Abstract
Viruses are ubiquitous and abundant throughout the biosphere. In marine systems, virus-mediated processes can have significant impacts on microbial diversity and on global biogeocehmical cycling. However, viral genetic diversity remains poorly characterized. To address this shortcoming, a metagenomic library was constructed from Chesapeake Bay virioplankton. The resulting sequences constitute the largest collection of long-read double-stranded DNA (dsDNA) viral metagenome data reported to date. BLAST homology comparisons showed that Chesapeake Bay virioplankton contained a high proportion of unknown (homologous only to environmental sequences) and novel (no significant homolog) sequences. This analysis suggests that dsDNA viruses are likely one of the largest reservoirs of unknown genetic diversity in the biosphere. The taxonomic origin of BLAST homologs to viral library sequences agreed well with reported abundances of cooccurring bacterial subphyla within the estuary and indicated that cyanophages were abundant. However, the low proportion of Siphophage homologs contradicts a previous assertion that this family comprises most bacteriophage diversity. Identification and analyses of cyanobacterial homologs of the psbA gene illustrated the value of metagenomic studies of virioplankton. The phylogeny of inferred PsbA protein sequences suggested that Chesapeake Bay cyanophage strains are endemic in that environment. The ratio of psbA homologous sequences to total cyanophage sequences in the metagenome indicated that the psbA gene may be nearly universal in Chesapeake Bay cyanophage genomes. Furthermore, the low frequency of psbD homologs in the library supports the prediction that Chesapeake Bay cyanophage populations are dominated by Podoviridae.
Viruses are the most numerically abundant biological entities in marine ecosystems (9, 49, 62), and the ecological importance of marine viruses is supported by a growing body of research (for reviews, see references 61, 67, and 73). Viruses are an important component of the marine microbial loop (7, 12, 30, 62, 69), and it is likely that all members of marine microbial communities (protists, microalgae, and prokaryotes) are prone to viral infection (17). Marine viral lysis impacts global carbon cycles by enhancing dissolved organic matter recycling, decreasing transfer of carbon to higher trophic levels, and exporting particulate organic carbon from the photic zone (30, 41, 42, 61). For example, viruses can significantly influence concentrations of micronutrients, such as iron, by cell lysis (47) or by acting as nucleation centers for iron adsorption and precipitation (26).
In addition to direct impacts on ocean biogeochemistry, the viral infection process may significantly alter the structure of microbial host communities. Virus-mediated changes in community genetic diversity are induced by selective infection and lysis of abundant community members (45, 63). Thus, host-selective viral infection may increase the overall clonal diversity of microbial host populations by removing numerically dominant community members from particular niches. Viruses, particularly bacteriophages, can also directly alter the phenotypes of host cells through genetic exchange (specific and generalized transduction) (44) or through the cryptic infectious state known as lysogeny (50), which has been observed in many marine ecosystems, especially under conditions which are unfavorable for host growth (40, 70). Because viral nucleic acids are incorporated into the host genome, lysogeny can lead to phage-mediated phenotypic conversion of prokaryotic hosts (44, 50).
The effects of viral infection in marine ecosystems emerge from the collective phage-host interactions in marine microbial communities. However, with the exception of a few well-known bacteriophages (e.g., T4, T7, λ, and P20), relatively little is known about the genetic capabilities and phenotypic characteristics of the vast majority of viruses. Whole-genome sequence data for a small collection of marine bacteriophage-host systems have revealed that the phages carry an unusually high proportion of unknown genes and have previously unexpected gene functions, such as involvement in phosphate uptake (e.g., phoH in Roseophage SIO1) (53) and photosynthesis (e.g., psbA in marine cyanophage) (38, 58, 59). Genomic investigations of phycoviruses have also revealed a high proportion of unknown genes and unusual functional genes, such as genes involved in the induction of apoptosis (71). Although investigations of single phage-host systems have provided unparalleled insights into these interactions, a broader understanding of virioplankton composition and diversity can come only from cultivation-independent approaches.
Characterization of whole viral assemblages based on pulsed-field gel electrophoresis (PFGE) has indicated that marine virioplankton community diversity varies dynamically in response to both seasonal and spatial gradients in ecosystem properties (74, 75) and that these changes mirror those in host bacterial communities (33). Higher-resolution surveys using marker genes (e.g., g20 and DNA polymerase genes) within specific lineages of virioplankton have uncovered extraordinary diversity (23, 77), yet some viral strains have been found in nearly all marine environments (15, 56, 57). While PFGE and marker genotyping approaches have been critical in the development of a foundation for synecological studies of virioplankton, they are limited either in resolution (PFGE) or breadth (marker genotyping).
Characterization of microbial communities using high-throughput DNA sequencing and bioinformatic approaches (i.e., metagenomics) addresses some of the limitations of these approaches and provides a high-resolution view of microbial diversity, as well as the potential functional capabilities within these assemblages. Based on the few metagenome surveys of marine viral communities completed to date, a consensus is emerging that virioplankton communities are extraordinarily diverse and contain a high proportion of unknown sequences (27). Against the typical backdrop of over 60% unknown sequences, viral metagenome libraries tend to contain a collection of gene homologs that are relatively distant from better-known representatives in bacterial genomes (13, 16). Despite the predisposition for horizontal gene transfer between bacteriophage genomes (32), there appears to be a specific “marine” aspect to virioplankton assemblages, which are dominated by genes from marine phages and cyanophages in particular (6). Analyses of a large database of short-read (∼100-base) sequences from four oceanic regions estimated that the compositions of virioplankton assemblages were extraordinarily even, that the assemblages contained between 500 and 130,000 genotypes, and that there were significant overlaps in genotype composition between disparate geographic regions (6).
