Abstract
For commonly encountered gram-negative bacilli, a MIC of cefepime of 8 μg/ml or less was defined by the Clinical and Laboratory Standards Institute as “susceptible” prior to the commercial release of the antibiotic. We assessed 204 episodes of bacteremia caused by gram-negative organisms for which patients received cefepime (typically 1 to 2 g every 12 h) as the primary mode of therapy. The cefepime MIC breakpoint derived by classification and regression tree (CART) software analysis to delineate the risk of 28-day mortality was 8 μg/ml. Patients infected with gram-negative organisms treated with cefepime at a MIC of ≥8 μg/ml had a mortality rate of 54.8% (17/31 died), compared to 24.1% (35/145 died) for those treated with a cefepime MIC of <8 μg/ml. The rate of mortality for those treated with a cefepime MIC of 8 μg/ml was 56.3% (9/16 died), compared to 53.3% (8/15 died) for those treated with cefepime at a MIC of >8 μg/ml. A multivariable analysis including severity of illness indices showed that treating patients with bacteremia due to gram-negative organisms with a cefepime MIC of ≥8 μg/ml was an independent predictor of mortality (P ≤ 0.001). There was no significant difference in outcome according to the dosage regimen utilized. Pharmacodynamic assessments that were presented previously would suggest that cefepime treatment (particularly a dosage of 1 g every 12 h) has a low probability of target attainment associated with successful in vivo outcome when the cefepime MIC is ≥8 μg/ml. It would appear reasonable, based on pharmacodynamic and clinical grounds, to lower the breakpoints for cefepime in countries where the cefepime dosage of 1 to 2 g every 12 h is the licensed therapy for serious infections, so that organisms with a cefepime MIC of 8 μg/ml are no longer regarded as susceptible to the antibiotic.
Breakpoints for differentiating between organisms that are susceptible or resistant to antimicrobial agents are determined by several different organizations. These organizations, including the U.S. Food and Drug Administration (FDA), the Clinical and Laboratory Standards Institute (CLSI), the European Committee on Antimicrobial Susceptibility Testing (EUCAST), and various national organizations, determine breakpoints for antimicrobial susceptibility at the time an antibiotic is undergoing approval for clinical use. Such breakpoints may also be revised when microbiologic, pharmacodynamic, or clinical information suggests a medical necessity to do so. Cefepime breakpoints for gram-negative bacilli were determined prior to the drug's commercial release more than a decade ago. The current breakpoints determined by the FDA and CLSI for the cefepime MIC against infection by the Enterobacteriaceae family, Pseudomonas aeruginosa, and Acinetobacter spp. are ≤8 μg/ml (susceptible), 16 μg/ml (intermediate), and ≥32 μg/ml (resistant). In contrast, EUCAST breakpoints for cefepime MIC against the Enterobacteriaceae family are ≤1 μg/ml (susceptible), 2 to 8 μg/ml (intermediate), and >8 μg/ml (resistant); and EUCAST breakpoints for cefepime MIC against Pseudomonas aeruginosa are ≤8 μg/ml (susceptible) and >8 μg/ml (resistant). No EUCAST breakpoints exist for cefepime against Acinetobacter spp.
Given these disparities in breakpoints for such a commonly used antibiotic as cefepime, we examined the clinical outcomes of patients with bacteremia caused by gram-negative organisms (gram-negative bacteremia) treated with cefepime to determine whether current breakpoints need to be revised or harmonized.
MATERIALS AND METHODS
Patients.
We reviewed our hospital's clinical microbiology database to identify patients with gram-negative bacteremia. Next, we identified those patients who received cefepime as the primary mode of therapy. This mode was defined as cefepime therapy which was started within 1 calendar day of the date on which blood cultures were found to be positive. We included both those patients who received cefepime monotherapy and those who received it as a part of combination therapy. A total of 284 episodes of bacteremia from 269 patients were treated with cefepime. Secondary to a lack of MIC data, we excluded 43 episodes, leaving us with 241 episodes from 229 patients. We further excluded all episodes of patients who had concomitant bloodstream infection from a gram-positive organism or fungus. This left us with 204 analyzable episodes of gram-negative bacteremia from 197 patients.
