Abstract
Small-colony variants (SCVs) of Staphylococcus aureus are a slow-growing subpopulation whose phenotypes can include resistance to aminoglycosides, defects in electron transport, and enhanced persistence in mammalian cells. Here we show that a subset of mutants selected as SCVs by reduced susceptibility to aminoglycosides are resistant to the antibiotic fusidic acid (FA) and conversely that a subset of mutants selected for resistance to FA are SCVs. Mutation analysis reveals different genetic classes of FA-resistant SCVs. One class, FusA-SCVs, have amino acid substitution mutations in the ribosomal translocase EF-G different from those found in classic FusA mutants. Most of these mutations are located in structural domain V of EF-G, but some are in domain I or III. FusA-SCVs are auxotrophic for hemin. A second class of FA-resistant SCVs carry mutations in rplF, coding for ribosomal protein L6, and are designated as FusE mutants. FusE mutants fall into two phenotypic groups: one auxotrophic for hemin and the other auxotrophic for menadione. Accordingly, we have identified new genetic and phenotypic classes of FA-resistant mutants and clarified the genetic basis of a subset of S. aureus SCV mutants. A clinical implication of these data is that FA resistance could be selected by antimicrobial agents other than FA.
Fusidic acid (FA) is an antibiotic used in the treatment of staphylococcal infections (6, 23, 30, 36, 55, 60, 61). The target of FA is the translation elongation factor EF-G in complex with the ribosome (13, 32). FA acts by preventing the release of EF-G from the ribosome after translocation, thus inhibiting further protein synthesis (53, 62). Although it was launched more than 4 decades ago, FA remains a useful antibiotic, not least because it is not cross-resistant with other antibiotics used to treat staphylococci. Although frequencies of resistance to FA have remained generally low, emerging resistance is a problem that could limit the therapeutic options available for treatment of staphylococcal infections (25, 39, 44, 47, 48). To maintain the usefulness of antibiotics, it is important to understand the mechanisms that cause relevant bacterial pathogens to become resistant. We have addressed this question with respect to FA resistance and Staphylococcus aureus.
Two mechanistic classes of resistance to FA have so far been described genetically (19). One class, FusA, is associated with mutations in fusA (11, 38), the gene encoding the ribosomal translocase, translation elongation factor EF-G. These mutations reduce the affinity of FA for its target, EF-G, on the ribosome. The second class of resistance mechanism, FusB, is usually associated with a 21-kb plasmid, pUB101, carrying the gene fusB (40), but is also found in the chromosome (43). The fusB gene encodes an inducible protein shown in vitro to protect EF-G against the inhibitory action of FA (42). Recently two homologues of fusB, designated fusC and fusD, have been identified in the chromosome of clinical isolates of S. aureus and Staphylococcus saprophyticus respectively (41).
We have previously reported the existence of another FA resistance class in S. aureus (38), here referred to as FusE. In this report, we show that mutants of the FusE class and some mutants of the FusA class have the classic characteristics of small-colony variants (SCVs) of S. aureus.
Bacteria with an SCV phenotype have been described for many species, including human pathogens such as Staphylococcus aureus (46). S. aureus SCVs are clinically important because they are associated with intracellular growth, increased persistence, and recurrent infections (1, 14, 45, 46, 51, 56, 58). In addition to their small colony size on solid media, SCVs can typically exhibit a variety of other characteristic phenotypes. These include reduced susceptibility to aminoglycosides; auxotrophy for any one of the compounds hemin, menadione, thiamine, or thymidine; reduced hemolytic activity; and reduced colony pigmentation. The phenotypes associated with S. aureus SCVs have been rationalized as consequences of a dysfunctional electron transport system (7, 27, 45, 46, 57, 58) or a defect in thymidine biosynthesis (18, 28, 29). The SCV phenotype is reported to be phenotypically unstable, with strains frequently reverting to a normal-colony phenotype (10, 59). The rate of emergence of selected SCVs is increased in a mutator strain, strongly suggesting a genetic rather than a regulatory basis for the phenotype (50). The actual genetic basis for the SCV phenotype in clinical isolates is in most cases unknown. However, selected or constructed mutations in hemB (9, 56), hemH (50), menD (9), and ctaA (20) produce the electron transport deficiency-associated phenotypes of SCVs. Recently, it was reported that the thymidine-auxotrophic SCVs from cystic fibrosis patients carry mutations in the thymidylate synthase gene, thyA (12).
Here we report that all FusE mutants and a subset of FusA mutants selected in S. aureus are SCVs. We identify particular mutations in EF-G and ribosomal protein L6 that separately are associated with the SCV phenotype. We also show that a subset of mutants selected in S. aureus as SCVs on the basis of resistance to aminoglycosides belong to either the FusA-SCV or FusE resistance class.
MATERIALS AND METHODS
Strains and antibiotics.
S. aureus 8325-4 was used as the standard drug-susceptible laboratory wild-type strain. Clinical S. aureus isolates from bacteremia patients (20 FA resistant and 10 FA susceptible) were supplied by Statens Serum Institut, Copenhagen, Denmark. Strains were grown in liquid Luria broth (LB) or on Luria agar plates (LA) unless otherwise stated. Liquid cultures were incubated overnight at 37°C on a shaker at 255 rpm. Strain stocks were stored frozen in LB plus dimethyl sulfoxide (7%) at −80°C. Antibiotics were dissolved in methanol (FA sodium salt at 5 mg/ml; Leo Pharma, Ballerup, Denmark) or double-distilled water (kanamycin [KAN] at 5 mg/ml; Sigma-Aldrich AB, Stockholm, Sweden) and stored at −20°C until used.
Selection of drug-resistant mutants.
A fresh overnight culture in LB was diluted, and approximately 100 CFU were inoculated into each of 20 tubes, each containing 2 ml LB, and grown into stationary phase. One hundred microliters from each overnight culture was plated on selective media. FA resistance was selected at 2 μg/ml. KAN resistance was selected at 8, 16, and 24 μg/ml. Plates were incubated 48 h at 37°C before picking colonies and purifying on selective media.
MIC.
The MIC was measured using Etest strips (AB Biodisk, Solna, Sweden) in accordance with the manufacturers’ description. A single colony was dissolved in 0.9% NaCl to a density of 0.5 McFarland unit. A cotton swab was used to spread the bacterial suspension over the surface of a Mueller-Hinton plate (Difco Becton Dickinson, MD). After application of the Etest strip, plates were incubated at 37°C for 18 to 24 h until growth of the SCV lawn was evident.
Growth rate.
Bacterial doubling times in LB were measured using the BioscreenC machine (Oy Growth Curves Ab Ltd., Helsinki, Finland). Cells were inoculated in 200 μl LB at a density of 104 to 105 cells/ml in 100-well honeycomb plates. Measurements of the optical density at 600 nm were taken each 10 min over a 24-h period at 37°C.
Auxotrophy complementation tests.
Auxotrophy was assayed by complementation with hemin, menadione sodium bisulfite, thiamine pyrophosphate, and thymidine (all from Sigma-Aldrich AB, Stockholm, Sweden). Five-millimeter-diameter filter discs (3MM paper; Whatman International, Maidstone, United Kingdom) were soaked in each solution at a concentration of 1,000 μg/ml (except menadione, which was used at 200 μg/ml) and placed onto LA plates spread with ∼105 CFU of the strain to be tested. Plates were incubated overnight at 37°C. An increase in colony size proximal to the cellulose disc was interpreted as a positive result (complementation of auxotrophy).
