to rupintrivir. The pathways for HRV resistance were characterized by the sequential accumulation of multiple mutations in diverse patterns. In general, these mutations were associated with only minimal to moderate reductions in susceptibility (fivefold or less for HRV 2, 39, and Hanks up to 62 days; sevenfold for HRV 14 up to 14 days). Subsequent serial passage of the HRV 14 variant for 72 days resulted in the accumulation of three additional mutations which conferred less than a threefold additional decrease in susceptibility to rupintrivir. Furthermore, the difficulties in increasing the selective pressure for two of the four virus strains (e.g., HRV Hanks and 39 replicated poorly or not at all in the presence of concentrations of rupintrivir greater than three- and sixfold above the EC50, respectively) imply that additional mutations necessary for subsequent virus replication are poorly tolerated. The general requirement for an accumulation of multiple substitutions over extended periods of time to achieve only moderate reductions in susceptibility suggests that there is a high genetic barrier to the development of resistance within the 3C protease for rupintrivir. Although the clinical relevance of these changes are not known, it is noteworthy that the time taken to generate initial HRV variants (14 to 40 days) exceeds the therapeutic dosing interval that would be anticipated for the treatment of acute HRV-induced illness (<7 days).
Although sequence analysis detected different genotypic patterns for each virus studied, the crystal structure of rupintrivir with HRV 2 that has been solved (19) provides important insights into the in vitro resistance profiles and highlights the common features among the different HRV serotypes described in this study. That study (19) described the interaction of rupintrivir with 23 different amino acid residues located in the substrate (inhibitor) binding pocket of the HRV 2 3C protease. Nine of the 23 amino acids interact with rupintrivir via the polypeptide backbone (including amino acid residues 22, 23, 24, 126, 143, 144, 145, 146, and 162). Substitutions at these amino acids would not be predicted to have significant effects on overall rupintrivir binding. In this regard, only one of the substitutions involved in the main chain interactions was selected in this study (e.g., T143P in HRV 14, corresponding to amino acid residue 144 in HRV 2). This is also consistent with data that showed no significant reductions in susceptibility to an HRV 14 variant constructed to contain the 143P change.
Fourteen of the 23 amino acid residues of the 3C protease involved in rupintrivir binding interact via their side chains (including 25, 40, 71, 125, 127, 128, 130, 142, 147, 161, 163, 164, 165, and 170). Presumably, amino acid substitutions at these residues would be predicted to have a greater potential impact on 3C protease-rupintrivir binding. Sequence analysis of the 3C proteases from 38 HRV serotypes and clinical isolates revealed that 13 of 14 amino acids with side chain interactions with rupintrivir are strictly conserved, further highlighting the significance of this subset of amino acids (4, 19, 20). In this regard, substitutions at two of these amino acid residues were identified during this study. Specifically, a substitution at residue 130 (T129A in HRV 14) occurred in three of the four HRVs studied (HRVs 14, 39, and Hanks), and a substitution at residue 165 occurred in one of the four HRVs studied (HRV 2). These mutations did not appear to be dictated by the HRV genetic subgroup, as shown by the presence of the change at residue 130 for both subgroup A (HRV 39, HRV Hanks) and subgroup B (HRV 14) serotypes and a change at residue 165 for a subgroup A serotype (HRV 2). Furthermore, the occurrence of one or the other but not both of these substitutions suggests that they may potentially be exclusive of each other. Analysis of resistance with a greater number of HRV serotypes would be required to investigate this relationship. Notably, neither change alone resulted in a significant reduction in susceptibility to rupintrivir. This could be explained by preservation of the binding interactions that occur with other amino acid residues located in the same substrate binding pockets. Specifically, residue 130 is located in the S2 pocket along with H40, E71, L127, and S128; and residue N165 resides in the S4 pocket along with I125, L126 or N126, L127, S128, G164, and F170 (19). Interestingly, the only 3C protease amino acid residue with side chain interactions with rupintrivir that is not absolutely conserved was 130 (4). Data from our study identifying this as a common substitution for three of the four strains implies that HRV variants may be more tolerant of substitutions at this residue. This is also consistent with data that show that changes can occur at this residue with little overall effect on susceptibility and broad-spectrum activity (4).
Other mutations that have not been described as having either main or side chain interactions with rupintrivir were identified during this study. The significance of these changes is unknown, although a role in compensating for the growth and the overall fitness of the virus could be presumed. This is consistent with little to no further reductions in susceptibility in the presence of additional mutations, as well as the findings of reversion studies, which demonstrated the stability of mutations in HRV variants passaged in the absence of drug (with the exception of a change at residue 103).
