Abstract
Antimicrobial peptides (AMPs) are among the leading candidates to replace antibiotics which have been rendered ineffective by the evolution of resistant bacterial strains. Concerns do exist, however, that the therapeutic administration of AMPs may also select for resistant strains but with much more dire consequences, as these peptides represent an endogenous and essential component of host immune defense. The recent demonstration that AMPs function as ligands for the bacterial sensory kinase PhoQ for the initiation of virulence and adaptive responses lends credence to these concerns. While the ability to serve as PhoQ ligands suggests that the therapeutic administration of AMPs could (i) exacerbate infections by promoting bacterial virulence and (ii) select resistant mutants by encouraging adaptive behaviors, it also provides a rational basis for AMP selection and optimization. Here, we demonstrate that derivatives of a representative AMP have differential abilities to serve as PhoQ ligands and that this correlates with the ability to induce bacterial adaptive responses. We propose that PhoQ-activating potential is a logical parameter for AMP optimization and introduce a novel strategy for the treatment of minimal bactericidal concentration data that permits the discrimination and quantification of the contributions of PhoQ-activating potential and direct antimicrobial activity to net antimicrobial efficiency.
The ability to treat bacterial infections with antibiotics has been a cornerstone of human medicine for over 60 years. However, the extensive application of antibiotics has served as a selection pressure to drive the evolution of human bacterial pathogens which are resistant to these treatments (41). The implications of this trend are socially and economically severe, as drug-resistant nosocomial bacterial infections cost an estimated 5 billion dollars a year and are responsible for more than 63,000 annual deaths in North America alone. Research efforts to develop novel antibiotics have not been in proportion to the magnitude of this growing problem, and there is a growing sense of urgency for the discovery or creation of novel antimicrobial compounds that are active against a broad range of pathogens, have minimal host toxicity, and perhaps most importantly, do not readily select for resistant mutants. Cationic antimicrobial peptides (AMPs) appear to meet all of these criteria (22, 27).
AMPs are a component of innate immune defense that is conserved in virtually all forms of life. While a diverse range of AMPs of various lengths and sequences have been identified, in general, these molecules tend to be small, of less than 50 amino acids in length, with a high percentage of cationic and hydrophobic residues (21). These peptides are either constitutively expressed or induced in response to infection, tissue damage, or other inflammatory stimuli (13, 42, 44). Genetic knockout of AMPs renders the host more susceptible to bacterial infections (32, 43), as does AMP deficiency through genetic disorders such as specific granule deficiency syndrome (15), atopic dermatitis (33), and morbus Kostmann syndrome (36). Conversely, overexpression (4, 38) or therapeutic administration (11, 16) of AMPs strengthens an animal's ability to counter infections and sepsis.
While the specific details of direct antimicrobial action have yet to be determined, it is hypothesized that the majority of AMPs, although not all (8, 39), kill bacteria by compromising the integrity of their membranes, which ultimately leads to cell lysis (29). That AMPs of such diverse sequences, as well as derivatives of individual AMPs, possess antimicrobial activity against a broad spectrum of bacteria suggests that the mechanism of action involves general characteristics of these peptides and their bacterial targets. Specifically, the positive charge of AMPs is hypothesized to be an essential feature in ensuring specificity for bacterial membranes, whose outer leaflet is rich in negatively charged phospholipids, as opposed to plant and animal membranes, which favor neutral lipids (29). This versatility of their structure-antimicrobial activity relationship highlights the potential for AMPs to serve as flexible templates for the construction of novel antibiotics.
Appropriate concerns have been raised that the therapeutic administration of AMPs could serve as a stimulus for the development of bacterial resistance, not unlike what has been observed for many conventional antibiotics. Since these molecules, unlike current antibiotics, are a part of our natural defensive arsenal, widespread resistance could be associated with much more dire consequences. Indeed, it has been speculated that bacterial resistance to AMPs could permanently compromise our ability to counter infections (5). This certainly represents a worst-case scenario, and it should be noted that compelling arguments against the likelihood and severity of this outcome have been presented. The most notable are (i) that these peptides have remained effective antimicrobials over the course of evolution; (ii) that the mechanism of AMP action involves conserved and relatively nonadaptable microbial targets, thus reducing the potential for developmental resistance; (iii) that direct antimicrobial activity (DAA) is only one facet of their antimicrobial action; and (iv) that resistant strains are very difficult to select (20).
While the evolutionary success of AMPs illustrates the apparent inability of bacteria to develop effective resistance mechanisms, it is also important to consider that the administration of these molecules outside of the physiological context might represent a unique milieu for the development of such strains. While many of the AMPs being designed for therapeutic application are for the modulation of innate immune responses rather than DAA, it is not the intent of the application that will determine the ability of bacteria to establish adaptive responses. Indeed, as the doses employed for immunomodulation may be sublethal for many bacterial strains, this may present an effective breeding ground for resistant strains.
While AMP resistance is difficult to select, such strains are observed in nature (34). Resistance is often achieved through modifications of the bacterial outer membrane to prevent AMP binding. The bacterial membrane is a dynamic structure whose composition can be modulated in response to various stressors, including AMPs. In particular, the lipid A moiety of lipopolysaccharide (LPS) has the potential to undergo modifications that result in AMP resistance (30). In Salmonella, modifications of lipid A are also associated with macrophage invasion (17), signifying their role in virulence, and modifications of lipid A have been shown to be equally important to the virulence of other bacteria (35). The PhoPQ two-component system has a central role in the regulation of LPS modifications (14, 19).
The PhoPQ two-component system consists of a transmembrane sensory kinase, PhoQ, and a cytosolic response regulator, PhoP. The activation of PhoQ results in its autophosphorylation, with subsequent phosphotransfer to PhoP (18). Once activated through phosphorylation, PhoP functions as a transcription factor to regulate the expression of a wide range of genes, including those associated with AMP resistance and virulence. The PhoPQ system is essential for the virulence of Salmonella in mice and humans (31) and has proven to be equally important for the virulence of a number of other gram-negative bacteria (12). From the perspective of this work, the significance of the PhoPQ system is that, in response to a single trigger, pathogenic bacteria initiate responses required for virulence and intracellular survival within host macrophages, leading to the establishment of chronic infections and decimation throughout the body.