PCR and sequencing.
Template DNA was prepared by dissolving a bacterial colony in 100 μl double-distilled water, adding ∼100 μl of 0.25-mm acid-washed glass beads (Sigma-Aldrich AB, Stockholm, Sweden), and vortexing for 10 s to disrupt the cells. Primers for amplifying and sequencing S. aureus genes were designed based on the publicly available genome sequences. After an initial denaturation step of 5 min at 95°C, 30 cycles of 15 s at 95°C, 15 s at 50°C, and ∼1 min/kb at 72°C were run. The last elongation step was prolonged to 5 min. Sequencing reactions were carried out at the DNA Sequencing Facility at Rudbeck Laboratory, Uppsala, Sweden, and at Macrogen Inc., Seoul, South Korea. Sequences were analyzed using BioEdit (version 7.0.1).
RESULTS
Isolation and classification of FA-resistant mutants in S. aureus 8325-4.
To isolate non-fusA chromosomal mutants, 20 independent cultures of S. aureus 8325-4 were grown into stationary phase and 108 CFU from each were spread on LA plates containing 2 μg/ml of FA. On average, 10 colonies appeared on each plate after an overnight incubation at 37°C. Twenty-eight FA-resistant colonies, including 7 small colonies from different cultures, were picked and purified. DNA sequencing revealed that the great majority of the mutants (n = 23) had point mutations in the fusA gene. Twenty-one of these were found in domain III (Table 1), in agreement with published data on the importance of this region of EF-G for FA resistance mutations (26, 38). The remaining two fusA mutations both had the substitution G664S in domain V of EF-G (Table 1). Only one FA-resistant mutation, A655P, has previously been identified in domain V of S. aureus EF-G, and that was in a clinical isolate that carried three additional mutations in domain III of EF-G (38). The remaining five FA-resistant mutants had no mutations in fusA and thus were classified as FusE. These five FusE mutants and the two FusA domain V mutants differ significantly from the 25 FusA domain III mutants in that they have a small-colony phenotype, requiring ≥48 h at 37°C to form 1-mm-diameter colonies on LA. The slow growth of the FusE and FusA domain V mutants is evident also in liquid culture (Table 1). We asked whether these slow-growing FA-resistant mutants might be similar to the previously described SCV type in S. aureus (46).
TABLE 1.
Mutants selected in 8325-4 as FA resistant and then screened for SCV phenotypes
| Strain and stock | Identified mutationa
|
MIC (μg/ml) of:
|
Doubling time (min)b | Auxotrophyc | nd | |||
|---|---|---|---|---|---|---|---|---|
| fusA (EF-G) | rplF (L6) | FA | STR | KAN | ||||
| 8325-4 | None | None | 0.125 | 3 | 1.5 | 24 | − | |
| FusA classic | ||||||||
| AH207 | Thr385Asn | None | 4 | 12 | 3 | 29 | − | |
| AH194 | Pro404Leu | None | 4 | 8 | 2 | 25 | − | 2 |
| AH196 | Pro406Leu | None | 6 | 8 | 3 | 31 | − | |
| AH198 | Gly452Ser | None | 32 | 6 | 4 | 38 | − | 2 |
| AH201 | Gly452Cys | None | 12 | 8 | 6 | 28 | − | |
| AH197 | Leu456Phe | None | 4 | 8 | 6 | 34 | − | |
| AH187 | His457Tyr | None | 96 | 6 | 3 | 29 | − | 9 |
| AH206 | Arg464Ser | None | 6 | 8 | 6 | 37 | − | |
| AH190 | Arg464His | None | 6 | 8 | 4 | 31 | − | 3 |
| FusA-SCV | ||||||||
| AH188 | Gly664Ser | None | 4 | 16 | 24 | 46 | Hemin | 2 |
| FusE | ||||||||
| AH193 | None | TAA stop at nt 229 | 4 | 24 | 8 | 54 | Hemin | |
| AH199 | None | TAA stop at nt 241 | 4 | 16 | 8 | 57 | Hemin | |
| AH202 | None | CTG start at nt 1 | 4 | 16 | 8 | 58 | Hemin | |
| AH204 | None | Δ of nt 203-231 | 4 | 24 | 8 | 57 | Hemin | |
| AH209 | None | Δ of nt 404-427 | 4 | 16 | 8 | 60 | Hemin | |
None, wild type. Underlined letters represent the mutated residues.
Doubling time (generation time) in minutes during exponential growth in LB.
Auxotrophy tested for hemin, menadione, thymine, and thymidine. −, nonauxotrophic.
Number of independent isolates with the same genotype and phenotype.
Characterization of slow-growing FA-resistant mutants isolated in 8325-4.
The classic SCV phenotypes described in S. aureus include, in various combinations, enhanced resistance to aminoglycosides; auxotrophy for thymidine; and auxotrophy for components of the electron transport system such as hemin, menadione, and thiamine (46). We assayed all 28 FA-resistant mutants for each of these phenotypes (Table 1). The 21 FusA domain III mutants vary widely in resistance to FA (MIC, 4 to 96 μg/ml), as expected, but are relatively uniform with respect to the other phenotypes tested (Table 1). Thus, they display relatively small increases in generation time (1.1- to 1.6-fold longer); small increases in MIC for the aminoglycosides KAN and streptomycin (STR) (1.3- to 4-fold increases); no significant change in MIC for the aminoglycoside gentamicin (not shown); and no auxotrophy for hemin, menadione, thiamine, or thymidine.
In contrast, the FusA domain V mutants and the FusE mutants have markedly different phenotypes (Table 1). Their FA MIC is uniform and at a medium level (MIC, 4 μg/ml), their generation times are significantly longer (2.3- to 3.6-fold longer), and their susceptibility to aminoglycosides is greatly reduced (KAN MIC, 5- to 16-fold increase; STR MIC, 5- to 8-fold increase; gentamicin MIC, 3- to 6-fold increase). In addition, the FusA domain V mutants and each of the members of the FusE class have an auxotrophy on LA that is complemented by hemin (Fig. 1 and Table 1). Each of these specific traits has been reported previously as a characteristic of the SCV phenotype (46). We concluded that these FA-resistant mutants (FusA domain V and FusE) should be classified within the SCV group of S. aureus.
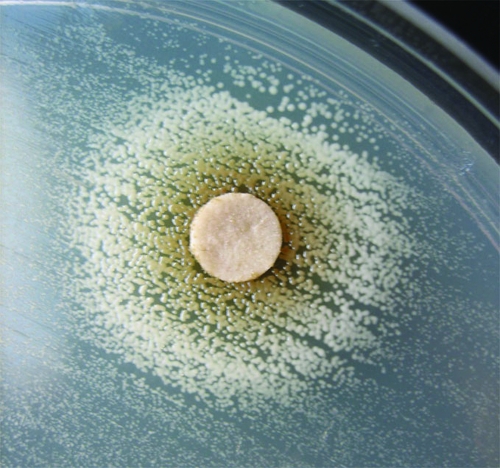
FIG. 1.
Complementation of hemin auxotrophy (8325-4 FusE-SCV).
Selection of Kanr SCV mutants of 8325-4.