Microbiologic analysis.
Susceptibility testing by broth microdilution (Trek Diagnostics, OH) was performed on a routine clinical basis by the hospital's clinical microbiology laboratory, using CLSI standards (5). Cases in which a polymicrobial bloodstream infection was present, in which all organisms were gram-negative bacilli, were classified according to the isolate with the highest MIC.
Clinical analysis.
We collected data including age, sex, presence of immunosuppression (neutropenia, history of solid-organ transplant, or AIDS), renal function (including the need for renal replacement therapy), and the source of bacteremia. The Acute Physiology and Chronic Health Evaluation (APACHE)-II score (6) was used to adjust for the severity of illness. The APACHE-II scores were stratified into quartiles in a manner that has been previously used in the literature (2).
Definitions.
Gram-negative bacteremia was defined as the presence of any aerobic gram-negative isolate in at least one blood culture. Cases were defined as discrete episodes of gram-negative bacteremia that were separated by at least 30 days. Polymicrobial infections were defined as those that consisted of two or more gram-negative isolates. Thus, the a priori primary endpoint was death from any cause by 28 days after the cefepime therapy was begun (2).
Cefepime dosages.
The recommended dosages at the institution were 1 to 2 g given intravenously every 12 h, for patients with creatinine clearance of ≥50 ml/min; 1 to 2 g every 24 h, for creatinine clearance of 29 to 50 ml/min; 0.5 to 1 g every 24 h, for creatinine clearance of 10 to 29 ml/min; 250 to 500 mg every 24 h, for creatinine clearance of less than 10 ml/min; and 500 mg every 24 h, for patients on dialysis.
Statistical analysis.
Analyses of each individual clinical outcome measure included only those cases in which a definitive endpoint could be identified. All variables were examined using PROC GENMOD (SAS software) in a univariate logistic regression. Factors that had a P value of less than 0.20 in the univariate analysis were eligible for entry into a multivariable, stepwise logistic regression model. Variables with a two-sided P value of <0.05 were considered significant.
The breakpoint in the distribution of cefepime MIC distribution was determined by classification and regression tree (CART; Salford Systems, San Diego, CA) analysis, a tool to identify breakpoints within ordinal and continuous variables where the outcome of interest is distinctly different between the resulting groups. Specifically, CART was used to identify the breakpoint in the cefepime MIC distribution that maximized the difference in 28-day mortality, thereby dividing the study population into two groups: those with a high likelihood of 28-day mortality and those with a low likelihood of 28-day mortality. Pruning and 10-fold cross-validation were used in the CART analysis to select the optimal nested subtree with the smallest misclassification cost.
RESULTS
Analysis was performed with 197 patients with gram-negative bacteremia who were treated with cefepime. Seven patients had two episodes of bacteremia so that 204 episodes were analyzed in total. Patients treated with cefepime were infected predominantly with P. aeruginosa (n = 50), Escherichia coli (n = 40), Klebsiella pneumoniae (n = 26), Serratia marcescens (n = 24), and Enterobacter cloacae (n = 21). Additionally, there were 24 cases of bacteremia caused by other gram-negative organisms and 19 cases of polymicrobial gram-negative organism infections (Table 1). The isolates were found to have the following MIC breakdown: 115 isolates with a MIC of ≤0.25, 11 with a MIC of 0.5, 14 with a MIC of 1, 19 with a MIC of 2, 11 with a MIC of 4, 17 with a MIC of 8, and 17 with a MIC of ≥16. The greatest number of isolates with a MIC against cefepime of ≤1 were Escherichia coli (39 isolates), Klebsiella species (30 isolates), Serratia species (23 isolates), and Enterobacter species (19 isolates). Pseudomonas aeruginosa was evenly distributed across the entire MIC spectrum (Table 1).
TABLE 1.