Studies describing the generation of in vitro resistance of picornaviruses to other antiviral agents have been limited. Most information to date has been generated for pleconaril, an orally bioavailable inhibitor of picornavirus that acts by binding to viral capsid and preventing attachment and/or uncoating (reviewed in reference 22). Pleconaril had progressed through phase III human clinical trials but was later denied for approval by the U.S. Food and Drug Administration, primarily on the basis of drug interactions, its marginal efficacy, and the possibility of transmission of resistant virus. In contrast to antiviral agents directed against the 3C protease (rupintrivir and compound 1 [4, 13, 23]), pleconaril and other agents that target capsid binding exhibit a wide range of susceptibilities and activities against most but not all picornavirus tested (14, 17, 23). In a recent study, Ledford et al. (17) demonstrated that 7 of 25 HRV B serotypes not inhibited by pleconaril (a >625 difference in susceptibility) had VP1 sequences that differed from those of susceptible strains at two positions (Y152F and V191L). In a subsequent study, V191L was shown to be sufficient for significant resistance to pleconaril (a 31-fold reduction in susceptibility) (16). The clinical relevance for these findings was recently demonstrated, whereby patients with baseline isolates that had greater susceptibility to pleconaril experienced lower rates of recovery of virus posttreatment and a greater reduction in the number of days to symptom resolution than patients with baseline isolates that were less susceptible to pleconaril. The latter patients experienced no benefit from drug treatment (25). In the same study, Pevear et al. (25) describe the emergence of HRV variants with a >10-fold reduction in susceptibility in 10.7% of patients during 3 to 5 days of treatment with pleconaril. The emergent resistance phenotype was associated with either I98M/F or I122F/L (17). These results are consistent with the findings from studies on the in vitro selection of coxsackievirus type B3 variants that exhibited a single substitution (I92M/L, corresponding to I98 in the HRV 16 numbering) sufficient to confer high levels of resistance (a >200-fold reduction in susceptibility) to pleconaril (12, 27).
In contrast to a compound that targets the capsid binding function (e.g., pleconaril), in which single amino acid substitutions are sufficient to confer significant reductions in or a complete loss of susceptibility, our results show that rupintrivir-resistant HRV variants contain single or multiple amino acid substitutions that in general confer little to only moderate reductions in susceptibility. These results are consistent with the strict conservation of those amino acids that have side chain interactions with rupintrivir and the broad-spectrum activity of rupintrivir against all picornaviruses tested to date. These data further highlight the 3C protease an attractive target for antiviral intervention.
Acknowledgments
We thank George Smith III for his critical review of the manuscript and Robert Love for his discussions on the structural implications of 3C protease mutations on interactions with rupintrivir.
Footnotes
Published ahead of print on 1 October 2007.
REFERENCES
- 1.Arden, K. E., P. McErlean, M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2006. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J. Med. Virol. 78:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arruda, E., A. Pitkaranta, T. J. Witek, Jr., C. A. Doyle, and F. G. Hayden. 1997. Frequency and natural history of rhinovirus infections in adults during autumn. J. Clin. Microbiol. 35:2864-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atmar, R. L., E. Guy, K. K. Guntupalli, J. L. Zimmerman, V. D. Bandi, B. D. Baxter, and S. B. Greenberg. 1998. Respiratory tract viral infections in inner-city asthmatic adults. Arch. Intern. Med. 158:2453-2459. [DOI] [PubMed] [Google Scholar]
- 4.Binford, S. L., F. Maldonado, M. A. Brothers, P. T. Weady, L. S. Zalman, J. W. Meador III, D. A. Matthews, and A. K. Patick. 2005. Conservation of amino acids in human rhinovirus 3C protease correlates with broad-spectrum antiviral activity of rupintrivir, a novel human rhinovirus 3C protease inhibitor. Antimicrob. Agents Chemother. 49:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collinson, J., K. G. Nicholson, E. Cancio, J. Ashman, D. C. Ireland, V. Hammersley, J. Kent, and C. O'Callaghan. 1996. Effects of upper respiratory tract infections in patients with cystic fibrosis. Thorax 51:1115-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couch, R. B. 2001. Rhinoviruses, p. 777-797. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology. Lippincott Williams & Wilkins, New York, NY.