There has been extensive research demonstrating that PhoQ is activated in vitro by cation-deficient environments and, specifically, that micromolar concentrations of magnesium activate PhoQ while millimolar levels repress the system (18). However, the physiological significance of PhoPQ activation by divalent cations has been called into question (1, 10). An alternative, and elegant, regulatory mechanism has recently been proposed with the demonstration that PhoQ can be directly activated by AMPs (3). In this hypothesis, bacteria utilize AMPs as indicators of the host environment to simultaneously initiate offensive and defensive maneuvers that adapt the bacteria to the host cell assault, including bestowing resistance to AMPs (2, 6). It has been demonstrated that the exposure of bacteria to either Mg2+-deficient environments or sublethal concentrations of AMPs induces adaptive patterns of expression that result in AMP-resistant phenotypes (2).
The ability to serve as PhoQ ligands and initiate adaptations that result in AMP resistance is in opposition to the antimicrobial objective of AMPs. As such, AMP optimization will benefit from consideration of AMP behavior from the two distinct perspectives of antimicrobial activity and PhoQ-activating potential (PAP). These two characteristics may function independently such that more-effective peptides can be generated through the optimization of either parameter. That all peptides do not share equal ability to activate PhoQ indicates that specific sequence and/or structural characteristics are associated with PhoQ activation potential (3). It may, therefore, be possible to design peptides with a reduced ability to activate PhoQ while maintaining or improving DAA. Peptides with reduced PAP would be anticipated to have improved therapeutic potential as they are less likely to induce bacterial responses that are deleterious to the host.
Here, we examine the hypothesis that AMPs have differential abilities to serve as PhoQ ligands and that this is predictive of their likelihood of inducing bacterial virulence and adaptive responses. Through two independent assay systems, we confirm that AMP derivatives have differential abilities to serve as PhoQ ligands and that this correlates with the induction of adaptive responses as determined by the development of AMP resistance. Based on these observations, we propose that PAP is a logical parameter for the selection and optimization of therapeutic AMPs and, toward this objective, introduce a novel strategy for the treatment of minimal bactericidal concentration (MBC) data, obtained against wild-type and PhoPQ mutant strains, that permits the discrimination and quantification of the contributions of PAP and DAA to net peptide antimicrobial efficiency. Understanding the relative contributions of each of these behaviors to antimicrobial efficiency will facilitate greater understanding of the structure-activity relationship of these multifunctional molecules and should facilitate the design of more-effective, and safer, therapeutic peptides.
MATERIALS AND METHODS
Peptide selection.
Bactenecin, also known as bovine dodecapeptide, is a small AMP cathelicidin discovered in bovine neutrophils (37). At 12 amino acids in length, bactenecin is one of the smallest active AMPs. Its small size and extensive characterization make it an ideal peptide model for the examination of the consequences of amino acid substitutions on antimicrobial efficiency and PAP. While the natural peptide is stabilized by an internal disulfide bridge, a linear variant, Bac2A (RLARIVVIRVAR-NH2), shows activity similar to that of the natural peptide against gram-negative bacteria and higher activity against gram-positive bacteria (45). Others have reported on the antimicrobial properties of a complete substitution library of Bac2A against Pseudomonas aeruginosa, as well as a partial screen against Salmonella enterica serovar Typhimurium (23). Based on the reported antimicrobial efficiencies against S. enterica serovar Typhimurium, we selected a series of Bac2A derivatives to represent peptides with improved, maintained, or compromised antimicrobial activity with respect to that of the parent peptide. The nomenclature for the peptide derivatives is maintained from the original publication (23). We also include a negative control peptide, NC, with the irrelevant sequence GATPEDLNQKLS-NH2.
Peptide synthesis.
All peptides were chemically synthesized on a Pioneer solid-phase peptide synthesizer (PerSeptive Biosystems, Foster City, CA) using Fmoc (9-fluorenylmethoxy carbonyl) chemistry. The peptide chains were synthesized from the carboxyl terminus to the amino terminus on [5-(4-Fmoc-aminomethyl-3,5-dimethyloxyphenoxy) valeric acid]-polyethylene glycol-polystyrene (PAL-PEG-PS) resin. Fmoc-protecting groups at the amino terminus were deprotected with piperidine. The peptides were cleaved from the resin with concurrent deprotection of the side chain-protecting groups by treating the resin-bound peptide with trifluoroacetic acid (TFA) (9.3 parts) in the presence of scavengers (anisole-ethyl-methyl sulfide-1,2-ethanedithiol [3:3:1]) for 7 h. The crude peptides were filtered from the resin, and the TFA was evaporated. Diethyl ether was added to the residues to precipitate the crude peptide. The peptides were isolated and purified by high-performance liquid chromatography on Vydac protein C4 columns (1.0 by 25 cm), being eluted with a linear gradient of 5% buffer A (H2O-0.1% TFA)-90% buffer B [acetonitrile-H2O (90/10)-0.01% TFA] for 40 min at a flow rate of 3 ml/minute. The purities and molecular weights of the respective peptides were confirmed by matrix-assisted laser desorption ionization-time of flight mass spectrometry on a PE Biosystems Voyager system 4068 (National Research Council, Plant Biotechnology Institute, Saskatoon, Canada) and by amino acid analysis.
Bacterial strains.
S. enterica serovar Typhimurium was selected as our model pathogen as the majority of investigations of the PhoPQ two-component system, as well as the initial characterization of the abilities of AMPs to serve as PhoQ ligands, have been performed with this bacterium. As an intracellular pathogen, S. enterica serovar Typhimurium is also physiologically relevant for AMP investigations as pathogenic strains of Salmonella encounter AMPs, including bactenecin, during the natural course of the infective cycle, in particular within the phagolysomes of neutrophils.
The strains used in this study were American Type Culture Collection strain ATCC 14028, a smooth virulent strain of S. enterica serovar Typhimurium; PhoQ− strain CS009 (phoQ101::MudJ); and PhoP− strain CS053 (phoP103:: MudJ). The ATCC 14028 strain represents the wild type and was used for the creation of each of the PhoP− and PhoQ− strains. We also employed a Salmonella PhoPQ reporter strain, CS102, which was a kind gift from Samuel Miller of the University of Washington.
Cloning and expression of the PhoPQ operon.
Plasmid pBBR1MCS-4-phoPQ was constructed through PCR amplification of the chromosomal phoPQ operon, including the promoter region, from wild-type S. enterica serovar Typhimurium with primers A, AAAGTCGGGCCAGTTAAG, and B, CGCCGGCAAATTATATCG, using S. enterica serovar Typhimurium chromosomal DNA as template. The resulting PCR product was cloned directly into the EcoRV site of pBBR1MCS-4 (24) to generate plasmid pBBR1MCS-4-phoPQ.