Distribution and cefepime MICs of 204 bloodstream isolates from cefepime-treated patients
| MIC (μg/ml) | No. of isolates of:
|
Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Acinetobacter spp. | E. coli | Enterobacter spp. | Klebsiella spp. | Serratia spp. | P. aeruginosa | Miscellaneousa | Polymicrobialb | ||
| ≤1 | 39 | 19 | 30 | 23 | 7 | 9 | 13 | 140 | |
| 2 | 1 | 1 | 1 | 1 | 12 | 3 | 19 | ||
| 4 | 2 | 1 | 7 | 1 | 11 | ||||
| 8 | 1 | 1 | 1 | 13 | 1 | 17 | |||
| ≥16 | 2 | 3 | 11 | 1 | 17 | ||||
| Total | 4 | 40 | 26 | 32 | 24 | 50 | 9 | 19 | 204 |
Miscellaneous comprises Citrobacter, Providencia, and Pantoea spp. (one isolate from each with a MIC of ≤1 μg/ml), and Proteus spp. (six isolates, all with a MIC of ≤1 μg/ml).
For polymicrobial infections, the highest cefepime MIC is recorded.
Clinical outcome by cefepime MIC: cefepime-treated patients.
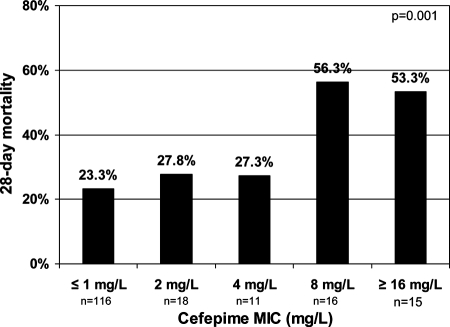
Twenty-one patients were discharged from the hospital within 28 days of culture-confirmed bloodstream infection and had no further contact with our hospital's health care system. We therefore could not analyze their outcomes at 28 days. The 28-day mortality rate for the remaining 176 patients with gram-negative bacteremia treated with cefepime was 29.5% (52/176). The rate of mortality varied by the cefepime MIC of the pathogen (Fig. 1); that is, the rate of mortality was 23.3% (27/116 died) for cefepime with a MIC of ≤1 μg/ml, 27.8% (5/18 died) with a MIC of 2 μg/ml, 27.3% (3/11 died) with a MIC of 4 μg/ml, 56.3% (9/16 died) with a MIC of 8, and 53.3% (8/15 died) with a MIC of ≥16 μg/ml.
FIG. 1.
Twenty-eight day mortality stratified by cefepime MIC.
The cefepime MIC breakpoint derived by CART analysis to delineate the risk of 28-day mortality was 8 μg/ml. Patients with cefepime MICs of ≥8 μg/ml had a twofold or greater increase in 28-day mortality over that of patients with MICs of <8 μg/ml (54.8% and 24.1%, respectively; P = 0.001). The 28-day mortality rates were similar for all groups with a MIC of <8 μg/ml, and higher 28-day mortality rates were observed when the cefepime MIC was ≥8 μg/ml (P = 0.001, using linear-by-linear association).
Other predictors of clinical outcome: univariate analysis.
Rising scores of severity of illness were highly correlated with 28-day mortality, as were renal impairment and the need for renal replacement therapy (Table 2). Specifically, those patients with an APACHE-II score of 3 to 19 had a mortality rate of 14.1%, whereas those with a score of 25 to 29 had a mortality rate of 54.5%, and those with a score of 30 to 53 had a rate of 75% (P values compared to 3 to 19 of 0.0002 and <0.0001, respectively). Patients who were receiving continuous renal replacement therapy had a mortality rate of 66.7% compared to 14.3% of those with a creatinine clearance of >100 ml/min (P = 0.0007). The univariate analysis of 28-day mortality in relation to the organism type showed that patients with bacteremia caused by P. aeruginosa infection had a trend toward an increased risk of dying (Table 2). There was no relationship between the organism type and the need for renal replacement therapy (data not shown). Neither age nor status of immune system was shown to be a predictor of death.
TABLE 2.