While viral communities in near-shore waters, sediments, and open oceans have been examined (6, 13, 16), the genomes of virioplankton in a highly productive estuarine ecosystem have not been described in detail. Previous investigations characterizing bacterioplankton diversity demonstrated that the variable estuarine environment selects for a unique bacterioplankton assemblage whose composition is temporally and spatially dynamic (25, 34). The earliest demonstrations that virioplankton exhibit seasonally dynamic patterns of abundance (72), diversity, and composition (74, 75) were obtained from studies of the Chesapeake Bay. This report describes an analysis of the first estuarine viral community metagenome from the Chesapeake Bay in terms of the viral community's taxonomic, functional, and genotypic diversity.
MATERIALS AND METHODS
Viral concentrate sample collection and processing.
On 27 and 28 September 2002 50-liter surface water samples were collected using a diaphragm pump at nine stations along the midstem of the Chesapeake Bay (stations 908 [39°08′N, 76°20′W], 858 [38°58′N, 76°23′W], 845 [38°45′N, 76°26′W], 834 [38°34′N, 76°26′W], 818 [38°18′N, 76°17′W], 804 [38°04′N, 76°13′W], 744 [37°44′N, 76°11′W], 724 [37°24′N, 76°05′W], and 707 [37°07′N, 76°07′W]). At station 858 a bottom water sample from 2 m above the sediment-water interface was also obtained using Niskin bottles on a conductivity-temperature-depth rosette. Water was passed through a 25-μm wound cartridge filter, and viruses were concentrated using a two-step tangential flow filtration process, resulting in an approximately 250-ml viral concentrate sample. In the first tangential flow filtration step a 0.22-μm-pore-size, 0.5-m2 Pellicon cartridge filter (Millipore Corp.) was used to remove all cells. Viruses in the 0.22-μm filtrate were concentrated to a final volume of 2 liters using 10-m2 30,000-molecular-weight cutoff spiral filters (Helicon; Millipore Corp.). In the final step a smaller (0.5-m2) 30,000-molecular-weight cutoff spiral filter (Pellicon; Millipore Corp.) was used to concentrate viruses in a final volume of 250 ml. On shore, 200 ml of each viral concentrate was further concentrated using 30,000-molecular-weight cutoff Centricon Plus-80 spin filters (Millipore Corp.) to a final volume of approximately 2 ml. Virus counts were obtained by epifluorescence microscopy (22), and approximately 109 viruses were suspended in 45 μl of SM buffer (without gelatin) (100 mM NaCl, 8 mM MgSO4·7H2O, 50 mM Tris-Cl [pH 7.5]) and added to an equal volume of a 1.5% In-cert agarose plug for each station. The plugs were digested with proteinase K prior to DNA separation by PFGE. The PFGE running conditions were as follows: buffer temperature, 14°C; voltage gradient, 60 V/cm; included angle, 120° and switch time increased from 1 to 15 s over 22 h. Subsamples (equal volumes) of each viral concentrate except that obtained from the bottom of station 858 were pooled and suspended in agarose plugs as described above. One plug was subsequently digested for construction of the metagenome library.
Enumeration of viruses, bacteria, and Synechococcus.
Water samples were collected from discrete depths using 10-liter Niskin bottles mounted on a conductivity-temperature-depth rosette. Subsamples were immediately collected in 50-ml centrifuge tubes, fixed with glutaraldehyde (final volume, 2.5%), and stored at 4°C in the dark for no more than 2 weeks prior to microscopy. Viral particles and bacterial cells were collected by gentle vacuum filtration onto a 25-mm-diameter 0.02-μm-pore-size Anodisk (Whatman) and stained with SYBR gold (Molecular Probes) as described by Chen et al. (22). Bacteria and viruses in 10 fields of view (a minimum of 200 total viruses and bacteria) were counted for each sample. For Synechococcus enumeration, bacterial cells were collected on a 25-mm-diameter 0.2-μm-pore-size black polycarbonate filter (Poretics) using gentle vacuum filtration and counted as described by Wang and Chen (66). At least 200 Synechococcus cells in 10 fields of view were selectively enumerated using green excitation (528 to 553 nm).
Metagenome library construction and sequencing.
A random shotgun library of pooled Chesapeake Bay viral concentrates (see above) was constructed using the linker amplified shotgun library method (16, 54) through the Nanoclone service provided by Lucigen Corporation. Transformation mixtures were plated on LB agar plates containing kanamycin and grown for 14 to 16 h at 37°C. A total of 3,072 colonies were picked and grown in 96-well plates in LB containing kanamycin (60 mg/ml) for 22 to 24 h. After growth, sterile 50% glycerol was added to each well (final concentration, 15%) and plates were frozen at −80°C.