Functional reconstitution of PhoP− and PhoQ−.
The vector containing the Salmonella PhoPQ operon, pBBR1MCS-4-PhoPQ, was transformed into S. enterica serovar Typhimurium PhoP− and PhoQ− strains to create strains PhoPrecon and PhoQrecon, respectively.
Determination of MBC.
The MBCs of the peptides were determined by using a modified broth microdilution method (45). Assays were performed in sterile, 96-well, round-bottomed polypropylene microtiter plates with an inoculum of 5 × 105 bacteria per ml in Luria-Bertani (LB) medium in the presence of various concentrations of the peptides. The plates were incubated at 37°C for 16 h, and the mixtures spotted onto LB agar plates. The MBC was taken as the concentration at which no growth was observed. The MBCs reported represent the averages of the results of at least three trials performed with a linear gradient of peptide concentrations.
Peptide stocks were made in distilled water at a concentration of 2 mg/ml and stored at −20°C. Appropriate dilutions of the peptides were made fresh prior to each MIC trial in 0.2% bovine serum albumin, 0.01% acetic acid and added to the culture following the addition of the bacterial inoculum.
Susceptibilities to conventional antibiotics.
For nonpeptide antibiotics, the MICs were determined by using the same modified broth microdilution method, and the MICs were taken as the concentration at which no growth was observed in the 96-well plate (45). The antibiotics were evaluated through a series of twofold dilutions, and the most frequently observed value from experiments performed in triplicate was reported as the MIC.
Quantification of adaptive responses.
The abilities of peptide derivatives to activate rapid adaptive responses were determined through a modification of the MBC assays. Bacterial cultures were preincubated in the 96-well plate for 4 h in LB medium supplemented with 5 mM MgCl2 in the presence of increasing but sublethal concentrations of AMPs (1 μg/ml at time zero, 1 μg/ml at 1 h, 2 μg/ml at 2 h, and a final addition of 4 μg/ml at 3 h). Following this adaptation period, the bacteria were examined through standard MBC assays to quantify shifts in AMP susceptibility. These results were compared to those for growth in LB medium with the inclusion of 5 mM MgCl2.
PhoPQ reporter assays.
S. enterica serovar Typhimurium strain CS102, which expresses a PhoQ-regulated fusion to Salmonella acid phosphatase (PhoN), was utilized for analysis of PhoPQ activation. The activity of this fusion protein was measured in LB medium containing various concentrations of the peptides and/or magnesium. For growth under PhoPQ-activating conditions, the growth medium was supplemented with 50 μM MgCl2. Growth under PhoPQ-repressive conditions was performed in the presence of 5 mM MgCl2, as were all of the assays of peptide responses. The assay conditions utilized are based on previously described protocols (7).
Calculation of DAA.
DAA is defined here as antimicrobial activity in the absence of PhoPQ-mediated protection, essentially, the sensitivity of a naïve bacteria to peptide killing. This is quantified by the MBCs against either the PhoQ− 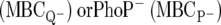 strains. The PhoP and PhoQ mutants were found to be functionally equivalent with respect to their AMP sensitivities, such that either can be used to define DAA. In this work, we chose to use the MBC against PhoQ− Salmonella (
strains. The PhoP and PhoQ mutants were found to be functionally equivalent with respect to their AMP sensitivities, such that either can be used to define DAA. In this work, we chose to use the MBC against PhoQ− Salmonella (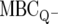 ) bacteria as the measure of DAA, although the MBC data against PhoP− would have been equally appropriate.
) bacteria as the measure of DAA, although the MBC data against PhoP− would have been equally appropriate.
For any peptide derivative (PD), the relative change in DAA with respect to the parent peptide (PP) can be quantified by the formula ΔDAA = 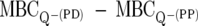 . The ΔDAA score provides a measure of how alterations to peptide sequence influence the ability to kill nonadapted bacteria. While this is anticipated to reflect the ability to disrupt bacterial membranes, the approach is valid regardless of the mechanism of peptide biological action. Negative DAA scores indicate peptide derivatives which are more-effective antimicrobials than the parent peptide.
. The ΔDAA score provides a measure of how alterations to peptide sequence influence the ability to kill nonadapted bacteria. While this is anticipated to reflect the ability to disrupt bacterial membranes, the approach is valid regardless of the mechanism of peptide biological action. Negative DAA scores indicate peptide derivatives which are more-effective antimicrobials than the parent peptide.
Calculation of PAP.
The PAP is the extent to which AMPs prompt bacteria to initiate phenotypic changes that decrease AMP sensitivity. In the context of this investigation, it more specifically refers to alterations mediated through the PhoPQ system. These adaptations reflect a bacterial response, as well as representing an intrinsic property of the peptide in its ability to initiate these adaptations by serving as a PhoQ ligand. For any given peptide, the PAP is calculated as the difference in MBCs between the wild type (wt) and the PhoQ− mutant such that PAP = MBCwt − MBCQ−.
Changes in the ability of a peptide derivative to activate bacterial defense mechanisms compared to the ability of the parent peptide are calculated by the formula ΔPAP = [MBCwt(PD) − 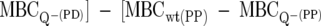 ]. A negative score indicates that the peptide derivative is a more-effective antimicrobial as a result of minimized induction of PhoQ-mediated adaptations.
]. A negative score indicates that the peptide derivative is a more-effective antimicrobial as a result of minimized induction of PhoQ-mediated adaptations.
The equations for the calculation of changes in DAA and PAP are designed such that negative scores indicate improved antimicrobials. This is to conceptually reflect that improved AMPs have lower MBCs.
Contributions of DAA and PAP to peptide efficiency.
Through our approach, changes in peptide efficiency (PE) can be more specifically defined in terms of the changes to DAA and PAP whereby ΔPE = ΔDAA + ΔPAP. Negative scores of peptide efficiency indicate more-effective antimicrobials, with the magnitude of the score reflecting the degree of improvement. Notably, the ΔPE score is numerically equivalent to the difference in MBCs between a parent peptide and its derivative against wild-type bacteria. The advantage of this approach is that it permits the discrimination and quantification of the relative contributions of each of these peptide behaviors toward net changes in peptide efficiency.
RESULTS
Disruption of PhoPQ does not increase sensitivity to non-AMP antibiotics.