Relationship between predictors of outcome and mortality at 28 days
| Patient dataa | No. of patient deaths/total n (%) | P value | OR | 95% CI |
|---|---|---|---|---|
| Organism types | ||||
| E. coli | 7/33 (21.2) | 0.25b | 0.6 | 0.2-1.5 |
| P. aeruginosa | 18/46 (39.1) | 0.10 | 1.8 | 0.9-3.7 |
| Enterobacter spp. | 5/23 (21.7) | 0.38 | 0.6 | 0.2-1.8 |
| Klebsiella spp. | 7/23 (30.4) | 0.92 | 1.05 | 0.4-2.7 |
| Proteus spp. | 1/4 (25) | 0.84 | 0.8 | 0.08-7.8 |
| Serratia spp. | 5/21 (23.8) | 0.54 | 0.7 | 0.3-2.1 |
| All others | 2/7 (28.6) | 0.95 | 0.95 | 0.2-5.1 |
| Polymicrobial | 7/19 (36.8) | 0.46 | 1.5 | 0.6-3.9 |
| APACHE-II scores | ||||
| 3-19 | 12/85 (14.1) | |||
| 20-24 | 10/38 (26.3) | 0.11c | 2.2 | 0.8-5.6 |
| 25-29 | 12/22 (54.5) | 0.0002 | 2.7 | 1.6-4.5 |
| 30-53 | 12/16 (75) | <0.0001 | 2.6 | 1.7-4.0 |
| Sources of bacteremia | ||||
| CVC | 1/14 (7.1) | 0.09d | 0.2 | 0.02-1.3 |
| UTI | 5/26 (19.2) | 0.22 | 0.5 | 0.2-1.5 |
| Pneumonia | 13/34 (38.2) | 0.22 | 1.6 | 0.8-3.6 |
| Other | 2/9 (22.2) | 0.62 | 0.7 | 0.1-3.3 |
| Unknown | 31/93 (33.3) | 0.24 | 1.5 | 0.8-2.8 |
| Creatinine clearance rates | ||||
| >100 ml/min | 4/28 (14.3) | |||
| 60-100 ml/min | 7/41 (17.1) | 0.76e | 1.2 | 0.3-4.7 |
| <60 ml/min | 21/57 (36.8) | 0.039 | 3.5 | 1.1-11.5 |
| CVVHD | 12/18 (66.7) | 0.0007 | 12.0 | 2.8-50.8 |
| HD | 7/30 (23.3) | 0.28 | 2.1 | 0.6-7.9 |
| Immune status | ||||
| Competent | 16/60 (26.7) | |||
| Compromised | 36/116 (31) | 0.55f | 1.2 | 0.6-2.5 |
| Ages | ||||
| ≤64 | 28/105 (26.7) | |||
| ≥65 | 24/71 (33.8) | 0.31g | 1.4 | 0.7-2.7 |
| Modes of therapy | ||||
| Monotherapy | 21/73 (28.8) | |||
| Combination therapy | 31/102 (30.4) | 0.82h | 1.08 | 0.56-2.09 |
Abbreviations: CVC, central venous catheter; UTI, urinary tract infection; CVVHD, continuous venovenous hemodialysis; HD, hemodialysis.
P value compared to that of all other organisms.
P value compared to that of score range 3-19.
P value compared to that of all other sources.
P value compared to that of creatinine clearance of >100 ml/min.
P value compared to competent status.
P value compared to those aged ≤64.
P value compared to that of monotherapy.
In order to determine the effect on mortality at 28 days of the use of combinations of antibiotics active against gram-negative bacilli plus cefepime, a comparison was made with monotherapy. A total of 73 patients received monotherapy with cefepime, and 102 received a combination therapy (Table 2). We found a 30.4% mortality rate with the combination therapy and a 28.8% rate with cefepime monotherapy (P value, 0.82; odds ratio[OR], 1.08; 95% confidence interval [CI], 0.56 to 2.09).
Predictors of adverse clinical outcome: multivariable analysis.
In our multivariable model of predictors of 28-day mortality, we included all items that had a P value of ≤0.2 on the univariate analysis. This consisted of having an APACHE-II score of ≥25, a creatinine clearance of <60 ml/min, the use of continuous renal replacement therapy, a cefepime MIC of ≥8 μg/ml, a central venous line as the source of bacteremia, and an infection with Pseudomonas aeruginosa. We found that the use of cefepime against an isolate with a MIC of ≥8 μg/ml remained an independent risk factor for 28-day mortality (P ≤ 0.001; adjusted OR, 8.2; 95% CI, 2.8 to 24.2). Other independent predictors of 28-day mortality on multivariable analysis included an APACHE-II score of ≥25 (P < 0.0001; OR, 5.9; 95% CI, 2.4 to 14.5), a creatinine clearance rate of <60 ml/min, and the use of continuous renal replacement therapy (P = 0.009; OR, 4.2; 95% CI, 1.4 to 11.4).