To test the robustness of our classification of some FA-resistant mutants as SCVs, we reversed the selection process. Thus, we selected Kanr mutants, screened for SCVs, and asked whether any displayed an FA-resistant phenotype. In a preliminary experiment, SCVs were selected both in a rifampin-resistant (Rifr) derivative of 8325-4 and in the original Rifs 8325-4. The use of a marked strain is an extra control that the selected SCVs originate from the strain plated. In those selections, 21/524 Rifr SCVs were FA resistant, while 51/3,300 Rifs SCVs were FA resistant. The low percentage of FA-resistant SCVs among the SCVs selected with KAN may explain why the FA-resistant class has not been identified previously by others. We concluded that some mutants selected with KAN as SCVs have the FA-resistant phenotype and proceeded to make a larger-scale experiment to obtain mutants for genetic and phenotypic analysis. Twenty independent cultures of S. aureus 8325-4 Rifr were grown overnight into the stationary phase. A total of 108 cells from each culture were spread on LA containing 8, 16, or 24 μg/ml of KAN. The plates were incubated 48 h at 37°C to allow slow-growing variants to form visible colonies. At 8 μg/ml of KAN, ∼103 colonies were visible, while at 16 and 24 μg/ml, the numbers of Kanr colonies were ∼200 and ∼30, respectively. Eight Kanr clones (two medium/large and six small) were picked from each culture and selection level, purified on selective plates, and then screened for FA resistance (LA plus FA, 1 μg/ml). Of the 360 SCV Kanr clones checked, 64 (18%) were resistant to FA. No FA-resistant mutants were found among the 120 medium/large colonies tested. The fusA gene (EF-G) was completely sequenced in each of these 64 FA-resistant SCV mutants. Mutations causing single-amino-acid substitutions in EF-G were identified in 38/64 mutants. When multiple identical isolates from the same culture were excluded, 27 independently selected mutations (12 different amino acid substitutions) were identified, distributed between structural domains I, III, and V of EF-G (Table 2). The majority of SCV mutations in EF-G occurred in domain V (20 independent, 6 different), followed by domain III (5 independent, 4 different), and finally domain I (2 different). The remaining 25 FA-resistant SCVs carried no mutations in fusA and were thus classified as FusE mutants.
TABLE 2.
Mutants selected in 8325-4 as Kanr SCVs and then screened for FA resistance
| Strain and stock (SCV) | Identified mutationa
|
MIC (μg/ml) of:
|
Doubling time (min)b | Auxotrophyc | nd | |||
|---|---|---|---|---|---|---|---|---|
| fusA (EF-G) | rplF (L6) | KAN | STR | FA | ||||
| 8325-4 | ||||||||
| AH001 | 1.5 | 3 | 0.125 | 24 | − | |||
| FusA-SCV | ||||||||
| AH366 (SCV-38) | Pro114His | None | 12 | 24 | 12 | 77 | Hemin | |
| AH369 (SCV-41) | Gln115Leu | None | 6 | 12 | 48 | 52 | Hemin | |
| AH364 (SCV-36) | Asp434Asn | None | 8 | 24 | 256 | 64 | Hemin | |
| AH334 (SCV-6) | Thr436Ile | None | 128 | 192 | 48 | 54 | Hemin + menadione | |
| AH336 (SCV-8) | His438Asn | None | 16 | 12 | 8 | 65 | Hemin | |
| AH338 (SCV-10) | Arg464Cys | None | 48 | 24 | 2 | 67 | Hemin | 2 |
| AH340 (SCV-12) | Gly617Asp | None | 12 | 24 | 4 | 52 | Hemin | 7 |
| AH332 (SCV-4) | Ala655Glu | None | 12 | 12 | 12 | 68 | Hemin | 2 |
| AH329 (SCV-1) | Arg659Cys | None | 12 | 24 | 12 | 63 | Hemin | 5 |
| AH331 (SCV-3) | Arg659His | None | 16 | 24 | 8 | 49 | Hemin | 3 |
| AH347 (SCV-19) | Arg659Ser | None | 12 | 32 | 12 | 91 | Hemin | |
| AH367 (SCV-39) | Gly664Ser | None | 12 | 16 | 12 | 46 | Hemin | 2 |
| FusE | ||||||||
| AH380 (SCV-52) | None | −1 FS at nt 158 | 4 | 24 | 6 | 52 | Hemin | |
| AH352 (SCV-24) | None | −1 FS at nt 106 | 6 | 24 | 4 | 53 | Hemin | |
| AH377 (SCV-49) | None | +1 FS at nt 23 | 6 | 24 | 4 | 49 | Hemin | |
| AH348 (SCV-20) | None | TGA stop at nt 244 | 8 | 16 | 8 | 62 | Hemin | |
| AH370 (SCV-42)e | None | TAA stop at nt 229 | 8 | 24 | 8 | 46 | Hemin | |
| AH346 (SCV-18) | None | −1 FS at nt 238 | 8 | 24 | 8 | 77 | Hemin | |
| AH333 (SCV-5) | None | TAA stop at nt 249 | 8 | 24 | 12 | 56 | Hemin | |
| AH392 (SCV-64) | None | −1 FS at nt 383 | 12 | 16 | 4 | 57 | Hemin | |
| AH379 (SCV-51) | None | TAA stop at nt 418 | 12 | 16 | 6 | 45 | Hemin | 2 |
| AH349 (SCV-21) | None | −1 FS at nt 404 | 16 | 64 | 4 | 67 | Hemin | |
| AH374 (SCV-46) | None | −5 FS at nt 236-240 | 32 | 96 | 8 | 59 | Hemin | |
| AH358 (SCV-30)f | None | −1 FS at nt 423 | 48 | 64 | 12 | 100 | Hemin | |
| AH339 (SCV-11) | None | −1 FS at nt 144 | 64 | 128 | 6 | 120 | Hemin | |
| AH337 (SCV-9)e | None | −1 FS at nt 220 | 96 | 256 | 12 | 130 | Hemin | |
| AH357 (SCV-29) | None | TAA stop at nt 301 | 96 | 256 | 12 | 79 | Hemin | |
| AH384 (SCV-56)g | None | −1 FS at nt 221 | 128 | 256 | 24 | 140 | Hemin | |
| AH378 (SCV-50)e,g | None | −1 FS at nt 221 | 16 | 256 | 8 | 200 | Menadione | |
| AH351 (SCV-23) | None | −4 FS at nt 45-48 | 64 | 256 | 16 | 180 | Menadione | |
| AH360 (SCV-32) | None | ATA start at nt 3 | 96 | 192 | 24 | 140 | Menadione | |
| AH383 (SCV-55) | None | TAA stop at nt 139 | 96 | 192 | 16 | 200 | Menadione | |
| AH362 (SCV-34) | None | TAA stop at nt 282 | 128 | 256 | 12 | 170 | Menadione | |
| AH330 (SCV-2)f | None | −1 FS at nt 423 | 128 | 96 | 12 | 200 | Menadione | |
| AH390 (SCV-62) | None | +1 FS at nt 84 | 256 | 64 | 16 | 170 | Menadione | |
| AH386 (SCV-58)e | None | −1 FS at nt 90 | 256 | 96 | 24 | 180 | Menadione | |
None, wild type. Underlined letters represent the mutated residue. FS, frameshift mutation.
Doubling time (generation time) in minutes during exponential growth in LB.
Auxotrophy tested for hemin, menadione, thymine, and thymidine. −, nonauxotrophic.