Two microliters of glycerol stock for each clone was used for TempliPhi (Amersham Biosciences) rolling circle amplification according to the manufacturer's instructions, with a extension step consisting of 16 h at 30°C. The completed TempliPhi reaction mixtures were diluted 1:1 with sterile H2O, and 6 μl of the dilutions was used in standard 20-μl sequencing reaction mixtures with Dynamic ET terminator chemistry (Amersham Biosciences). Each clone was sequenced bidirectionally, using a modified version of the forward primer (SL1; 5′ CAGTCCAGTTACGCTGGAGTC 3′) and reverse primer (SR1; 5′ CTTTCTGCTATGGAGGTCAGGTATG 3′) recommended for the pSMART-HCK vector (Lucigen Corp.). During protocol optimization, 768 clones were sequenced twice. Sequencing reaction mixtures were cleaned by ethanol precipitation and resuspended in the loading solution provided with a Dynamic ET chemistry kit (Amersham Biosciences). The products were separated with a MegaBACE 4000 capillary electrophoresis instrument (Amersham Biosciences) using low voltage (6 kV) and long run times (240 min) to obtain 550- to 650-base read lengths for over 85% of 6,912 total sequencing runs. Initial base calling and quality assessment were done using the Sequence Analyzer program (Amersham Biosciences).
Metagenome sequence analysis.
Sequences were screened for vector sequence, linker sequence, and minimum base quality using Phred and Crossmatch (28). After screening, all sequences smaller than 50 bases were removed and 6,478 sequences were carried forward for further analysis. The clones which were sequenced twice during protocol optimization were compared, and the shorter of each pair of corresponding sequences was removed, leaving 5,641 nonredundant sequences. These sequences were translated in six frames and compared (as amino acids) to six databases using tBLASTx version 2.2.8 (for nucleotide databases) or BLASTx version 2.2.9 (for protein databases) (3, 4). The GenBank databases used were updated on 1 July 2004 prior to all BLAST comparisons and included the nonredundant nucleotide (nt) and protein (nr) databases, as well as environmental nucleotide (env-nt) and environmental protein (env-nr) databases. Two additional viral metagenome sequence databases were used in tBLASTx homology searches. The first of these databases included viral sequences from a California near-shore water column and sediment, as well as viral sequences from human feces (13, 14, 16). The second database was composed of viral sequences generated from a Delaware agricultural soil sample (K. E. Wommack, S. R. Bench, and K. E. Williamson, unpublished data). At the time of analysis the GenBank environmental databases were composed of environmental microbial metagenome sequences from an acid mine drainage biofilm (64) and the Sargasso Sea (65). The databases were grouped into three categories according to the origin of their sequences: (i) traditional sequences, generally derived from cultivated organisms (GenBank nt and nr databases); (ii) microbial metagenome sequences (env-nt and env-nr databases); and (iii) viral metagenome sequences (vir-mg databases). Further comparisons of viral metagenome sequence data against single viral genomes were performed using tBLASTx with metagenome reads as queries and the nucleotide sequence of each viral genome as a single sequence subject database. Translated BLAST alignments to viral genome sequences with E values below 10−6 were considered significant.
The composition of the metagenome sequence library was determined based on BLAST sequence homology, using only alignments with E values less than 10−3. Each sequence was categorized based on the alignment quality, organism, and gene function of its most similar BLAST homolog. Taxonomic origins and functions were proposed for the subset of sequences with a significant BLAST homolog in one or both of the nt and nr databases. For bacterial species, categories were based on the NCBI taxonomy (8, 68) of the organism supplying the top homolog. For viruses, taxonomy was established as described by the International Committee on Taxonomy of Viruses (ICTV) (18) and also by using the phage proteomic tree (27, 52). Functional gene assignments were grouped according to the TIGR-CMR functional categories (46), which were originally derived from functional information for Escherichia coli genes (51). In the event of conflicts between databases for assigning taxonomy and function, priority was given to the alignment with the lowest E value.
Construction of PsbA phylogenetic tree.
A collection of 99 nonredundant PsbA protein sequences were collected from public sequence databases and used as a comparison set for the nine unique PsbA sequences that were sufficient length (>187 amino acids) identified in the Chesapeake Bay metagenomic sequence data. Clones identified as psbA gene homologs were sequenced as described above, using internal primers to obtain coverage of a larger portion of the gene. Multiple-sequence alignment was performed by using the ClustalW algorithm in MEGA (36). The sequence data were also evaluated by ProtTest (1) to establish the most appropriate amino acid substitution matrix to reconstruct phylogenetic relationships among the sequences. The final tree was constructed by the neighbor joining method in MEGA using the JTT matrix allowing for rate variation between sites with a gamma distribution over four rate categories of 0.441, as suggested by the ProtTest analysis with pairwise deletion of gapped positions. Bootstrapping was performed for 500 trials, and the results were displayed as a percentage of the trees containing the node specified. Alternative tree topologies, including collapse of the tree on the nodes, were examined to verify nodes with bootstrap values below 50. All of the alternative trees agreed with the grouping of the Chesapeake Bay PsbA sequences presented below.
Estimates of viral community diversity.
Metagenome library sequences were assembled using Sequencher (Gene Codes Corporation) according to parameters described by Bretibart et al. (16), and the number of resulting contiguous sequences (i.e., the contig spectrum) was used to predict possible virioplankton population structure at the time of sampling. Three assemblies were generated and analyzed: one with all library sequences, one with only forward sequence reads, and one with only reverse sequence reads. The online PHACCS tool was used for assessing viral community diversity with the power law model (5, 27). The contig spectra used as input contained values for the first 12 contig types (i.e., up to contigs containing 12 sequences) and were as follows: assembly of all 5,641 sequences = [5435 100 2 0 0 0 0 0 0 0 0 0]; assembly of 2,798 forward sequences = [2712 43 0 0 0 0 0 0 0 0 0 0]; and assembly of 2,843 reverse sequences = [2758 41 1 0 0 0 0 0 0 0 0 0 0].