In a secondary analysis, patients with cefepime MICs of 8 and ≥16 μg/ml were included at model entry as distinct variables. Both cefepime MICs of 8 μg/ml (P = 0.002; adjusted OR, 9.1; 95% CI, 2.2 to 37.5) and ≥16 μg/ml (P = 0.004; adjusted OR, 7.5; 95% CI, 1.9 to 29.2) were independently associated with 28-day mortality when scores were adjusted for the other aforementioned univariate predictor variables.
Outcomes of patients infected with P. aeruginosa.
Twenty-eight-day outcome data were available for 46 patients infected with P. aeruginosa as the sole bloodstream isolate. Mortality was higher from P. aeruginosa bacteremia treated with cefepime when isolates had a cefepime MIC of 8 μg/ml (66.7%; 8/12 died) than when isolates had a cefepime MIC of ≤4 μg/ml (20.8%; 5/24 died) (P = 0.01; OR = 7.6; 95% CI, 1.7 to 34.5) and higher when the mortality rate for those with a cefepime MIC of ≥8 μg/ml (59.1%; 13/22 died) was compared to that of a cefepime MIC of ≤4 μg/ml (20.8%; 5/24 died) (P = 0.008). Specifically, the 28-day mortality rate for patients with bacteremia due to P. aeruginosa infection was 33% (2/6 died) with a cefepime MIC of ≤1 μg/ml, 18% (2/11 died) with a cefepime MIC of 2 μg/ml, 14% (1/7 died) with a cefepime MIC of 4 μg/ml, 67% (8/12 died) with a cefepime MIC of 8 μg/ml, and 50% (5/10 died) with a cefepime MIC of ≥16 μg/ml. There were no differences between the proportion of patients with P. aeruginosa infection who received combination therapy (52%; 26/50 died) and that of patients infected with other bacteria (61%; 93/153 died; P = 0.32).
Outcomes of patients with beta-lactamase-producing Enterobacteriaceae infection.
Ten patients infected with organisms known to be capable of hyperproducing AmpC (e.g., Enterobacter and Serratia spp., etc.) died within 28 days of developing bacteremia. These patients had cefepime MICs of 0.25 μg/ml (nine patients) and 4 μg/ml (one patient). (One additional patient who died had a mixed infection with Enterobacter cloacae [a cefepime MIC of 8 μg/ml] and Pseudomonas aeruginosa [a cefepime MIC of 1 μg/ml]). Only one patient was infected at baseline with an organism which was resistant to ceftazidime; 0/9 patients with baseline ceftazidime MICs in the susceptible range had a documented selection of a mutant isolate resistant to ceftazidime.
Eleven patients were infected with extended-spectrum beta-lactamase (ESBL)-producing organisms (seven patients were infected with E. cloacae, one with Klebsiella oxytoca, one with Enterobacter aerogenes, and one with E. coli). A total of 5 of 10 (50%) patients for whom 28-day mortality was known died within 28 days of developing bacteremia. The cefepime MICs of the infecting organisms and patient outcomes were as follows: 2/3 died (MIC of 2 μg/ml), 2/3 died (MIC of 4 μg/ml), 1/2 died (MIC of 8 μg/ml), and 0/2 died (MIC of 16 μg/ml).
Outcomes of patients with regard to cefepime dosing.
The dosing schedules given to patients whose infecting isolates had a MIC of 8 were 500 mg every 12 h (one patient with an unknown 28-day mortality), 1 g with dialysis (1/1 died), 1 g every 24 h (3/4 died), 2 g every 24 h (1/1 died), 1 g every 12 h (0/4 died), 2 g every 12 h (2/4 died), and 2 g every 8 h (2/2 died). No correlation was observed between dosing schedule and mortality rate in this group. However, the numbers were too limited for formal analysis. Finally, there were no significant differences in dosing regimens between patients with isolates whose MICs were less than 8, equal to 8, or greater than 16 (data not shown).