Genetic disruption of PhoPQ may increase its sensitivity to antibiotics independent of their direct association or involvement with this two-component system. First, genetic manipulations, independent of the biological function of the targeted gene, have the potential to increase antibiotic susceptibility as a result of a general reduction in bacterial viability. Alternatively, as the PhoPQ system is a central mechanism for the regulation of bacterial membrane modifications, disruption of this system may affect membrane permeability in a manner that renders the bacterium more sensitive to antibiotics.
To verify that the mutant strains do not have a nonspecific increase in antibiotic sensitivity, we determined the MICs for six conventional antibiotics against the PhoQ− mutant strain. No significant differences in susceptibilities were observed for these nonpeptide antibiotics, which confirms that the alterations to peptide susceptibility result specifically from the loss of the PhoPQ system (Table 1).
TABLE 1.
MICs of nonpeptide antibiotics against wild-type and PhoQ mutants of Salmonellaa
| Antibiotic | MIC (μg/ml) for:
|
|
|---|---|---|
| Wild type | PhoQ− | |
| Ampicillin | 10.0 | 8.0 |
| Carbenicillin | 10.0 | 8.0 |
| Gentamicin | 16.0 | 16.0 |
| Tetracycline | 3.0 | 3.0 |
| Nalidixic acid | 3.0 | 3.0 |
| Trimethoprim | 1.0 | 1.0 |
A series of nonpeptide antibiotics were screened against wild-type, PhoQ−, and PhoP− strains of Salmonella enterica serovar Typhimurium. The most frequently observed MIC value from triplicate experiments is reported.
PhoP− and PhoQ− strains are sensitized to AMPs.
Peptides were selected for this investigation based upon a previous report in which a substitution library of Bac2A derivatives was quantified with respect to antimicrobial activity (23). Specific peptides were selected from this library to represent derivatives with either improved (R3 and Sub3), maintained (G12), or compromised (P7 and K7) antimicrobial activity. MBCs were determined for all of these peptides against wild-type, PhoQ−, and PhoP− Salmonella strains.
The PhoPQ mutant strains were considerably sensitized to the antimicrobial effects of all of the peptides. This emphasizes the importance of this system in facilitating protection against AMPs (Table 2). The MBCs for individual peptides against the PhoQ− and PhoP− strains are comparable, indicating that these mutants are functionally equivalent in their inability to initiate protective responses to AMPs. This is not unexpected, as the inability to detect or respond to a stimulus as a result of the loss of the histidine kinase or response regulator, respectively, would be anticipated to result in an equivalent phenotype.
TABLE 2.
MBCs of select Bac2A peptide derivativesa
| Sequence | Peptide | MBC (μg/ml) for:
|
|||
|---|---|---|---|---|---|
| Wild type | PhoQ− | PhoP− | PhoQrecon | ||
| GATPEDLNQKLS-NH2 | NC | >500 | >500 | >500 | >500 |
| RLARIVVIRVAR-NH2 | Bac2a | 32.8 ± 4.1 | 6.2 ± 1.6 | 6.5 ± 1.5 | 42.5 ± 3.5 |
| RLRRIVVIRVAR-NH2 | R3 | 11.5 ± 3.2 | 4.2 ± 1.3 | 3.8 ± 0.9 | 30.0 ± 5.0 |
| RLARIVKIRVAR-NH2 | K7 | 68.8 ± 3.7 | 13.1 ± 2.0 | 13.3 ± 2.1 | 69.0 ± 1.4 |
| RLARIVPIRVAR-NH2 | P7 | >500 | 46.7 ± 4.1 | 45.4 ± 3.2 | >500 |
| RLARIVVIRVAG-NH2 | G12 | 31.4 ± 3.5 | 8.6 ± 1.3 | 10.1 ± 1.9 | 34.0 ± 4.2 |
| RRWRIVVIRVRR-NH2 | Sub3 | 10.0 ± 2.0 | 5.7 ± 1.0 | 6.4 ± 0.9 | 19.0 ± 3.8 |
A series of peptides were screened against wild-type, PhoQ−, and PhoP− strains of Salmonella enterica serovar Typhimurium, as well as the functionally reconstituted PhoQrecon strain. The MBCs represent the averages ± standard deviations of the results of triplicate experiments.
Functional reconstitution of the PhoP− or PhoQ− strains with a plasmid expressing the PhoPQ operon, creating the PhoPrecon and PhoQrecon strains, returns the MBCs to levels comparable to those in the wild-type bacterium (Table 2). However, a consistent result for all of the peptides is that the MBCs against the PhoQrecon strain were slightly higher than for the wild-type bacterium, indicating a lower AMP susceptibility. This may be a consequence of higher levels of expression of PhoP and PhoQ from the plasmid than from the endogenous operon.
Contributions of DAA to antimicrobial efficiency.
Among the derivatives, we observed examples with improved (Sub3 and R3), maintained (G12), or compromised (K7 and P7) direct antimicrobial efficiencies. Relative to the Bac2A parent peptide, the ΔDAA scores for the derivatives ranged from −2.4 to +40.5 μg/ml (Table 3). Quantitatively, the most significant alterations were associated with derivatives that had reduced efficiencies. It is not unexpected that the alteration of the sequence of an evolutionarily selected peptide or protein would be more likely to result in a loss of, rather than improvement of, biological activity. That the magnitudes of the changes to DAA are insufficient to account for the range of antimicrobial efficacies observed against wild-type bacteria indicates that alterations to other aspects of peptide behavior also contribute to the changes in efficiency.
TABLE 3.
Calculation of changes in DAA and activation of PAPsa
| Peptide | DAA | ΔDAA | PAP | ΔPAP | ΔPE |
|---|---|---|---|---|---|
| Bac2a | 6.2 ± 1.6 | −26.6 ± 4.4 | |||
| R3 | 4.2 ± 1.3 | −2.0 ± 2.1 | 7.3 ± 3.5 | −19.3 ± 5.4 | −21.3 ± 5.8 |
| K7 | 13.1 ± 2.0 | 6.9 ± 26 | 55.7 ± 4.2 | 29.1 ± 5.8 | 33.4 ± 6.4 |
| P7 | 46.7 ± 3.1 | 40.5 ± 3.5 | |||
| G12 | 8.6 ± 1.3 | 2.4 ± 2.1 | 22.8 ± 3.7 | −3.8 ± 5.5 | −1.4 ± 5.9 |
| Sub3 | 5.7 ± 1.0 | −0.5 ± 1.9 | 34.3 ± 2.2 | −22.3 ± 4.7 | −22.8 ± 5.1 |
The peptides R3 and Sub3 are improved antimicrobials as indicated by the decreased MBC against wild-type Salmonella. This effect is largely mediated through the decreased tendency to activate PhoQ-dependent PAPs, as indicated by the ΔPAP scores. The scores are the averages ± standard deviations of the results.