Number of independent isolates with the same genotype and phenotype.
SCV-42, -9, -50, and -58 were sequenced for all ribosomal proteins and rRNA operons to determine whether any other mutations in the ribosome could distinguish between the hemin- and menadione-auxotrophic classes.
One of a pair of mutants with the same mutation in rplF but different phenotypes.
One of a pair of mutants with the same mutation in rplF but different phenotypes.
Phenotypes of Kanr-selected SCVs of 8325-4.
Each of the 64 FA-resistant SCV strains was assayed for MIC, doubling time, and complementation of auxotrophy by hemin, menadione (Fig. 2), thiamine, and thymidine. The results are summarized in Table 2, showing a single representative of each of the 12 different FusA mutants and all 25 FusE mutants. All of the mutants had increased MICs for KAN (3- to 170-fold wild type), STR (3- to 80-fold wild type), and FA (32 to 384-fold wild type). In addition, the MIC for the aminoglycoside gentamicin was increased by 4- to 48-fold (data not shown). Doubling times in liquid media were increased significantly for all SCV mutants, as expected. Among the 12 different FusA SCVs, 11/12 were complemented by hemin alone, while 1 was complemented by both hemin and menadione. Among the 25 FusE SCVs, 17 were complemented by hemin, while the remaining 8 were complemented by menadione (Table 2). None of the 37 SCV mutants was complemented by thiamine or thymidine.
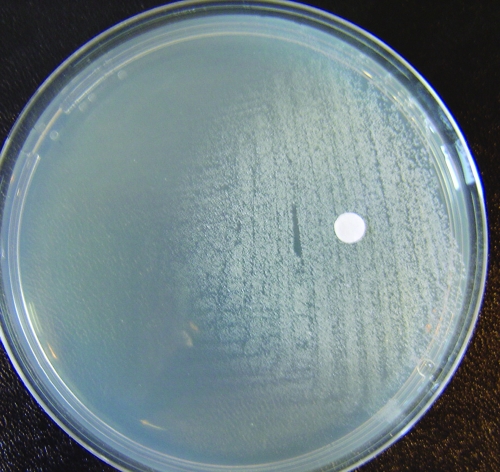
FIG. 2.
Complementation of menadione auxotrophy (8325-4 FusE-SCV).
We also tested 19 SCVs selected independently in 8325-4 as Kanr but not showing the FA resistance phenotype on plates. Each of these had a MIC for KAN in the range of 24 to 256 μg/ml and a MIC for FA that was unchanged from the wild type (0.125 μg/ml). Thus, only a specific subgroup of aminoglycoside-resistant SCVs have an increased MIC for FA.
We asked whether FA-resistant SCVs might be mutator strains: i.e., whether the two phenotypes FA resistance and SCV might have arisen independently because of a hypermutator phenotype. Eleven FA-resistant SCVs, independently isolated in 8325-4, were tested for mutation frequency by screening five independent cultures of each strain for spontaneous Rifr mutants on LA with 100 μg/ml RIF. The parental strain 8325-4 was used as a control on mutation frequency. The Rifr mutation frequencies ranged from 10−8 to 6 × 10−7. In addition, we measured the mutation frequency to Strr for the FA-resistant SCVs listed in Table 2. (Strr was assayed because these mutants are already RIF resistant). No significant increase in mutation frequency relative to the wild type was observed for any of the mutants. The normal range of mutation frequencies argues against hypermutation as the origin of FA resistance among SCVs.
Identification of mutations in ribosomal protein L6 specific to FusE mutants.
We rationalized that the FA-resistant phenotype of the FusE mutants might be caused by a mutation affecting the ribosome. Accordingly we choose two hemin-auxotrophic (SCV-9 and SCV-42) and two menadione-auxotrophic (SCV-50 and SCV-58) FusE mutants and determined, by PCR and DNA sequencing, their genotypes for each of the rRNA operons and for all 56 annotated ribosomal protein genes, including those that have been documented to interact with EF-G (8). In all four strains, we identified mutations in only one of the sequenced genes, rplF, the gene coding for ribosomal protein L6. We then sequenced rplF in all SCVs analyzed in this paper. We found that all FusE mutants had mutations in rplF (Tables 1, 2, and 3), whereas this gene was wild type in all FusA-SCVs. The mutations identified in rplF are predicted to knock out the function of the gene. They each mutate the coding sequence and include nucleotide substitutions altering the start codon, insertion, or deletion mutations that alter the reading frame and nucleotide substitution mutations that introduce premature termination codons. Mutations were located throughout rplF, from as early as the start codon to as late as nucleotide (nt) 418 in the 534-nt coding sequence. We also sequenced rplF in 15 of the FA-resistant SCVs isolated in the preliminary selections in 8325-4 and the 8325-4 Rifr variant and identified L6 mutations in both sets of strains (seven different frameshift mutations in rplF [data not shown]). Finally, we sequenced fusA and rplF in 19 independently isolated FA-susceptible SCVs (selected in 8325-4 for KAN resistance) and found no mutations in either gene. We conclude that knockout mutations in rplF are specific to the FusE mutants and are not a general characteristic of all SCVs. We also predict that one or both of the FusE subgroups, either the hemin auxotrophs or the menadione auxotrophs, must carry an additional mutation, to explain this difference in auxotrophy. From our sequence analysis we conclude that the additional mutation is not in a ribosomal protein or rRNA gene.
TABLE 3.
Mutants selected as Kanr SCVs in FA-susceptible clinical isolates and then screened for FA resistance
| Strain and stock | Sourcea | Identified mutationb
|
Antibiotic MIC (μg/ml) of:
|
Doubling time (min)c | Auxotrophyd | ne | |||
|---|---|---|---|---|---|---|---|---|---|
| fusA (EF-G) | rplF (L6) | KAN | STR | FA | |||||
| IN476-IN494 | None | None | 2 | 6 | 0.047 | 24 | − | ||
| FusA-SCV | |||||||||
| AH421 | IN488 | Pro404Arg | None | 8 | 12 | 3 | 36 | − | |
| AH288 | IN476 | Gly617Asp | None | 16 | 32 | 6 | 53 | Hemin | 2 |
| AH294 | IN490 | Gly628Val | None | 16 | 8 | 2 | 40 | − | |
| AH287 | IN476 | Ala655Glu | None | 16 | 48 | 12 | 64 | Hemin | |
| AH291 | IN484 | Arg659His | None | 24 | 24 | 4 | 43 | − | |
| AH297 | IN492 | Ser660Pro | None | 24 | 2 | 53 | Hemin | ||
| AH296 | IN490 | Gly664Ala | None | 16 | 8 | 3 | 39 | − | |
| AH292 | IN484 | Gly666Val | None | 32 | 16 | 6 | 36 | − | |
| AH298 | IN494 | Gly666Val | None | 32 | 6 | 6 | 37 | − | |
| FusE | |||||||||
| AH422 | IN488 | None | +2 FS at nt 222 | 128 | 32 | 12 | 81 | Menadione | |
| AH290 | IN478 | None | Duplicate nt 202-293 | 256 | 64 | 8 | 200 | Menadione | |
| AH420 | IN484 | None | Δ of nt 72-80 | 256 | 256 | 12 | 150 | Menadione | |
| AH417 | IN480 | None | −1 FS at nt 422 | 256 | 256 | 24 | 180 | Menadione | |
| AH416 | IN476 | None | Δ of nt 395-398 | 256 | 256 | 48 | 170 | Menadione | |
Each of the original FA-susceptible clinical isolates had similar MICs for KAN, STR, and FA and similar doubling times in LB.