Nucleotide sequence accession numbers.
The nonredundant set of 5,641 metagenome sequences has been deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/) under genome project number 16522; the accession numbers are EI103240 to EI108880.
RESULTS AND DISCUSSION
This is the first metagenomic study that focused on estuarine virioplankton, and several characteristics of the metagenome obtained are similar to characteristics of viral metagenomes from other environments. The Chesapeake Bay metagenome contained 5,641 nonredundant sequences with an average read length of 695 bp for a total of 3.92 Mb of sequence data. The G+C content of each sequence was calculated, and the values were normally distributed between 24 and 65%, with a mean of 46% for the library. With a maximum E value criterion of 10−3, 30% of the sequences in our metagenome library did not have a BLAST homolog and another 31% (1,731 sequences) were similar to only environmental sequences of unknown function (Table 1). Thus, 61% of Chesapeake Bay virioplankton metagenome sequences have not been observed in currently cultivated or well-studied organisms. Similarly low levels of known sequences, 21 to 41%, were reported in five other Sanger sequenced viral metagenome libraries constructed using approaches similar to the one reported here (13, 14, 16, 20). A significantly smaller fraction of the sequences (4.7%) of a 181-Mb library of short-read (∼100-bp) virioplankton sequences from 68 sites in four ocean regions were categorized as known sequences (6). This lower frequency of known sequences is likely related to the sequence lengths of the libraries. Extensive comparisons of short-read (∼100-bp) and long-read (>600-bp) sequence data sets generated in silico, starting with the Chesapeake Bay metagenome sequence collection, indicated that short-read sequences fail to detect more than 60% of the BLAST homologs detected by long-read data, even with 6- to 10-fold oversampling relative to the long-read data set (K. E. Wommack, J. Bhavsar, and J. Ravel, submitted for publication). These observations suggest that short-read sequences are less appropriate than long-read sequences for functional characterization of viral metagenomes based on BLAST homology searches.
TABLE 1.
Metagenome sequence BLAST homology by database and domain (E value, <10−3)
| Database or domain | Avg log10 E value | Median log10 E value | No. of sequences | % of totala |
|---|---|---|---|---|
| GenBank nt or nr database | −21.5 | −13.7 | 2,195 | 39 |
| Archaea | −11.4 | −8.2 | 25 | 0.4 (1) |
| Bacteria | −17.1 | −10.1 | 1,031 | 18 (47) |
| Eukaryote | −8.1 | −4.5 | 149 | 2.6 (7) |
| Virus | −28.4 | −21.2 | 962 | 17 (44) |
| Environmental databaseb | −24.5 | −17.1 | 1,731 | 31 |
| No homology | NAc | NA | 1,715 | 30 |
| Total | NA | NA | 5,641 | 100 |
For domains the value in parentheses is the percentage of the 2,195 sequences displaying similarity to nt or nr database sequences.
The sequence was similar to at least one environmental database sequence (env-nt, env-nr, or viral metagenome) but showed no similarity to the nt or nr database.
NA, not applicable.
The frequency of known BLAST homologs within four previously reported long-read viral metagenome libraries did not change significantly over 2 years despite a doubling in the size of the GenBank nr database (27). A reanalysis of 1,000 randomly selected sequences from the Chesapeake Bay virioplankton library also showed that the proportion of known sequences changed little over 18 months following the original BLAST search used in this analysis (Wommack et al., submitted). Because this low level of homology to GenBank sequences was not observed in RNA viral metagenomes from human feces (91% of sequences were homologous) (76), it is likely that the extant diversity of double-stranded (dsDNA) viruses is poorly represented in GenBank.
Earlier reports of viral metagenome libraries focused on BLAST searches against only GenBank nt and nr databases. However, the release of data for 1,360 Mb of microbial metagenome sequences from the Sargasso Sea (65) and of data for 76 Mb from an acid mine drainage microbial community (64) enabled homology searches against strictly environmental sequences. Comparing quality scores of BLAST alignments from environmental databases to quality scores from the GenBank nt and nr databases revealed that the Chesapeake Bay metagenome was more similar to environmental sequences. For example, the average and median E values were approximately 3 logs lower for homologs to environmental sequences than for sequences with GenBank nt/nr database homologs (Table 1).
This type of comparison also revealed that Chesapeake Bay sequences were more similar to known viral sequences than to other known sequences. The vast majority (91%) of the 2,195 sequences with similarity to GenBank nt and nr database sequences were most similar to sequences from prokaryotes or viruses. Alignments to viral sequences were the highest in quality, with median and average E values more than 11 logs lower than the values for BLAST alignments to bacterial sequences (Table 1). Alignments to environmental sequences were the second highest quality, with E values 7 logs lower than the values for bacterial alignments. The 149 Chesapeake Bay viral sequences that were most similar to eukaryotic sequences had the lowest quality alignments (the E values were up to 22 logs higher than the values for virus sequence alignments), suggesting that eukaryotic viruses were rare in the Chesapeake Bay at the time of sampling (Table 1).