While the changes in DAA for most of the derivatives were relatively minor, the P7 derivative offers an example of a peptide in which a single amino acid substitution results in dramatic alteration to antimicrobial activity. This peptide is inactive against the wild-type bacterium and has only minimal DAA against the sensitized PhoPQ mutant strains. As the formation of helical structure is thought to be a prerequisite for the antimicrobial activity of many AMPs, the fact that the introduction of a proline in the central region of Bac2A abolishes this function is consistent with this hypothesis.
Contribution of PAP to antimicrobial efficiency.
Among the derivatives, there were examples of peptides with both increases and decreases to the calculated ΔPAPs. For example, the ΔPAP scores for the R3 and Sub3 peptides were −19.3 μg/ml and −22.3 μg/ml, respectively, implying that these peptides are more-effective antimicrobials than the parent peptide as a consequence of their decreased ability to serve as PhoQ ligands. In contrast, the K7 peptide appears to be a less-effective antimicrobial as a consequence of being a more-potent PhoQ ligand, with a ΔPAP score of 29.1 μg/ml. The G12 substitution does not appear to affect the ability to serve as a PhoQ ligand as the PAP score is comparable to that of Bac2A (Table 3).
Contribution of DAA and PAP to antimicrobial efficiency.
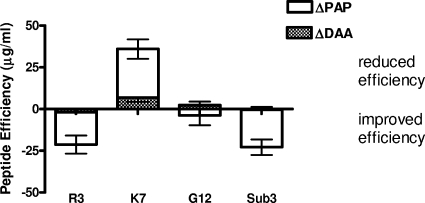
Two of the peptides, R3 and Sub3, are significantly improved as antimicrobials, with decreases in their MBCs relative to those of the parent peptide of −21.3 and −22.8 μg/ml, respectively, against wild-type Salmonella. The magnitudes of these improvements closely correspond with their calculated ΔPAP scores of −19.3 and −22.3 μg/ml, indicating that, for these peptides, the change in the PAP is the principal, although not exclusive, determinant of the improved antimicrobial efficiency (Fig. 1). Presumably, for these derivatives, the ability to avoid detection by the PhoPQ system allows them to act more effectively against membranes that have not been adapted for AMP resistance.
FIG. 1.
Relative contributions of changes in DAA and tendencies to activate PAPs toward the change in net peptide efficiency of a peptide derivative compared to that of the parent peptide. Peptides with negative scores indicate more effective antimicrobials. Error bars show standard deviations.
In contrast, as demonstrated by the results for the K7 peptide, alterations to PAP also have the ability to compromise antimicrobial efficiency. K7 has compromised antimicrobial activity, as demonstrated by an MBC of 68.8 μg/ml against wild-type Salmonella compared to 32.8 μg/ml for the parent peptide. This reduced antimicrobial activity appears to result predominantly from an increased ability to act as a PhoQ ligand, as indicated by the ΔPAP score of 29.1 μg/ml. This suggests that effective recognition of K7 by PhoQ permits the bacteria the opportunity to initiate adaptations that protect against AMPs, thereby increasing the MBC.
While theoretically possible, we did not observe any examples where changes to DAA and PAP were in functional opposition to each other. While the G12 derivative appears to show both a modest improvement with respect to PAP and an equally small loss of DAA, the magnitudes of these alterations are not statistically significant.
Verification of differential PhoPQ activation by peptide derivatives.
The underlying assumption of the mathematical model is that the PAP score reflects differential abilities to activate PhoPQ-dependent responses. However, the PhoPQ regulon is complex, with the ability to modulate a wide variety of bacterial behaviors, as well as to influence other two-component systems, such as pmrAB, which has also been implicated in resistance to certain AMPs (2, 28, 40). We therefore sought to validate through an independent methodology that the calculated ΔPAP scores accurately predict the ability of specific AMPs to activate PhoQ.
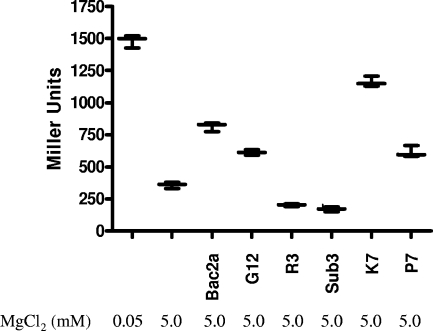
Salmonella strain CS102, in which the expression of an acid phosphatase reporter enzyme is driven by activated PhoP, permits the quantification of PhoPQ activity. This system has demonstrated utility in measuring PhoQ activation by various ligands and conditions, including AMPs (3). Employing this system, we confirmed the differential abilities of the peptides to activate PhoQ. In general, Bac2A derivatives are potent PhoQ ligands, as low levels (1 μg/ml) of these peptides achieve degrees of activation comparable to what is observed under micromolar concentrations of magnesium (Fig. 2). This is consistent with previous reports of other AMPs that are similarly effective in stimulating PhoPQ (3).
FIG. 2.
Activation of PhoPQ by AMPs. The abilities of Bac2A peptide derivatives to initiate the activation of a PhoPQ-dependent reporter were examined by the incubation of bacteria in the presence of sublethal (1 μg/ml) concentrations of the indicated peptides, as well as 1 mM magnesium. The values reported represent the averages of the results for three replicate experiments. The activation and repression of PhoPQ by low (50 μM) and high (5 mM) magnesium concentrations, respectively, are also reported. Error bars show standard deviations.
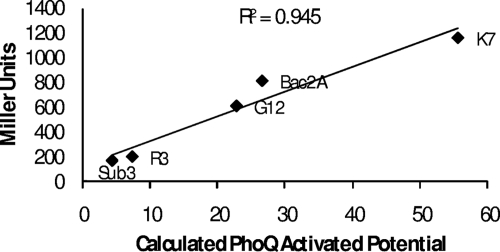
Importantly, a strong correlation was observed between the calculated PAP scores and the degree of PhoPQ activation observed in the live bacterial reporter strain. Peptides such as K7 which had a higher PAP score than the parent peptide were more-effective ligands for the reporter strain, while peptides such as Sub3 and R3, whose PAP scores are lower than the score for Bac2A, were less-effective PhoPQ activators. A linear regression plot confirms the relationship between the calculated PAP scores and the directly measured PhoQ activation, verifying the validity of our mathematical approach (Fig. 3).