None, wild type. FS, frameshift mutation.
Doubling time (generation time) in minutes during exponential growth in LB.
Auxotrophy tested for hemin, menadione, thymine, and thymidine. −, nonauxotrophic.
Number of independent isolates with same genotype and phenotype.
Selection of Kanr SCVs in FA-susceptible clinical isolates of S. aureus.
Two independent cultures of each of 10 different FA-susceptible (MIC, ≤0.25 μg/ml) clinical bacteremia isolates were grown; used to select for Kanr at 8, 16, or 24 μg/ml; and then screened for SCVs. Similar numbers of Kanr colonies were selected at each level as previously found with strain 8325-4. Eight small colonies were picked from each selection plate (480 colonies total), purified at the same level of KAN, and then screened for FA resistance (LA plus FA, 1 μg/ml). Fifteen FA-resistant mutants were isolated from 8 of the 10 clinical isolates (Table 3). DNA sequencing of fusA showed that 10 mutants belonged to the FusA-SCV class (mutant fusA, EF-G) with the remaining 5 being FusE (mutant rplF, L6). Among the FusA-SCV mutants, 9 of 10 were in domain V, while 1 mutation was mapped in domain III (Pro404Arg, which has not been described before). Four of the FusA-SCVs were auxotrophic for hemin, while the remaining six showed no auxotrophy. These six FusA SCVs were the fastest growing of those isolated, and the lack of obvious complementation probably reflects their intrinsically faster growth. All five of the FusE mutants were complemented by menadione. All 15 FA-resistant SCVs selected from the clinical isolates also showed loss of pigmentation. We concluded that SCVs with an FA-resistant phenotype, of both the FusA-SCV and FusE classes, can be selected from clinical isolates of S. aureus.
DISCUSSION
Genetics and phenotypes of FA-resistant SCVs.
Recently the genetic basis of the SCV phenotype in thymidine-auxotrophic clinical isolates of S. aureus was identified as knockout mutations in thyA, the gene coding for thymidylate synthase (12). The genetic causes of other clinical or spontaneous SCVs are still unknown, but the hemin- and menadione-auxotrophic SCV phenotypes have been reproduced in laboratory strains constructed by disrupting hemB (56) and menD (9), genes essential for cytochrome biosynthesis.
Here we have identified a novel subset of S. aureus SCVs that are resistant to FA. Thus, we have shown that some mutants selected in S. aureus for resistance to the antibiotic FA are also SCV mutants. Using the reverse approach, we have shown that some mutants selected in S. aureus as SCVs, on the basis of resistance to aminoglycosides, are resistant to FA. The FA-resistant SCVs fall into distinct genetic and phenotypic classes and carry mutations in fusA or in rplF. (i) FusA-SCV mutants have amino acid substitutions in EF-G (most in structural domain V, but some in domain I or III) and are auxotrophic for hemin. (ii) FusE mutants have knockout mutations in rplF (ribosomal protein L6) and are auxotrophic for hemin or for menadione. The different auxotrophies associated with the FusE class (some of which have identical mutations in rplF [see Table 2]) suggest that one or the both of these subgroups must carry an additional mutation. This interrelatedness of the FA resistance and SCV phenotypes was demonstrated both in the standard laboratory wild-type strain, 8325-4, and in a variety of clinical S. aureus isolates.
Clinical relevance of FA-resistant SCVs.
FA-resistant SCV mutants have several features that make them interesting from a clinical perspective. First, they arise at a high spontaneous frequency relative to other FA-resistant mutants. When FA resistance is selected at ∼1 μg/ml of the drug, approximately 50% of the resistant mutants belong to the SCV classes (Table 1) (38). Second, the MICs of the resulting FA-resistant SCVs are in the same range as the great majority of classical FusA mutants and of FusB strains. Third, although their characteristic slow growth might be expected to result in a strong Darwinian selection against them in the absence of antibiotic selective pressure, there is evidence to suggest that this interpretation is too simplistic. Thus, S. aureus SCVs have been associated with an ability to establish and maintain chronic infections as shown directly in animal models and indirectly by the isolation of SCVs from patients suffering chronic infections (12, 46). This feature has not been shown directly for FA-resistant SCVs as they have not previously been described. However, we earlier described an FA-resistant clinical isolate that carries multiple mutations in fusA causing amino acid substitutions in structural domains III and V of EF-G (38). The domain V residue mutated in that clinical isolate, A655P, is also mutated in FusA-SCVs selected in this study in 8325-4 (Table 2) and in a susceptible clinical isolate (Table 3), although the substitution, A655E, is different. The multiply mutated FA-resistant clinical isolate carried, in addition to A655P, two substitutions in domain III (L461F and D463G), in a region previously associated with mutations causing FA resistance (38). One attractive possibility to explain this genotype is that that the domain V mutation arose as an SCV and that the two mutations in domain III were subsequently selected for improved growth fitness and/or increased FA resistance. Because FA-resistant SCVs arise spontaneously at a high frequency, they might be a significant source of FA-resistant mutants in clinical settings. This remains to be studied.
FA resistance mutations in EF-G and L6 in terms of structure and position.
The major function of EF-G is to promote the translocation, in a coordinated movement, of two tRNA molecules and the mRNA, through the central cavity formed between the two subunits of the ribosome. The distribution of FA resistance mutations in EF-G is strikingly nonrandom. All of the mutations, both classical FusA and FusA-SCV, map in one of the EF-G structural domains I, III, or V. The domain boundaries of EF-G have been defined for the structure of the homologous protein from Thermus thermophilus (2) transposed to S. aureus (32). Structural domain I (the guanine nucleotide-binding domain) extends from residues 1 to 280, domain III from residues 404 to 483, and domain V from residues 606 to 693. Classical FA resistance mutations selected in S. aureus in this study and those selected previously (38) map overwhelmingly in structural domain III (80 independent isolates), with a few found in domain I (5 independent isolates). In contrast, the FusA-SCV mutations map mostly in structural domain V (31 independent isolates), with fewer in domains III (5 independent isolates) and I (2 independent isolates). Clearly, the FusA-SCV mutants are strongly associated with domain V, whereas the classical FusA mutants are predominantly associated with alterations in domain III. The interaction of EF-G with the ribosome has been mapped in the posttranslocational state by site-directed hydroxyl radical probing, including three residues in domain V (64). This showed that amino acid residues in domain V of EF-G are in very close proximity to specific regions of 23S rRNA on the large ribosomal subunit, in particular to helices 43, 44, 89, and 95 (known also as the sarcin-ricin loop). This unambiguously positions domain V of EF-G relative to 23S rRNA. When additional hydroxyl radical probing data, together with the available structural data (3-5), were used to dock EF-G onto the ribosome, it supported this positioning of domain V and further suggested that structural domain III of EF-G lies in close proximity to ribosomal protein S12 on the 30S ribosomal subunit (63). Additional structural studies of EF-G and its interactions with the ribosome may provide information on the functional significance of these mutational preferences and their association with the different phenotypes associated with classical FusA and FusA-SCV mutants.