- 7.Dragovich, P. S., T. J. Prins, R. Zhou, T. O. Johnson, Y. Hua, H. T. Luu, S. K. Sakata, E. L. Brown, F. C. Maldonado, T. Tuntland, C. A. Lee, S. A. Fuhrman, L. S. Zalman, A. K. Patick, D. A. Matthews, E. Y. Wu, M. Guo, B. C. Borer, N. K. Nayyar, T. Moran, L. Chen, M. C. Guzman, E. Z. Dovalsantos, S. Lee, K. McGee, M. Mohajeri, A. Liese, J. Tao, M. B. Kosa, B. Liu, M. R. Batugo, J. R. Gleeson, Z. P. Wu, J. Liu, J. W. Meador III, and R. A. Ferre. 2003. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 8. Pharmacological optimization of orally bioavailable 2-pyridone-containing peptidomimetics. J. Med. Chem. 46:4572-4585. [DOI] [PubMed] [Google Scholar]
- 8.Dragovich, P. S., T. J. Prins, R. Zhou, S. E. Webber, J. T. Marakovits, S. A. Fuhrman, A. K. Patick, D. A. Matthews, C. A. Lee, C. E. Ford, B. J. Burke, P. A. Rejto, T. F. Hendrickson, T. Tuntland, E. L. Brown, J. W. Meador III, R. A. Ferre, J. E. Harr, M. B. Kosa, and S. T. Worland. 1999. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 4. Incorporation of P1 lactam moieties as l-glutamine replacements. J. Med. Chem. 42:1213-1224. [DOI] [PubMed] [Google Scholar]
- 9.Dragovich, P. S., S. E. Webber, R. E. Babine, S. A. Fuhrman, A. K. Patick, D. A. Matthews, C. A. Lee, S. H. Reich, T. J. Prins, J. T. Marakovits, E. S. Littlefield, R. Zhou, J. Tikhe, C. E. Ford, M. Wallace, J. W. Meador III, R. A. Ferre, E. L. Brown, S. L. Binford, J. E. V. Harr, D. M. DeLisle, and S. T. Worland. 1998. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 1. Michael acceptor structure-activity studies. J. Med. Chem. 41:2806-2818. [DOI] [PubMed] [Google Scholar]
- 10.Dragovich, P. S., S. E. Webber, R. E. Babine, S. A. Fuhrman, A. K. Patick, D. A. Matthews, S. H. Reich, J. T. Marakovits, T. J. Prins, R. Zhou, J. Tikhe, E. S. Littlefield, T. M. Bleckman, M. Wallace, T. Little, C. E. Ford, J. W. Meador III, R. A. Ferre, E. L. Brown, S. L. Binford, D. M. DeLisle, and S. T. Worland. 1998. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 2. Peptide structure-activity studies. J. Med. Chem. 41:2819-2834. [DOI] [PubMed] [Google Scholar]
- 11.Dragovich, P. S., R. Zhou, D. J. Skalitzky, S. A. Fuhrman, A. K. Patick, C. E. Ford, J. W. Meador III, and S. T. Worland. 1999. Solid-phase synthesis of irreversible human rhinovirus 3C protease inhibitors. 1. Optimization of tripeptides incorporating N-terminal amides. Bioorg. Med. Chem. 7:589-598. [DOI] [PubMed] [Google Scholar]
- 12.Groake, J. M., and D. C. Pevear. 1999. Attenuated virulence of pleconaril-resistant coxsackievirus B3 variants. J. Infect. Dis. 179:1538-1541. [DOI] [PubMed] [Google Scholar]
- 13.Hayden, F. G., R. B. Turner, J. M. Gwaltney, K. Chi-Burris, M. Gersten, P. Hsyu, A. K. Patick, G. J. Smith III, and L. S. Zalman. 2003. Phase II, randomized, double-blind, placebo-controlled studies of rupintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob. Agents Chemother. 47:3907-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaiser, L., C. E. Crump, and F. G. Hayden. 2000. In vitro activity of pleconaril and AG7088 against selected serotypes and clinical isolates of human rhinoviruses. Antivir. Res. 47:215-220. [DOI] [PubMed] [Google Scholar]
- 15.Krausslich, H. G., and E. Wimmer. 1988. Viral proteinases. Annu. Rev. Biochem. 57:701-754. [DOI] [PubMed] [Google Scholar]
- 16.Ledford, R. M., M. S. Collett, and D. C. Pevear. 2005. Insights into the genetic basis for natural phenotypic resistance of human rhinoviruses to plconaril. Antivir. Res. 68:135-138. [DOI] [PubMed] [Google Scholar]