FIG. 3.
Verification that calculated PAPs accurately predict the ability of a peptide to serve as a PhoQ ligand. The close correlation between calculated and observed PAPs is demonstrated by a linear regression plot.
PhoQ ligand potential predicts induction of AMP resistance.
It has been observed that exposure to sublethal concentrations of AMPs promotes adaptive responses that include resistance to a broad spectrum of AMPs (2). These adaptations may be of physiological significance, as it has been demonstrated that bacteria taken from the lungs of cystic fibrosis patients have MICs which are orders of magnitude higher than those observed for nonadapted strains (28). While it has yet to be definitively proven that the resistance of these bacteria to AMPs represents an adaptive response, the lung surface does present a suitable location for ongoing exposure to AMPs.
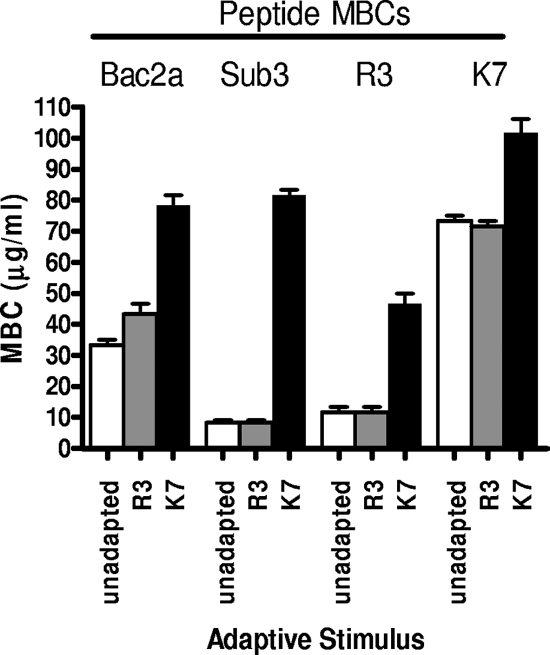
To verify that exposure to sublethal concentrations of AMPs initiates bacterial defensive responses and that the magnitude of these responses is predicted by the PAP scores, bacteria were preincubated with increasing sublethal concentrations of either the K7 or R3 peptides over a 4-h time period. These derivatives were selected as they represent the extremes of PhoQ ligand efficiencies, K7 with the greatest and R3 with the least PAP, as determined through both the calculated PAP scores and direct measurement of PhoPQ activation. Following this preadaptive period, the bacteria were subjected to a standard MBC assay to quantify the extent of any protective effect. Consistent with the hypothesis that PhoQ ligand potential predicts the ability to induce bacterial adaptations, K7 had the most dramatic shift in MBC scores, corresponding to up-to-10-fold increases over the values observed for naïve bacteria. In contrast, bacteria preincubated with the R3 peptide, which is a poor PhoQ ligand, had limited protection against AMPs, as the associated MBCs were comparable to those for the naïve, unadapted bacteria (Fig. 4).
FIG. 4.
Activation of AMP responses following exposure to sublethal concentrations of AMPs. The abilities of peptide derivatives to induce adaptive responses were determined by preincubation of bacteria for 4 h in the presence of sublethal concentrations of peptides prior to standard MBC assays. The values reported represent the averages of the results of three repeated experiments. Error bars show standard deviations. All assays were performed in the presence of 1 mM Mg2+.
DISCUSSION
Given the potential for AMPs to function as novel antibiotics, there is considerable value in elucidating their structure-activity relationship so that more-effective molecules can be rationally designed. Such efforts typically involve screening a panel of AMP derivatives for antimicrobial activity and subsequently rationalizing changes in efficacy with respect to changes in sequence/structure. However, the recent demonstrations that AMPs activate PhoQ-dependent responses, bestowing AMP resistance, and that individual AMPs have unique abilities to initiate these responses, limit the ability to interpret antimicrobial data in such a linear fashion, as AMP sequence alterations may influence the net antimicrobial efficiency through alterations of either DAA or PAP. As changes to the abilities of AMP derivatives to activate PhoQ will also determine their propensities to induce undesired bacterial responses, the mechanisms underlying changes to antimicrobial efficiency may be as significant as the magnitudes and direction of these changes.
To facilitate a more-descriptive analysis of the structure-function relationship of AMPs, we have established a methodology based upon a novel treatment of MBC data for wild-type and PhoPQ mutant strains that permits the discrimination and quantification of alterations in DAA and PAP. This methodology offers numerous advantages over traditional approaches. First, our strategy allows the simultaneous evaluation of both DAA and PhoQ ligand potential within a single experiment. Second, as the input for the equations is derived from live bacteria, the information obtained is more likely than that obtained from traditional phosphotransfer assays, which employ semipurified proteins and are also further disadvantaged by their dependence on radioactive materials, to accurately reflect physiological ligand binding by PhoQ. Our system can be utilized for any bacteria for which either a PhoP or PhoQ mutant strain exists, and while we have utilized MBC data, MIC data would be equally appropriate, as the mechanism of AMPs is bacteriocidal such that MICs and MBCs usually coincide (27).
Most of the bactenecin derivatives within our investigation fell within a limited range of DAA. This may arise due to the inability of the sensitized strains to discriminate the efficiencies of these peptides, but it more likely indicates that the majority of these substitutions permit sufficient maintenance of structure and properties to retain antimicrobial action. This is anticipated, as the ability for AMP to serve as flexible templates for the creation of novel antibiotics is an attractive feature of this class of molecules for antibiotic design. In contrast to the limited effect of the derivatives on DAA, more dramatic alterations were observed for the PAP. This would suggest, at least for these derivatives, that the association of peptides with the bacterial membranes for the purpose of antimicrobial activity is more tolerant of sequence alteration than interactions with PhoQ. While both of these interactions are anticipated to be driven largely by electrostatic interactions, it is not unexpected that associations with proteins would have greater structural stringency than interactions with membranes.
The differential abilities of the various peptides to serve as PhoQ ligands were confirmed through the use of a Salmonella PhoPQ reporter strain, and a high degree of correlation was verified between the observed and calculated PAPs. This validates the utility of the mathematical approach for determining AMP PhoQ ligand potentials. The physiological significance of AMP-mediated PhoQ activation is supported by the observation that peptides with higher PAP scores are more effective in inducing AMP-resistant phenotypes. AMPs, and in particular those with high PAP scores, are potent inducers of adaptive responses, as the incubation of bacteria with sublethal concentrations of these peptides increases AMP resistance by up to an order of magnitude in as short a duration as 4 h.