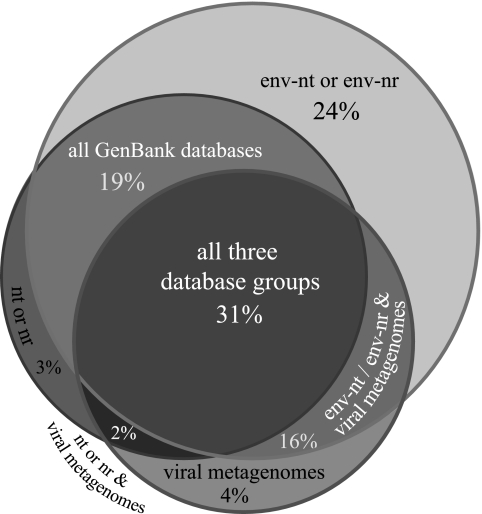
The overlap of homology between databases also suggested the nature of the source DNA used for library construction. Among viral metagenome sequences with BLAST homologs, the largest fraction (1,235 sequences or 31%) showed similarity to at least one sequence in each of the three database categories (Fig. 1, central region) (see Materials and Methods for descriptions of database categories.). One-half (51%) of the Chesapeake Bay metagenome sequences had a homolog in the env-nt database or the env-nr database or both, and more than one-half of these (30% of the total) had homologs only in the environmental databases and no homology to any sequence in the GenBank nt or nr database. The majority of the matches were to sequences from the Sargasso Sea metagenome library (65), indicating that there were overall similarities between the microbial and viral communities in diverse marine environments. This type of signal was not detectable in short-read virioplankton libraries, where even marine-derived viral sequences had a low BLAST homolog rate (4%) with the env-nt and env-nr databases, similar to the rate observed when the GenBank nr and nt databases were queried (6). Sequences with matches to both the GenBank nt/nr and env-nt/env-nr databases occurred at a frequency (19%) similar to the frequency of matches with homologs in both the env-nt/env-nr and vir-mg databases (16%). Chesapeake Bay virioplankton sequences with homology solely to the GenBank nt/nr or vir-mg database or to both the GenBank nt/nr and vir-mg databases were rare, with 88 to 163 sequences per category (Fig. 1).
FIG. 1.
Distribution of translated BLAST (tBLASTx against nucleotide databases and BLASTx against protein databases) matches between all database “types.” The upper, largest circle represents matches to the env-nt and/or env-nr database comprised mostly of Sargasso Sea bacterial metagenomic data. The leftmost circle represents matches to either of the traditional GenBank (nt and nr) databases. The bottom right circle represents matches to any of a series of small viral metagenomes from terrestrial and marine environments (see Materials and Methods for details). Intersections of circles represent sequences that had BLAST homology to more than one database type, and the center area represents sequences with homology to all three types.
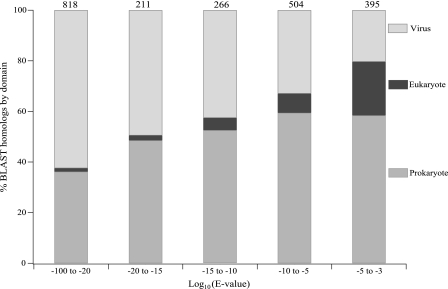
As a further demonstration of the utility of BLAST alignment statistics for general observations about the nature of the source DNA in a metagenome library, sequences that could be taxonomically categorized were binned according to the log10 of BLAST E values (Fig. 2). For simplicity, archaeal and bacterial homologs were combined in the prokaryote category. As BLAST alignment quality increased from a log10 of −3 to a log10 of less than −20, the frequency of sequences with best homologs to viral sequences tripled from 20% to more than 60%, again showing that the highest-quality alignments were between metagenome sequences and known virus sequences. Across the same range, the frequency of homologs to eukaryotic sequences declined from 21% to less than 2%, and the proportion of homologs to prokaryotes ranged from less than 40% in the highest-quality bin to nearly 60% in the three lowest-quality bins (Fig. 2). Nearly equal fractions of homologs to the bacterial and viral domains have been observed in other marine viral metagenome libraries (13, 16), and this is strong evidence supporting the established theory that marine virioplankton assemblages are dominated by bacteriophages (67, 72).
FIG. 2.
Distribution of translated BLAST sequence matches across taxonomic domains sorted by match quality. Sequences were placed in nonredundant bins according to quality (i.e., E value), and relative domain percentages were calculated for each bin. The smallest E values represent the highest-quality matches on the left. The least confident matches are on the right, with a maximum E value of 10−3. The numbers of sequences in the bins are indicated above the bars.
Four lines of evidence argue that the frequency of bacterial BLAST homologs is due to a predominance of phages within the Chesapeake Bay metagenome and is not the result of contaminating bacterial DNA. First, the proportion of bacterial homologs in bins grouped by BLAST alignment quality (E value) was lowest (<40%) in the highest-quality bin, while the frequency of viral homologs increased with increasing quality (Fig. 2). Second, inspection of the genomic context surrounding many of the bacterial BLAST homologs revealed that the virioplankton sequences were likely homologs of prophage sequences. Third, because viruses are obligate parasites which rely on the host cell transcription and translation systems, viral genes often have sequence characteristics (e.g., G+C contents and tetranucleotide usage frequencies [48]) similar to those of their hosts. Thus, a background of homology between viral genes and genes of the hosts that the viruses infect is to be expected, and the distribution of virioplankton sequences among bacterial taxa may actually reflect host preferences within the virioplankton at the time of sampling. The prevalence of cyanophage and cyanobacterial homologs within the library (see below) strongly supports this assertion. Finally, no significant matches were detected when various 16S rRNA sequences were used in searches against the Chesapeake Bay metagenome by nucleotide BLAST (see the supplemental material for a list of subject 16S rRNA sequences). If there were significant contamination by bacterial or archaeal genomic DNA in our 3.9-Mb sequence library, identification of three or four 16S rRNA genes would be expected based on the observed frequency of 1.1 copies of 16S rRNA genes per Mb in bacterial and archaeal genomes (29).