- 17.Ledford, R. M., N. R. Patel, T. M. Demenczuk, A. Watanyar, T. Herbertz, M. S. Collett, and D. C. Pevear. 2004. VP1 sequencing of all human rhinovirus serotypes: insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J. Virol. 78:3663-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makela, M. J., T. Puhakka, O. Ruuskanen, M. Leinonen, P. Saikku, M. Kimpimaki, S. Blomqvist, T. Hyypiä, and P. Arstila. 1998. Viruses and bacteria in the etiology of the common cold. J. Clin. Microbiol. 36:539-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews, D. A., P. S. Dragovich, S. E. Webber, S. A. Fuhrman, A. K. Patick, L. S. Zalman, T. F. Hendrickson, R. A. Love, T. J. Prins, J. T. Marakovits, R. Zhou, J. Tikhe, C. E. Ford, J. W. Meador, R. A. Ferre, E. L. Brown, S. L. Binford, M. A. Brothers, D. M. DeLisle, and S. T. Worland. 1999. Structure-assisted design of mechanism based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl. Acad. Sci. USA 96:11000-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meador, J. W., III, H. Ngo, C. E. Ford, A. K. Patick, R. A. Ferre, D. A. Matthews, and S. T. Worland. 1998. PCR amplification and determination of the RNA sequences for the P3 coding region of human rhinoviral serotypes. Antivir. Res. 37:A72. [Google Scholar]
- 21.Patick, A., and K. Potts. 1998. Protease inhibitors as antiviral agents. Clin. Microbiol. Rev. 11:614-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patick, A. K. 2006. Rhinovirus chemotherapy. Antivir. Res. 71:391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patick, A. K., S. L. Binford, M. A. Brothers, R. L. Jackson, C. E. Ford, M. D. Diem, F. Maldonado, P. S. Dragovich, R. Zhou, T. J. Prins, S. A. Fuhrman, J. W. Meador, L. S. Zalman, D. A. Matthews, and S. T., Worland. 1999. Antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 43:2444-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patick, A. K., M. A. Brothers, F. Maldonado, S. Binford, O. Maldonado, S. Fuhrman, A. Petersen, G. J. Smith III, L. S. Zalman, L. A. Burns-Naas, and J. Q. Tran. 2005. In vitro antiviral activity and single-dose pharmacokinetics in humans of a novel, orally bioavailable inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 49:2267-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pevear, D. C., F. G. Hayden, T. M. Demenczuk, L. A. McKinlay, and M. S. Collett. 2005. Relationship of pleconaril susceptibility and clinical outcome in treatment of common colds caused by rhinoviruses. Antimicrob. Agents Chemother. 49:4492-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitkaranta, A., and F. G. Hayden. 1998. Rhinoviruses: important respiratory pathogens. Ann. Med. 30:529-537. [DOI] [PubMed] [Google Scholar]
- 27.Schmidtke, M., E. Hammerschmidt, S. Schüler, R. Zell, E. Birch-Hirschfeld, V. A. Makarov, O. B. Riabova, and P. Wutzler. 2005. Susceptibility of coxsackievirus B3 laboratory strains and clinical isolates to the capsid function inhibitor pleconaril: antiviral studies with virus chimeras demonstrate the crucial role of amino acid 1092 in treatment. J. Antimicrob. Chemother. 56:648-656. [DOI] [PubMed] [Google Scholar]
- 28.Scudiero, D. A., R. H. Shoemaker, K. D. Paull, A. Monks, S. Tierney, T. H. Nofziger, M. J. Currens, D. Seniff, and M. R. Boyd. 1988. XTT evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 48:4827-4833. [PubMed] [Google Scholar]
- 29.Seemungal, T. A. R., R. Harper-Owen, A. Bhowmik, D. J. Jeffries, and J. A. Wedzicha. 2000. Detection of rhinovirus in induced sputum at exacerbation of chronic obstructive pulmonary disease. Eur. Respir. J. 16:677-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skern, T., W. Sommergruber, D. Blaas, P. Gruendler, F. Fraundorfer, C. Pieler, I. Fogy, and E. Kuechler. 1985. Human rhinovirus 2: complete nucleotide sequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res. 13:2111-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanway, G., P. J. Hughes, R. C. Mountford, P. D. Minor, and J. W. Almond. 1984. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 12:7859-7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weislow, O. S., R. Kiser, D. L. Fine, J. Bader, R. H. Shoemaker, and M. R. Boyd. 1989. New soluble-formazan assay for HIV-1 cytopathic effects: application to high-flux screening of synthetic and natural products for AIDS-antiviral activity. J. Natl. Cancer Inst. 81:577-586. [DOI] [PubMed] [Google Scholar]
- 33.Zalman, L. S., M. A. Brothers, P. S. Dragovich, R. Zhou, T. J. Prins, S. T. Worland, and A. K. Patick. 2000. Inhibition of human rhinovirus-induced cytokine production by AG7088, a human rhinovirus 3C protease inhibitor. Antimicrob. Agents Chemother. 44:1236-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]