It might be anticipated that the same sequence and/or structural characteristics that define effective antimicrobials would also predict effective PhoQ ligands. That is to say that the PhoQ sensory system would have evolved to recognize peptides representing the greatest threat to the bacteria. It would appear, however, that the relationship between antimicrobial efficiency and PhoQ ligand potential is not that easily defined. For example, the P7 derivative, which has abolished antimicrobial activity, remains equally effective as a PhoQ ligand as the parent peptide. This indicates that distinct, but undoubtedly overlapping, structural features govern antimicrobial efficiency and PhoQ ligand potential. This is not limited to AMPs, as recent work has demonstrated the ability of cationic polyamines, such as spermidine and spermine, also to induce PhoPQ-dependent bacterial adaptations that result in AMP resistance (25, 26). Clearly, more information is required to elucidate the structural features that define effective antimicrobials and PhoQ ligands.
The availability of the crystallographic structure of the sensory domain of S. enterica serovar Typhimurium PhoQ permits some rationalization of the mechanism of activation by AMPs (9). While structural information is not yet available for complexes of PhoQ with peptide ligands, the peptide binding region has been predicted and verified through a variety of experimental approaches (3). In the proposed model, a planar acidic surface of PhoQ runs parallel to the bacterial membrane. Electrostatic repulsion between the negatively charged bacterial membrane and the acidic surface of PhoQ is neutralized by the coordination of cations by metal binding sites of PhoQ. These electrostatic bridges dock the sensory domain to the membrane in an inactive conformation. In the absence of these counterions, electrostatic repulsion displaces the sensory domain from the membrane, resulting in the activation of the protein. In the crystallographic structure, this region was found to coordinate calcium ions and is suggested by a variety of techniques to represent the region of the protein responsible for binding metal ions and cationic peptides (9).
The abilities of AMPs to activate PhoQ is dependent upon their ability to provide suitable electrostatic interactions to replace those provided by bound cations. This ability is likely conserved to various degrees for all AMPs by virtue of their shared cationic nature, which offers an explanation for the ability of this sensor to be activated by a diverse but structurally related range of molecules. While the interaction is undoubtedly influenced by electrostatic interactions, caution must also be exerted in assuming that peptides with a more-extensive positive charge will be more-effective PhoQ ligands. We observe that the extent of the positive charge associated with an AMP does not predict its ligand potential. For example, the K7 and R3 derivatives, which both have a 1-unit increase in positive charge relative to that of Bac2A, represent the most- and least-effective PhoQ ligands, respectively. As such, it is premature to speculate on the specifics of why various derivatives are better able to serve as ligands.
Acknowledgments
This work was supported by funding from the Natural Sciences and Engineering Research Council to S.N. as well as funding from the Advancing Canadian Agriculture and Agri-Food (ACAAF) program.
Footnotes
Published ahead of print on 15 October 2007.
Published with permission of the director of VIDO as journal series number 464.
REFERENCES
- 1.Alpuche Aranda, C. M., J. A. Swanson, W. P. Loomis, and S. I. Miller. 1992. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc. Natl. Acad. Sci. USA. 89:10079-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bader, M. W., W. W. Navarre, W. Shiau, H. Nikaido, J. G. Frye, M. McClelland, F. C. Fang, and S. I. Miller. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 50:219-230. [DOI] [PubMed] [Google Scholar]
- 3.Bader, M. W., S. Sanowar, M. E. Daley, A. R. Schneider, U. Cho, W. Xu, R. E. Klevit, H. L. Moual, and S. I. Miller. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461-472. [DOI] [PubMed] [Google Scholar]
- 4.Bals, R., D. J. Weiner, A. D. Moscioni, R. L. Meegalla, and J. M. Wilson. 1999. Augmentation of innate host defense by expression of a cathelicidin antimicrobial peptide. Infect. Immun. 67:6084-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell, G., and P. H. Gouyon. 2003. Arming the enemy: the evolution of resistance to self-proteins. Microbiology 149:1367-1375. [DOI] [PubMed] [Google Scholar]
- 6.Bishop, J. L., and B. B. Finlay. 2005. Friend or foe? Antimicrobial peptides trigger pathogen virulence. Trends Mol. Med. 12:3-6. [DOI] [PubMed] [Google Scholar]
- 7.Brickman, E., and J. Beckwith. 1975. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J. Mol. Biol. 96:307-316. [DOI] [PubMed] [Google Scholar]
- 8.Brotz, H., G. Bierbaum, K. Leopold, P. E. Reynolds, and H. G. Sahl. 1998. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 42:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, U. S., M. W. Bader, M. F. Amaya, M. E. Daley, R. E. Klevit, S. I. Miller, and W. Xu. 2006. Metal bridges between the PhoQ sensor domain and the membrane regulate transmembrane signaling. J. Mol. Biol. 356:1193-1206. [DOI] [PubMed] [Google Scholar]
- 10.Christensen, K. A., J. T. Myers, and J. A. Swanson. 2002. pH-dependent regulation of lysosomal calcium in macrophages. J. Cell Sci. 115:599-607. [DOI] [PubMed] [Google Scholar]
- 11.Cirioni, O., A. Giacometti, R. Ghiselli, C. Bergnach, F. Orlando, C. Silvestri, F. Mocchegiani, A. Licci, B. Skelavaj, M. Rocchi, V. Saba, M. Zanetti, and G. Scalise. 2006. LL-37 protects rats against lethal sepsis caused by gram-negative bacteria. Antimicrob. Agents Chemother. 50:1672-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derzelle, S., E. Turlin, E. Duchaud, S. Pages, F. Kunst, A. Givaudan, and A. Danchin. 2004. The PhoP-PhoQ two-component system of Photorhabdus luminescens is essential for virulence in insects. J. Bacteriol. 186:1270-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duits, L. A., B. Ravensbergen, M. Rademaker, P. S. Hiemstra, and P. H. Nibbering. 2002. Expression of beta-defensin 1 and 2 mRNA by human monocytes, macrophages and dendritic cells. Immunology 106:517-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fields, P. I., E. A. Groisman, and F. Heffron. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059-1062. [DOI] [PubMed] [Google Scholar]