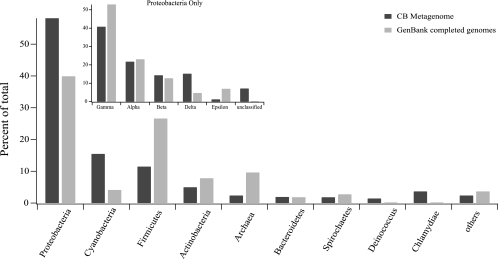
The majority (nearly 60%) of the 1,056 Chesapeake Bay virioplankton sequences with best BLAST matches to prokaryote sequences were most similar to Proteobacteria (Fig. 3). The Gammaproteobacteria subphylum accounted for the largest portion (40%) of the proteobacterial homologs, while the Alpha-, Beta-, and Deltaproteobacteria were less common, accounting for only 15 to 20% of this group (Fig. 3, inset). BLAST homologs to Cyanobacteria and Firmicutes accounted for another 26% of the prokaryotic sequences (15 and 11%, respectively). One caveat of these data is the potential influence of the subject database on the taxonomic distribution of metagenome homologs. To estimate the amount of taxonomic bias introduced by database contents, we compared the taxonomic distribution of the metagenome sequences to the phylogenetic composition of prokaryotic genome sequences in GenBank. This comparison revealed that four of the eight most common phyla occurred at similar frequencies in the query (Chesapeake) and subject (GenBank) databases (Fig. 3). However, the frequency of BLAST homologs among the remaining four phyla differed from the GenBank distribution by more than 5%.
FIG. 3.
Distribution of translated BLAST prokaryotic homolog sequences. The data are organized according to prokaryotic phyla. Data for completed prokaryote genomes in GenBank (at the time of metagenome sequence comparison) are shown to illustrate groups that are overrepresented (e.g., cyanobacteria and Proteobacteria) or underrepresented (e.g., Firmicutes) in the metagenome relative to the subject database. CB, Chesapeake Bay.
Interannual observations of picophytoplankton abundance have shown that there is a summer peak in cyanobacterial abundance in the Chesapeake Bay (2, 66). At the time of sampling in September 2002, picocyanobacteria accounted for an average of 7% (range, 3 to 14%) of the bacterial abundance in surface waters (see Fig. S1 in the supplemental material). Thus, the presence of abundant cyanobacterial sequence homologs within the Chesapeake Bay virioplankton is consistent with the abundance of this host group at the time of sampling. In a similar way, the increased frequency of proteobacterial homologs may reflect previously reported data demonstrating that there was a late summer peak in the abundance of Gammaproteobacteria in the Chesapeake Bay (31). These observations support the supposition that the taxonomic affiliation of BLAST homologs broadly reflects host preferences among the virioplankton and may actually overcome database biases.
Archaea and Firmicutes were underrepresented in the Chesapeake Bay metagenome library, while Proteobacteria and Cyanobacteria were overrepresented with respect to the GenBank database (Fig. 3). The underrepresentation of archaeal homologs likely reflected the known low abundance of archaea previously observed in the Chesapeake Bay (11) and the lack of genomic sequence information for mesophilic marine archaea (35). Although members of the Firmicutes and Actinobacteria are known to occur in the Chesapeake Bay (34), the underrepresentation of these groups in the virioplankton metagenome may reflect a bias towards terrestrial strains in the GenBank databases. Overall, these discrepancies illustrate real taxonomic biases in the composition of the Chesapeake Bay host community and the fact that available sequence databases are not ideally suited for characterization of metagenomic libraries because many environmentally important groups of prokaryotes are underrepresented in these databases.
Similar to other viral metagenomes, when sequences were classified using the taxonomic scheme outlined by the ICTV (18), the vast majority of Chesapeake Bay metagenome sequences with viral homologs were categorized as bacteriophages. Over 90% of these sequences were most similar to the tailed bacteriophage order Caudovirales (Table 2), and 83% of these sequences were most similar to members of the Myoviridae and Podoviridae families (42 and 41%, respectively). In contrast, the third major family, Siphoviridae, accounted for only 6% of the viral homolog sequences, and another 3% were unclassified below the order level. Viruses that infect algae (Phycodnaviridae; e.g., viruses PbCV-1 and EsV-1) comprised only 1% of the virus BLAST homologs to Chesapeake Bay virioplankton sequences, further demonstrating that viruses with eukaryotic hosts were rare in this sample (Table 2). The relative percentages of the Caudovirales families observed in the Chesapeake Bay library contrast strikingly with the results of previous studies of a variety of sample types and locations, which estimated that between 28 and 76% of the sequences could be classified as Siphoviridae (13, 14, 16, 20). The uniquely low proportion of Siphoviridae found in the Chesapeake Bay library suggests that most bacteriophages in this estuary are virulent as a large proportion of temperate phages belong to the Siphoviridae. By extension, it also suggests that there is a relatively low rate of lysogeny within Chesapeake Bay bacterioplankton host populations in late summer and is consistent with results for Tampa Bay that showed that the lowest proportion of lysogenic hosts within estuarine bacterioplankton occurred in warm productive months (40, 70).