Mutants selected in Escherichia coli for reduced susceptibility to the aminoglycoside gentamicin have been shown to be mutated in ribosomal protein L6 (15). These mutants also have reduced susceptibility to other aminoglycosides. The L6 mutations restrict the errors of translation (31). Two of these L6 mutations have been characterized genetically. They carry a 7-nt duplication or an 11-nt deletion, respectively, in each case predicted to result in a reading frame shift and premature termination in the C-terminal half of the protein (21). These truncated mutant proteins have been shown to be present within the ribosomes of gentamicin-resistant E. coli (15). The increased accuracy of translation by these mutant ribosomes may be caused by increased proofreading of ternary complexes, because the L6 mutations are predicted to distort elongation factor binding on the large subunit (21). L6 is situated at the subunit interface close to the GTPase center in the region of the L7/L12 stalk (8) and has been cross-linked to EF-G (54). L6 may modulate the RNA structure that forms the elongation factor binding site (37). The L6 mutants isolated in the present study in S. aureus are distributed throughout the rplF coding sequence and are also predicted to cause premature termination of L6 synthesis. In two extreme cases, we have identified mutations altering the initiation codon (Table 1 and Table 2). Our conclusion from the nature and distribution of the mutations in rplF is that the SCV phenotype of FusE mutants can be associated with a ribosome completely lacking L6 protein. This does not rule out the possibility that for some of the FusE mutants a truncated L6 is bound to the ribosome. An unusual feature of protein L6 in the ribosome is that, unlike most ribosomal proteins, it interacts significantly with several other ribosomal proteins. The members of this protein cluster include L3, L6, L13, and L14, and all are found close to the elongation factor binding site (8). The existence of this protein cluster motivated us to ask whether a second mutation in one of these proteins might explain the two different phenotypes, hemin or menadione auxotrophy, associated with FusE mutants. Accordingly, we sequenced the genes for each of these ribosomal proteins from two hemin-auxotrophic and two menadione-auxotrophic FusE mutants. We extended this analysis by sequencing the genes for all 56 annotated ribosomal proteins based on sequences in the S. aureus genome and for each of the rRNA genes. We found no additional mutations in any of the four mutants and thus cannot yet explain the basis for the two phenotypic variants of FusE mutants.
Our tentative conclusion from the genetic analysis of FusA-SCV and FusE mutants is that fusA and rplF mutations cause FA resistance by altering, directly or indirectly, the structural conformations of EF-G on the ribosome and thus its sensitivity to inhibition by FA. The documented aminoglycoside resistance caused by L6 mutations in E. coli (15) and the interactions of L6 and EF-G on the ribosome (8) make it plausible that the reduced aminoglycoside susceptibility we have measured in FusA-SCV and FusE mutants is also, at least in part, caused at the level of antibiotic binding to the ribosome.
Similarities between FA-resistant SCVs and fusA1 in Salmonella enterica serovar Typhimurium.
An FA-resistant mutant in Salmonella enterica serovar Typhimurium shares some phenotypic characteristics with the FA-resistant SCVs described here. The fusA1 allele has the mutation P414L in domain III of EF-G (26). fusA1 causes slow growth both on rich solid medium and in liquid medium. The mutant P414L EF-G has an increased Km for GTP and is inhibited in translation in vitro by ppGpp (35). ppGpp is a global transcriptional regulator molecule produced in a RelA-dependent reaction at the factor binding site of the ribosome (17). ppGpp is normally produced at a low basal level in fast-growing cells and is strongly induced when cells enter starvation or stationary phase (17). However, strains with the mutant P414L EF-G make only one-third the normal basal level of ppGpp and have a very low level of induction upon starvation (35). ppGpp is a positive regulator of the RNA polymerase sigma factor RpoS (22), itself a global regulator of stress response genes (24). Strains with the mutant P414L EF-G are defective in RpoS induction (34) and as a consequence produce reduced levels of heme, have increased sensitivity to oxidative stress, and have a reduced rate of aerobic respiration (33). Addition to the growth medium of precursors of heme biosynthesis alleviates the sensitivity to oxidative stress (33). Thus, this FA-resistant mutant in S. enterica serovar Typhimurium shares with FA-resistant SCV mutants in S. aureus an auxotrophy in the heme biosynthesis pathway. It was earlier shown that mutations that directly affect heme biosynthesis, hemL in S. enterica serovar Typhimurium (16) or hemB in E. coli (49), result in SCVs that cause persistent and recurrent infections. In the case of fusA1, it is clear that the SCV-like phenotypes are indirect consequences of alterations in gene expression resulting from an aberrant EF-G-ribosome interaction. Accordingly, the FA-resistant SCVs in S. aureus may also have their SCV phenotypes as a result of altered gene expression patterns resulting from aberrant ribosome activities, mediated through either mutant EF-G or mutant L6. We have also shown that mutations altering EF-G or ribosomal protein L6 are specific to FA-resistant SCVs, and they were not found in any of 19 independently isolated FA-susceptible SCVs.
Potential for coselection of FA resistance in S. aureus.
SCVs of S. aureus frequently show increased resistance to other antibiotics (46) and can even be selected by triclosan (52). A clinically relevant conclusion from our data is that FA resistance in S. aureus can be selected by aminoglycosides. If this is also so in the clinical setting, then the dynamics of antimicrobial drug use driving the selection and spread of FA resistance will have to be considered in a wider context.
Acknowledgments
This work was supported by grants from the European Union 6th Framework Programme (EAR project), the Swedish Research Council (Vetenskapsrådet), and the Leo Pharma Forskningsfond to D.H.
Footnotes
Published ahead of print on 8 October 2007.