Outcome of patients with gram-negative bacteremia treated with other antibiotics.
In order to determine whether bloodstream infection with an organism with a cefepime MIC of 8 μg/ml is in itself a marker for poor clinical outcome, we compared the outcome of patients treated with cefepime versus those treated with other antibiotics to which the bloodstream isolate was susceptible. For this comparison, we identified 53 bacteremic patients during the period January 2001 to April 2005 whose bacterial isolates showed a cefepime MIC of 8 and were treated with an antibiotic other than cefepime. We excluded cases in which the isolate was resistant to the chosen therapy or in which the isolate had no susceptibility result for the antibiotic chosen and patients who had a concomitant bloodstream infection with a gram-positive isolate or fungus. This left us with 19 cases from the same number of patients.
This study group consisted of 10 patients who were treated with either piperacillin or piperacillin-tazobactam (a piperacillin MIC of 4 μg/ml in one patient; a MIC of 32 μg/ml in seven patients; a MIC of 64 μg/ml in one patient; and one with no MIC but a disk diffusion result of susceptible), 3 who were treated with a quinolone (a ciprofloxacin MIC of 0.25 μg/ml in one patient; a levofloxacin MIC of ≤0.5 μg/ml in one patient; and a MIC of 1 μg/ml in one patient), 4 who were treated with an aminoglycoside (a tobramycin MIC of ≤1 μg/ml in two patients, an amikacin MIC of 16 μg/ml in one patient, and one with no MIC but a susceptible disk diffusion result), and 2 who were treated with a carbapenem (both with a MIC of 2 μg/ml). There were no significant differences between the population treated with cefepime and the population treated with another agent, in terms of organism type, APACHE-II scores, sources of bacteremia, creatinine clearance rates, immune status, age, or receipt of monotherapy versus combination therapy. The 28-day mortality rate was higher in those treated with cefepime (56.3%) than those treated with alternative antibiotics (38.9%), although this difference was not statistically significant (P = 0.31; OR, 2.0; 95% CI, 0.5 to 7.9).
DISCUSSION
Antibiotic susceptibility breakpoints are determined typically by the integration of a variety of microbiologic, pharmacokinetic/pharmacodynamic (PK/PD), and clinical data (4). In the optimal situation, each of these data components show consistent results and strongly support a particular breakpoint. However, it is potentially naïve to think that such a situation will always occur or that all pieces of data will be both robust and consistent. This is particularly so when breakpoints are reconsidered after a particular antibiotic has been in clinical use for some years. In such a situation, new resistance mechanisms may have arisen, causing a “spread” of MICs away from wild-type distributions. Randomized clinical trials are difficult to perform after a drug has undergone requirements for registration as an approved drug. We believe that an examination of PK/PD and clinical data supports an alteration of the breakpoints for cefepime and gram-negative bacilli or a reexamination of dosing regimens of the drug, even though such data do not come from recently performed randomized trials.
Since the commercial release of cefepime, new mechanisms of antibiotic resistance have been detected. These include the production of ESBLs and metalloenzymes, many of which do hydrolyze cefepime (7, 11). While some of these organisms may have very high cefepime MICs (for example, more than 32 μg/ml), numerous examples now exist whereby such beta-lactamase-producing organisms have elevated cefepime MICs compared to that of wild-type organisms, yet the MICs are still in the susceptible range (“hidden resistance”) (3, 8, 9). The CLSI currently recommends that ESBL-producing organisms be reported as resistant to cefepime. Small case series have suggested that the outcome for cefepime-treated patients is poor for serious infections with ESBL-producing organisms regardless of the MIC (10). In this study, we have too small a number to address this question specifically for ESBL producers, although mortality was substantial (50% [5/10] died).