- 15.Ganz, T., J. A. Metcalf, J. I. Gallin, L. A. Boxer, and R. I. Lehrer. 1988. Microbicidal/cytotoxic proteins of neutrophils are deficient in two disorders: Chediak-Higashi syndrome and “specific” granule deficiency. J. Clin. Investig. 82:552-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghiselli, R., A. Giacometti, O. Cirioni, G. Dell'Acgua, C. Bergnach, F. Orlando, F. Mocchegiani, C. Silvestri, B. Skallavj, A. Licci, N. Balaban, M. Zanetti, G. Scalise, and V. Saba. 2006. RNAIII-inhibiting peptide in combination with the cathelicidin BMAP-28 reduces lethality in mouse models of staphylococcal sepsis. Shock 26:296-301. [DOI] [PubMed] [Google Scholar]
- 17.Gibbons, H. S., S. R. Kalb, R. J. Cotter, and C. R. Raetz. 2005. Role of Mg2+ and pH in the modification of Salmonella lipid A after endocytosis by macrophage tumour cells. Mol. Microbiol. 55:425-440. [DOI] [PubMed] [Google Scholar]
- 18.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, L., K. B. Lim, J. S. Gunn, B. Bainbridge, R. P. Darveau, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 20.Hancock, R. E. W. 2003. Concerns regarding resistance to self-proteins. Microbiology 149:3343-3344. [DOI] [PubMed] [Google Scholar]
- 21.Hancock, R. E. W., and G. Diamond. 2000. The role of cationic antimicrobial peptides in innate host defenses. Trends Microbiol. 8:402-410. [DOI] [PubMed] [Google Scholar]
- 22.Hancock, R. E., and H. G. Sahl. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24:1551-1557. [DOI] [PubMed] [Google Scholar]
- 23.Hilpert, K., R. Volkmer-Engert, T. Walter, and R. E. Hancock. 2005. High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotechnol. 23:1008-1012. [DOI] [PubMed] [Google Scholar]
- 24.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 25.Kwon, D. H., and C. D. Lu. 2006. Polyamines increase antibiotic susceptibility in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50:1623-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon, D. H., and C. D. Lu. 2007. Polyamine effects of antibiotic susceptibility in bacteria. Antimicrob. Agents Chemother. 51:2070-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marr, A. K., W. J. Gooderham, and R. E. W. Hancock. 2006. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr. Opin. Pharmacol. 6:468-472. [DOI] [PubMed] [Google Scholar]
- 28.McPhee, J. B., S. Lewenza, and R. E. Hancock. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205-217. [DOI] [PubMed] [Google Scholar]
- 29.McPhee, J. B., and R. E. Hancock. 2005. Function and therapeutic potential of antimicrobial peptides. J. Pept. Sci. 11:677-687. [DOI] [PubMed] [Google Scholar]
- 30.Miller, S. I., R. K. Ernst, and M. W. Bader. 2005. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 3:36-46. [DOI] [PubMed] [Google Scholar]
- 31.Miller, S. I., A. M. Kukral, and J. L. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nizet, V., T. Ohtake, X. Lauth, J. Trowbridge, J. Rudisill, R. A. Dorshcner, V. Pestonjamasp, J. Piraino, K. Huttner, and R. L. Gallo. 2001. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414:454-457. [DOI] [PubMed] [Google Scholar]
- 33.Ong, P. Y., T. Ohtake, C. Brandt, I. Strickland, M. Boguniewicz, T. Ganz, R. L. Gallo, and D. Y. Leung. 2002. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N. Engl. J. Med. 347:1151-1160. [DOI] [PubMed] [Google Scholar]
- 34.Peschel, A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10:179-186. [DOI] [PubMed] [Google Scholar]
- 35.Preston, A., E. Maxim, E. Toland, E. J. Pishko, Harvill, E. T. M. Caroff, and D. J. Maskell. 2003. Bordetella bronchiseptica PagP is a Bvg-regulated lipid A palmityl transferase that is required for persistent colonization of the mouse respiratory tract. Mol. Microbiol. 48:725-736. [DOI] [PubMed] [Google Scholar]
- 36.Putsep, K., G. Carlsson, H. G. Boman, and M. Andersson. 2002. Deficiency of antibacterial peptides in patients with morbus Kostmann: an observation study. Lancet 360:1144-1149. [DOI] [PubMed] [Google Scholar]
- 37.Romeo, D., B. Skerlavaj, M. Bolognesi, and R. Gennaro. 1988. Structure and bactericidal activity of an antibiotic dodecapeptide purified from bovine neutrophils. J. Biol. Chem. 263:9573-9575. [PubMed] [Google Scholar]
- 38.Salzman, N. H., D. Ghosh, K. M. Huttner, Y. Paterson, and C. L. Bevins. 2003. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422:522-526. [DOI] [PubMed] [Google Scholar]
- 39.Subbalakshmi, C., and N. Sitaram. 1998. Mechanism of antimicrobial action of indolicidin. FEMS Microbiol. Lett. 160:91-96. [DOI] [PubMed] [Google Scholar]
- 40.Tamayo, R., S. S. Ryan, A. J. McCoy, and J. S. Gunn. 2002. Identification and genetic characterization of PmrA-regulated genes and genes involved in polymyxin B resistance in Salmonella enterica serovar Typhimurium. Infect. Immun. 70:6770-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tenover, F. C. 2001. Development and spread of bacterial resistance to antimicrobial agents: an overview. Clin. Infect. Dis. 33:S108-S115. [DOI] [PubMed] [Google Scholar]
- 42.Vora, P., A. Youdim, L. S. Thomas, M. Fukata, S. Y. Tesfay, K. Lukasek, K. S. Michelsen, A. Wada, T. Hirayama, M. Arditi, and M. T. Abreu. 2004. Beta-defensin-2 expression is regulated by TLR signaling in intestinal epithelial cells. J. Immunol. 173:5398-5405. [DOI] [PubMed] [Google Scholar]
- 43.Wilson, C. L., A. J. Oullette, D. P. Satchell, T. Ayabe, Y. S. Lopez-Boado, J. L. Stratmen, S. J. Hultgren, L. M. Matrisian, and W. C. Parks. 1999. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 286:113-117. [DOI] [PubMed] [Google Scholar]
- 44.Wolk, K., S. Kunz, E. Witte, M. Friedrich, K. Asadullah, and R. Sabat. 2004. IL-22 increases the innate immunity of tissues. Immunity 21:241-254. [DOI] [PubMed] [Google Scholar]
- 45.Wu, M., and R. E. Hancock. 1999. Improved derivatives of bactenecin, a cyclic dodecameric antimicrobial cationic peptide. Antimicrob. Agents Chemother. 43:1274-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]