REFERENCES
- 1.Abele-Horn, M., B. Schupfner, P. Emmerling, H. Waldner, and H. Goring. 2000. Persistent wound infection after herniotomy associated with small-colony variants of Staphylococcus aureus. Infection 28:53-54. [DOI] [PubMed] [Google Scholar]
- 2.Aevarsson, A., E. Brazhnikov, M. Garber, J. Zheltonosova, Y. Chirgadze, S. al-Karadaghi, L. A. Svensson, and A. Liljas. 1994. Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J. 13:3669-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agrawal, R. K., A. B. Heagle, P. Penczek, R. A. Grassucci, and J. Frank. 1999. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat. Struct. Biol. 6:643-647. [DOI] [PubMed] [Google Scholar]
- 4.Agrawal, R. K., J. Linde, J. Sengupta, K. H. Nierhaus, and J. Frank. 2001. Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation. J. Mol. Biol. 311:777-787. [DOI] [PubMed] [Google Scholar]
- 5.Agrawal, R. K., P. Penczek, R. A. Grassucci, and J. Frank. 1998. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc. Natl. Acad. Sci. USA 95:6134-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atkins, B., and T. Gottlieb. 1999. Fusidic acid in bone and joint infections. Int. J. Antimicrob. Agents 12(Suppl. 2):S79-S93. [DOI] [PubMed] [Google Scholar]
- 7.Balwit, J. M., P. van Langevelde, J. M. Vann, and R. A. Proctor. 1994. Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J. Infect. Dis. 170:1033-1037. [DOI] [PubMed] [Google Scholar]
- 8.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 9.Bates, D. M., C. von Eiff, P. J. McNamara, G. Peters, M. R. Yeaman, A. S. Bayer, and R. A. Proctor. 2003. Staphylococcus aureus menD and hemB mutants are as infective as the parent strains, but the menadione biosynthetic mutant persists within the kidney. J. Infect. Dis. 187:1654-1661. [DOI] [PubMed] [Google Scholar]
- 10.Becker, K., N. Al Laham, W. Fegeler, R. A. Proctor, G. Peters, and C. von Eiff. 2006. Fourier-transform infrared spectroscopic analysis is a powerful tool for studying the dynamic changes in Staphylococcus aureus small-colony variants. J. Clin. Microbiol. 44:3274-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Besier, S., A. Ludwig, V. Brade, and T. A. Wichelhaus. 2003. Molecular analysis of fusidic acid resistance in Staphylococcus aureus. Mol. Microbiol. 47:463-469. [DOI] [PubMed] [Google Scholar]
- 12.Besier, S., A. Ludwig, K. Ohlsen, V. Brade, and T. A. Wichelhaus. 2007. Molecular analysis of the thymidine-auxotrophic small colony variant phenotype of Staphylococcus aureus. Int. J. Med. Microbiol. 297:217-225. [DOI] [PubMed] [Google Scholar]
- 13.Bodley, J. W., F. J. Zieve, L. Lin, and S. T. Zieve. 1969. Formation of the ribosome-G factor-GDP complex in the presence of fusidic acid. Biochem. Biophys. Res. Commun. 37:437-443. [DOI] [PubMed] [Google Scholar]
- 14.Brouillette, E., A. Martinez, B. J. Boyll, N. E. Allen, and F. Malouin. 2004. Persistence of a Staphylococcus aureus small-colony variant under antibiotic pressure in vivo. FEMS Immunol. Med. Microbiol. 41:35-41. [DOI] [PubMed] [Google Scholar]
- 15.Buckel, P., A. Buchberger, A. Bock, and H. G. Wittmann. 1977. Alteration of ribosomal protein L6 in mutants of Escherichia coli resistant to gentamicin. Mol. Gen. Genet. 158:47-54. [DOI] [PubMed] [Google Scholar]
- 16.Cano, D. A., M. G. Pucciarelli, M. Martinez-Moya, J. Casadesús, and F. Garcia-del Portillo. 2003. Selection of small-colony variants of Salmonella enterica serovar Typhimurium in nonphagocytic eucaryotic cells. Infect. Immun. 71:3690-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 18.Chatterjee, I., M. Herrmann, R. A. Proctor, G. Peters, and B. C. Kahl. 2007. Enhanced post-stationary-phase survival of a clinical thymidine-dependent small-colony variant of Staphylococcus aureus results from lack of a functional tricarboxylic acid cycle. J. Bacteriol. 189:2936-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chopra, I. 1976. Mechanisms of resistance to fusidic acid in Staphylococcus aureus. J. Gen. Microbiol. 96:229-238. [DOI] [PubMed] [Google Scholar]
- 20.Clements, M. O., S. P. Watson, R. K. Poole, and S. J. Foster. 1999. CtaA of Staphylococcus aureus is required for starvation survival, recovery, and cytochrome biosynthesis. J. Bacteriol. 181:501-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies, C., D. E. Bussiere, B. L. Golden, S. J. Porter, V. Ramakrishnan, and S. W. White. 1998. Ribosomal proteins S5 and L6: high-resolution crystal structures and roles in protein synthesis and antibiotic resistance. J. Mol. Biol. 279:873-888. [DOI] [PubMed] [Google Scholar]
- 22.Gentry, D. R., V. J. Hernandez, L. H. Nguyen, D. B. Jensen, and M. Cashel. 1993. Synthesis of the stationary-phase sigma factor σs is positively regulated by ppGpp. J. Bacteriol. 175:7982-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godtfredsen, W. O., S. Jahnsen, H. Lorck, K. Roholt, and L. Tybring. 1962. Fusidic acid: a new antibiotic. Nature 193:987. [DOI] [PubMed] [Google Scholar]
- 24.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σs (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howden, B. P., and M. L. Grayson. 2006. Dumb and dumber—the potential waste of a useful antistaphylococcal agent: emerging fusidic acid resistance in Staphylococcus aureus. Clin. Infect. Dis. 42:394-400. [DOI] [PubMed] [Google Scholar]
- 26.Johanson, U., and D. Hughes. 1994. Fusidic acid-resistant mutants define three regions in elongation factor G of Salmonella typhimurium. Gene 143:55-59. [DOI] [PubMed] [Google Scholar]
- 27.Jonsson, I. M., C. von Eiff, R. A. Proctor, G. Peters, C. Ryden, and A. Tarkowski. 2003. Virulence of a hemB mutant displaying the phenotype of a Staphylococcus aureus small colony variant in a murine model of septic arthritis. Microb. Pathog. 34:73-79. [DOI] [PubMed] [Google Scholar]
- 28.Kahl, B., M. Herrmann, A. S. Everding, H. G. Koch, K. Becker, E. Harms, R. A. Proctor, and G. Peters. 1998. Persistent infection with small colony variant strains of Staphylococcus aureus in patients with cystic fibrosis. J. Infect. Dis. 177:1023-1029. [DOI] [PubMed] [Google Scholar]
- 29.Kahl, B. C., G. Belling, R. Reichelt, M. Herrmann, R. A. Proctor, and G. Peters. 2003. Thymidine-dependent small-colony variants of Staphylococcus aureus exhibit gross morphological and ultrastructural changes consistent with impaired cell separation. J. Clin. Microbiol. 41:410-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, H. B., H.-C. Jang, H. J. Nam, Y. S. Lee, B. S. Kim, W. B. Park, K. D. Lee, Y. J. Choi, S. W. Park, M.-d. Oh, E.-C. Kim, and K. W. Choe. 2004. In vitro activities of 28 antimicrobial agents against Staphylococcus aureus isolates from tertiary-care hospitals in Korea: a nationwide survey. Antimicrob. Agents Chemother. 48:1124-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhberger, R., W. Piepersberg, A. Petzet, P. Buckel, and A. Bock. 1979. Alteration of ribosomal protein L6 in gentamicin-resistant strains of Escherichia coli. Effects on fidelity of protein synthesis. Biochemistry 18:187-193. [DOI] [PubMed] [Google Scholar]
- 32.Laurberg, M., O. Kristensen, K. Martemyanov, A. T. Gudkov, I. Nagaev, D. Hughes, and A. Liljas. 2000. Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site. J. Mol. Biol. 303:593-603. [DOI] [PubMed] [Google Scholar]