TABLE 2.
Distribution of top BLAST homologs to viral sequences (E value, <10−3) organized by ICTV taxonomy or phage proteomic tree cluster
| Family or cluster | No. of sequences | % of total | No. of different species |
|---|---|---|---|
| ICTV viral families | |||
| Caudovirales | 886 | 92 | 135 |
| Myoviridae | 404 | 42 | 66 |
| Podoviridae | 394 | 41 | 25 |
| Siphoviridae | 57 | 6 | 35 |
| Unclassified | 31 | 3 | 9 |
| Unclassified bacteriophage | 14 | 2 | 11 |
| Phycodnaviridae | 13 | 1 | 4 |
| Other | 49 | 5 | 14 |
| Total | 962 | 100 | 164 |
| Phage proteomic tree cluster | |||
| T7-like podophage | 292 | 75 | 7 |
| Corndog-like siphophage | 25 | 6 | 13 |
| λ-Like siphophage | 8 | 2 | 5 |
| Sfi11-like siphophage | 5 | 1 | 2 |
| Sfi21-like siphophage | 3 | 1 | 2 |
| D3-like siphophage | 3 | 1 | 3 |
| Unclassified | 46 | 12 | 7 |
| Other | 6 | 2 | 5 |
| Total | 388 | 100 | 44 |
While distinctions of phage life cycles based on morphological family are not absolute (19), the prevailing view (based primarily on marine viruses) is that virulent phages with broad host ranges are members of the Myoviridae, while the virulent phages with narrower host ranges belong to the Podoviridae (60, 61). Sequences of members of both of these phage groups were equally represented in the Chesapeake Bay metagenome (Table 2). This observation is supported by the high levels of bacterial production that occur in the Chesapeake Bay during late summer and by the idea that virulent phage populations are sustained through high reproduction rates and short generation times (61). The dominance of lytic phage groups may also be an environmental signal resulting from viral termination of the summer picoplankton bloom. Previous studies suggested that cold seasons select for a higher incidence of inducible temperate phages (40, 70). If this trend occurs in the Chesapeake Bay, then metagenome data from the late fall or winter months, when picoplankton are less abundant (2, 66), would be expected to show a higher proportion of sequences homologous to Siphoviridae. Nevertheless, the apparent taxonomic composition of the Chesapeake Bay virioplankton in the early fall of 2002 indicates that the proposed paradigm that “Siphophages might be the most abundant genome arrangement on Earth” (27) is not universal.
Classification of metagenome sequences using the second version of the phage proteomic tree (27, 52) showed that cyanophage P60 was the phage most commonly detected (>50% of all proteomic tree homologs), followed by homologs to the closely related Pseudomonas aeruginosa phage PaP3 and Roseophage SIO1 (Table 2). Virioplankton sequence homologs were distributed throughout the P60, PaP3, and SIO1 genomes, indicating that intact phage genomes similar to these species, rather than particular genes or regions, were abundant in the metagenome library. These three phages are in the T7-like Podophage clade of the phage proteomic tree (27) and are listed within the “Cyanophage P60 group” in NCBI taxonomy (8, 68). Because P60 was isolated from an estuarine environment and is known to infect Synechococcus species (21) that were also abundant in the Chesapeake Bay at the time of sampling (see Fig. S1 in the supplemental material), the similarity to P60-like phage is not surprising. Recent analyses of the taxonomic structure of marine phage communities based on the phage proteomic tree indicated that members of the T7-like Podophage clade are also common in a broad range of oceanic environments (6). In contrast to the ICTV taxonomic distribution, the phage proteomic tree comparison identified a slightly higher proportion (11%) of virioplankton sequences as most similar to Siphophage families. However, at the time of this analysis the phage proteomic tree contained 167 phages whose genomes had been sequenced (27) and did not include the cyanomyophages P-SSM2 and P-SSM4 or the cyanopodophage P-SSP7. The percentage of siphophage homologs might be closer to the ICTV percentage if the metagenome were compared to a proteomic tree which included these cyanophages.