Several studies have now assessed the PK/PD profile of cefepime and would support a change in cefepime breakpoints or an elimination of all but a dosage regimen of 2 g every 8 h for empirical therapy of serious infections. A 10,000-subject Monte Carlo simulation using published mean pharmacokinetic parameter estimates and PK/PD targets derived from a murine infection model has been presented (1). The FDA has not given a specific label for the use of cefepime in the treatment of bloodstream infections. However, for moderate to severe pneumonia due to P. aeruginosa, K. pneumoniae or Enterobacter spp. infection, the recommended dosage is 1 to 2 g every 12 h; the empirical therapy for febrile, neutropenic patients is 2 g every 8 h. According to the model just described (1), the dosage regimen of 1 g of cefepime every 12 h has just a 35.9% probability of resulting in a percentage of time above a MIC of greater than 50% if the MIC is 8 μg/ml. Thus, these models would predict that a dose of 1 g every 12 h would most likely fail if the cefepime MIC is 8 μg/ml. However, this particular model would predict that a dosage regimen of 2 g every 12 h or 2 g every 8 h would have a greater than 90% probability of resulting in a percentage of time above a MIC of greater than 50% if the MIC is 8 μg/ml. In contrast, an alternative model showed a probability of the percentage of time above 50% if the MIC was 8 μg/ml of 2% for cefepime at 1 g every 12 h, 21% for 2 g every 12 h, and 88% for 2 g every 8 h (10). It would appear that the preponderance of evidence from PK/PD analyses suggests that the breakpoint of 8 μg/ml is too high for dosages of cefepime of 1 g every 12 h and quite possibly also for 2 g every 12 h.
From a clinical perspective, we have evaluated the outcome of almost 200 patients who received cefepime empirically for the treatment of gram-negative bacteremia. We found that the 28-day mortality of patients whose organisms had a MIC of 8 μg/ml (56.3%) approximated that of patients with MICs outside of the susceptible range (53.3%) and far exceeded that of patients whose organisms had a MIC of <8 μg/ml (24.1%). We chose 28-day mortality a priori as our endpoint since this was the definition in a large trial of patients with sepsis published in the New England Journal of Medicine (2). In order to account for important variables such as severity of illness, which may potentially confound this result, we used a multivariable analysis. This analysis showed that having a cefepime MIC of 8 μg/ml was an independent predictor of 28-day mortality in patients treated with cefepime for gram-negative bacteremia. Although 28-day mortality is widely used in other studies, a particular criticism of this endpoint is that variables other than antibiotic use may be responsible for the patient's death. In order to add another layer of rigor to our analysis, we compared outcomes of patients with bacteremia with cefepime MICs of 8 μg/ml treated with cefepime to those of patients treated with other antibiotics to which the organism was susceptible. This was done in order to exclude the hypothesis that some unforeseen variable leads to inferior outcomes for patients with bacteremia due to cefepime MICs of 8 μg/ml. If that hypothesis were correct, patients with bacteremia caused by an organism with a cefepime MIC of 8 μg/ml would have poor outcomes regardless of which antibiotic was chosen. In contrast, we found that patients infected with organisms at this MIC that were treated with alternative antibiotics had a trend toward superior outcomes compared to those treated with cefepime, suggesting that this potential hypothesis was incorrect.
Despite our rigorous clinical analysis of retrospective data, we would have preferred to have performed a prospective trial in which patients suspected of having gram-negative bacteremia were randomized to cefepime and an alternative antibiotic. Ideally, such a trial would include a pharmacokinetic analysis to determine if suboptimal cefepime “exposure” could be correlated with suboptimal clinical outcome. Unfortunately, the sample size of many hundreds of patients required to enroll sufficient patients with confirmed gram-negative bacteremia with organisms with a cefepime MIC of 8 μg/ml precludes the initiation of such a study. We are performing a more limited prospective, pharmacokinetic analysis of patients with serious gram-negative infections treated with cefepime.