- 33.Macvanin, M., A. Ballagi, and D. Hughes. 2004. Fusidic acid-resistant mutants of Salmonella enterica serovar Typhimurium have low levels of heme and a reduced rate of respiration and are sensitive to oxidative stress. Antimicrob. Agents Chemother. 48:3877-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macvanin, M., J. Björkman, S. Eriksson, M. Rhen, D. I. Andersson, and D. Hughes. 2003. Fusidic acid-resistant mutants of Salmonella enterica serovar Typhimurium with low fitness in vivo are defective in RpoS induction. Antimicrob. Agents Chemother. 47:3743-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macvanin, M., U. Johanson, M. Ehrenberg, and D. Hughes. 2000. Fusidic acid-resistant EF-G perturbs the accumulation of ppGpp. Mol. Microbiol. 37:98-107. [DOI] [PubMed] [Google Scholar]
- 36.Mason, B. W., and A. J. Howard. 2004. Fusidic acid resistance in community isolates of methicillin susceptible Staphylococcus aureus and the use of topical fusidic acid: a retrospective case-control study. Int. J. Antimicrob. Agents 23:300-303. [DOI] [PubMed] [Google Scholar]
- 37.Melançcon, P., W. E. Tapprich, and L. Brakier-Gingras. 1992. Single-base mutations at position 2661 of Escherichia coli 23S rRNA increase efficiency of translational proofreading. J. Bacteriol. 174:7896-7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagaev, I., J. Bjorkman, D. I. Andersson, and D. Hughes. 2001. Biological cost and compensatory evolution in fusidic acid-resistant Staphylococcus aureus. Mol. Microbiol. 40:433-439. [DOI] [PubMed] [Google Scholar]
- 39.Norén, T., T. Åkerlund, M. Wullt, L. G. Burman, and M. Unemo. 2007. Mutations in fusA associated with posttherapy fusidic acid resistance in Clostridium difficile. Antimicrob. Agents Chemother. 51:1840-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Brien, F. G., C. Price, W. B. Grubb, and J. E. Gustafson. 2002. Genetic characterization of the fusidic acid and cadmium resistance determinants of Staphylococcus aureus plasmid pUB101. J. Antimicrob. Chemother. 50:313-321. [DOI] [PubMed] [Google Scholar]
- 41.O'Neill, A. J., F. McLaws, G. Kahlmeter, A. S. Henriksen, and I. Chopra. 2007. Genetic basis of resistance to fusidic acid in staphylococci. Antimicrob. Agents Chemother. 51:1737-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Neill, A. J., and I. Chopra. 2006. Molecular basis of fusB-mediated resistance to fusidic acid in Staphylococcus aureus. Mol. Microbiol. 59:664-676. [DOI] [PubMed] [Google Scholar]
- 43.O'Neill, A. J., A. R. Larsen, A. S. Henriksen, and I. Chopra. 2004. A fusidic acid-resistant epidemic strain of Staphylococcus aureus carries the fusB determinant, whereas fusA mutations are prevalent in other resistant isolates. Antimicrob. Agents Chemother. 48:3594-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osterlund, A., G. Kahlmeter, S. Haeggman, and B. Olsson-Liljequist. 2006. Staphylococcus aureus resistant to fusidic acid among Swedish children: a follow-up study. Scand. J. Infect. Dis. 38:332-334. [DOI] [PubMed] [Google Scholar]
- 45.Proctor, R. A., P. van Langevelde, M. Kristjansson, J. N. Maslow, and R. D. Arbeit. 1995. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 20:95-102. [DOI] [PubMed] [Google Scholar]
- 46.Proctor, R. A., C. von Eiff, B. C. Kahl, K. Becker, P. McNamara, M. Herrmann, and G. Peters. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4:295-305. [DOI] [PubMed] [Google Scholar]
- 47.Rayner, C., and W. J. Munckhof. 2005. Antibiotics currently used in the treatment of infections caused by Staphylococcus aureus. Intern. Med. J. 35(Suppl. 2):S3-S16. [DOI] [PubMed] [Google Scholar]
- 48.Rennie, R. P. 2006. Susceptibility of Staphylococcus aureus to fusidic acid: Canadian data. J. Cutan. Med. Surg. 10:277-280. [DOI] [PubMed] [Google Scholar]
- 49.Roggenkamp, A., A. Sing, M. Hornef, U. Brunner, I. B. Autenrieth, and J. Heesemann. 1998. Chronic prosthetic hip infection caused by a small-colony variant of Escherichia coli. J. Clin. Microbiol. 36:2530-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaaff, F., G. Bierbaum, N. Baumert, P. Bartmann, and H. G. Sahl. 2003. Mutations are involved in emergence of aminoglycoside-induced small colony variants of Staphylococcus aureus. Int. J. Med. Microbiol. 293:427-435. [DOI] [PubMed] [Google Scholar]
- 51.Schroder, A., R. Kland, A. Peschel, C. von Eiff, and M. Aepfelbacher. 2006. Live cell imaging of phagosome maturation in Staphylococcus aureus infected human endothelial cells: small colony variants are able to survive in lysosomes. Med. Microbiol. Immunol. (Berlin) 195:185-194. [DOI] [PubMed] [Google Scholar]
- 52.Seaman, P. F., D. Ochs, and M. J. Day. 2007. Small-colony variants: a novel mechanism for triclosan resistance in methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 59:43-50. [DOI] [PubMed] [Google Scholar]
- 53.Seo, H. S., S. Abedin, D. Kamp, D. N. Wilson, K. H. Nierhaus, and B. S. Cooperman. 2006. EF-G-dependent GTPase on the ribosome: conformational change and fusidic acid inhibition. Biochemistry 45:2504-2514. [DOI] [PubMed] [Google Scholar]
- 54.Skold, S. E. 1982. Chemical cross-linking of elongation factor G to both subunits of the 70-S ribosomes from Escherichia coli. Eur. J. Biochem. 127:225-229. [DOI] [PubMed] [Google Scholar]
- 55.Spelman, D. 1999. Fusidic acid in skin and soft tissue infections. Int. J. Antimicrob. Agents 12(Suppl. 2):S59-S66. [DOI] [PubMed] [Google Scholar]
- 56.von Eiff, C., C. Heilmann, R. A. Proctor, C. Woltz, G. Peters, and F. Götz. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179:4706-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Eiff, C., P. McNamara, K. Becker, D. Bates, X.-H. Lei, M. Ziman, B. R. Bochner, G. Peters, and R. A. Proctor. 2006. Phenotype microarray profiling of Staphylococcus aureus menD and hemB mutants with the small-colony-variant phenotype. J. Bacteriol. 188:687-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.von Eiff, C., G. Peters, and K. Becker. 2006. The small colony variant (SCV) concept—the role of staphylococcal SCVs in persistent infections. Injury 37(Suppl. 2):S26-S33. [DOI] [PubMed] [Google Scholar]
- 59.von Eiff, C., R. A. Proctor, and G. Peters. 2000. Small colony variants of staphylococci: a link to persistent infections. Berl. Muench. Tieraerztl. Wochenschr. 113:321-325. [PubMed] [Google Scholar]
- 60.Whitby, M. 1999. Fusidic acid in septicaemia and endocarditis. Int. J. Antimicrob. Agents 12(Suppl. 2):S17-S22. [DOI] [PubMed] [Google Scholar]
- 61.Whitby, M. 1999. Fusidic acid in the treatment of methicillin-resistant Staphylococcus aureus. Int. J. Antimicrob. Agents 12(Suppl. 2):S67-S71. [DOI] [PubMed] [Google Scholar]
- 62.Willie, G. R., N. Richman, W. P. Godtfredsen, and J. W. Bodley. 1975. Some characteristics of and structural requirements for the interaction of 24,25-dihydrofusidic acid with ribosome-elongation factor G complexes. Biochemistry 14:1713-1718. [DOI] [PubMed] [Google Scholar]
- 63.Wilson, K. S., and R. Nechifor. 2004. Interactions of translational factor EF-G with the bacterial ribosome before and after mRNA translocation. J. Mol. Biol. 337:15-30. [DOI] [PubMed] [Google Scholar]
- 64.Wilson, K. S., and H. F. Noller. 1998. Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell 92:131-139. [DOI] [PubMed] [Google Scholar]