In summary, our data add to the weight of data supporting a change of breakpoint for cefepime in countries where the cefepime dosage regimen of 1 to 2 g every 12 h is the licensed therapy for serious infections. First, two different PK/PD models strongly show that a cefepime dose of 1 g every 12 h has a low probability of reaching important PK/PD targets when the cefepime MIC is 8 μg/ml. It could be argued that higher doses are frequently used, but (i) some models question even the utility of 2 g every 12 h in treating organisms with a MIC of 8 μg/ml, and (ii) there are practical concerns about the communication of “dose-specific” breakpoints to prescribers. Second, our clinical data show that 28-day mortality, a widely used outcome measure in studies of sepsis, is higher in cefepime-treated patients with gram-negative bacteremia due to organisms with a cefepime MIC of 8 μg/ml than in patients infected with organisms with lower cefepime MICs. While inadequacies in this clinical study, such as its limited sample size and arbitrary outcome measures, are present, we believe the weight of data does support lowering the cefepime breakpoints so that a cefepime MIC of 8 μg/ml is no longer regarded as susceptible (if 1 to 2 g every 12 h is a licensed dosing regimen for serious infections, such as it is in the United States). We would propose that clinical data of the treatment of serious gram-negative infections with other antibiotics (for example, piperacillin-tazobactam) should also be investigated to determine if the breakpoints or dosing regimens of other commonly used antibiotics should also be changed.
Acknowledgments
D.P. received research support from Elan and AstraZeneca and is a consultant to Merck and Acureon. B.C. and B.P. are on the speakers’ bureau for Wyeth.
Footnotes
Published ahead of print on 15 October 2007.
REFERENCES
- 1.Ambrose, P. G., S. M. Bhavnani, R. N. Jones, W. A. Craig, and M. N. Dudley. 2004. Use of pharmacokinetic-pharmacodynamic and Monte-Carlo simulation as decision support for the reevaluation of NCCLS cephem susceptibility breakpoints for Enterobacteriaceae. Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother., abstr. A-138.
- 2.Bernard, G. R., J. L. Vincent, P. F. Laterre, S. P. LaRosa, J. F. Dhainaut, A. Lopez-Rodriguez, J. S. Steingrub, G. E. Garber, J. D. Helterbrand, E. W. Ely, and C. J. Fisher, Jr., for the Recombinant Human Activated Protein C Worldwide Evaluation in Severe Sepsis (PROWESS) study group. 2001. Efficacy and safety of recombinant human activated protein C for severe sepsis. N. Engl. J. Med. 344:699-709. [DOI] [PubMed] [Google Scholar]
- 3.Bratu, S., D. Landman, R. Haag, R. Recco, A. Eramo, M. Alam, and J. Quale. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch. Intern. Med. 165:1430-1435. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute/NCCLS. 2001. Development of in vitro susceptibility testing criteria and quality control parameters. Approved guideline, 2nd ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Knaus, W. A., D. P. Wagner, E. A. Draper, J. E. Zimmerman, M. Bergner, P. G. Bastos, C. A. Sirio, D. J. Murphy, T. Lotring, A. Damiano, et al. 1991. The APACHE III prognostic system. Risk prediction of hospital mortality for critically ill hospitalized adults. Chest 100:1619-1636. [DOI] [PubMed] [Google Scholar]
- 7.Paterson, D. L., and R. A. Bonomo. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paterson, D. L., W. C. Ko, A. Von Gottberg, J. M. Casellas, L. Mulazimoglu, K. P. Klugman, R. A. Bonomo, L. B. Rice, J. G. McCormack, and V. L. Yu. 2001. Outcome of cephalosporin treatment for serious infections due to apparently susceptible organisms producing extended-spectrum beta-lactamases: implications for the clinical microbiology laboratory. J. Clin. Microbiol. 39:2206-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peleg, A. Y., C. Franklin, J. M. Bell, and D. W. Spelman. 2005. Dissemination of the metallo-beta-lactamase gene blaIMP-4 among gram-negative pathogens in a clinical setting in Australia. Clin. Infect. Dis. 41:1549-1556. [DOI] [PubMed] [Google Scholar]
- 10.Reese, A. M., C. R. Frei, and D. S. Burgess. 2005. Pharmacodynamics of intermittent and continuous infusion piperacillin/tazobactam and cefepime against extended-spectrum beta-lactamase-producing organisms. Int. J. Antimicrob. Agents 26:114-119. [DOI] [PubMed] [Google Scholar]
- 11.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-beta-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]