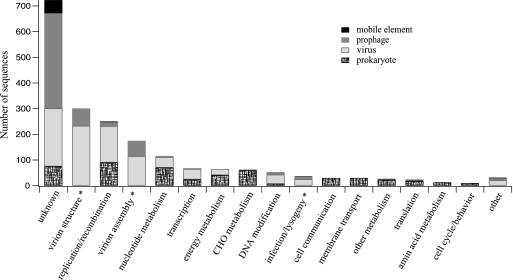
Chesapeake Bay metagenome sequences were divided into 17 functional categories according to the gene function of the highest-quality BLAST alignment. Each functional category was further divided based on the likely taxonomic position of the BLAST homolog (viral, bacterial [no evidence of prophage in the genome region of the best BLAST homolog], prophage [bacterial genome match to an annotated or suspected prophage], or mobile element [transposon or plasmid]) (Fig. 4). Of the 2,195 sequences with a BLAST homolog in the GenBank nt/nr databases, 86% (2,010 sequences) were annotated. Among the 39% “known” virioplankton sequences, functional categories for virion structure, replication/recombination, virion assembly, and nucleotide metabolism each represented between 6 and 15% of the known BLAST homologs. Only a small fraction of sequences were homologous to functional groups outside those directly related to viruses (assembly, structure, and lysogeny) or nucleotide metabolism (biosynthesis, DNA modification, replication, recombination, and transcription), and these sequences originated almost entirely from viruses or bacterial genomes with no evidence of prophage in the genomic neighborhood of the match. However, unknown or hypothetical proteins were the most dominant functional class (36% of the functionally classified sequences) in the Chesapeake Bay virioplankton library (Fig. 4). These findings are similar to those reported for other long-read viral metagenome libraries (13, 14, 16) and support the “unknown” nature of extant DNA virus diversity.
FIG. 4.
Viral metagenome translated BLAST homologs sorted according to annotated functional gene category. Each sequence was assigned to a presumptive functional category based on the highest-quality sequence homolog. The most likely phylogenetic affiliations (virus, bacteria, prophage, and mobile element) for each sequence category are indicated. Asterisks indicate categories that could not contain prokaryote sequences because they are purely viral functions.
Over one-half of the unknown functional category BLAST homologs were classified as prophages based on the genomic context in which the sequences were found (Fig. 4). Cross-referencing phylogenetic affiliation with functional annotation has not been reported previously for viral metagenome sequences, and our analysis supports the idea that prophages account for a large proportion of hypothetical and unknown functional genes in bacterial genomes (10). Because a substantial proportion of virioplankton sequences were homologous to prophage-derived open reading frames of unknown function in bacterial genomes, these sequences may provide an important resource for defining the contributions of bacteriophages to the processes of horizontal gene transfer and bacterial genome evolution.
The relative abundance of prophage-like sequences contrasts with the low number (36 sequences) of lysogeny-related functional genes identified in the Chesapeake Bay metagenome library. Furthermore, the small number of Siphoviridae-like sequences identified also suggested that lysogeny was not prevalent in the Chesapeake Bay at the time of sampling, as discussed above. One possible explanation for this inconsistency may be the subject databases and methods used for functional assignments. Specifically, the vast majority of annotated genes in the GenBank nr database come from cellular organisms and not viral genome sequences. As a result, BLAST similarity searches of viral metagenome sequences would be more likely to find a homolog in the larger cellular sequence set. Thus, even virus-derived metagenome sequences may appear to be most similar to prokaryotic (i.e., host) sequences, because their exact homologs are not represented by a viral sequence in the subject database. Another explanation may be that bacteriophage groups other than Siphoviridae contribute more to bacterial lysogeny in this environment. If this is the case, then a clear signal of lysogenic frequency may be difficult to discern from viral metagenome sequence data. To discern between unrecognized, genuine lysogeny and overannotation of prophage sequences, future viral metagenomic investigations should be directly coupled with induction experiments to measure the level and ascertain the identity of inducible prophages in the community at the time of sampling.
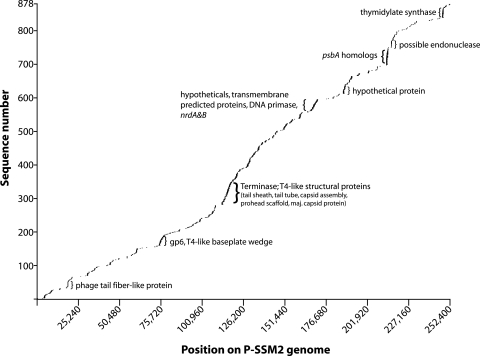
Comparison of Chesapeake Bay metagenome sequences with specific phage genomes revealed that nearly 14% of the library had significant homology to cyanophages, while a much smaller fraction of the library was homologous to known noncyanophage viruses (Table 3). Mapping of metagenome sequences onto the genome of P-SSM2 revealed that the vast majority of identified open reading frames in this cyanophage had homologs in the Chesapeake Bay library (Fig. 5). The position map revealed particular functions that were found with increased frequency in the metagenome library, such as phage structural genes, nucleotide metabolism and replication genes, and a gene (psbA) encoding the core photosystem II D1 protein.
TABLE 3.
Frequency of Chesapeake Bay metagenome BLAST homologs to virus genomes
| Family | Subgroup | Virus | GenBank accession no. | Genome size (kb) | No. of significant homologsa | % of librarya | Median log10 E value |
|---|---|---|---|---|---|---|---|
| Myoviridae | Cyanophage | P-SSM2 | AY939844 | 252 | 878 | 13.6 | −31.2 |
| Cyanophage | P-SSM4 | AY940168 | 178 | 723 | 11.2 | −27.2 | |
| T-even phage | RB69 | AY303349 | 168 | 178 | 2.7 | −23.2 | |
| T-even phage | T4 | AF158101 | 167 | 177 | 2.7 | −22.7 | |
| Schizo-T-even | KVP40 | AY283928 | 245 | 197 | 3.0 | −23.2 | |
| Schizo-T-even | Aeh1 | AY266303 | 233 | 191 | 2.9 | −24.4 | |
| Pseudo-T-even | RB49 | AY343333 | 164 | 181 | 2.8 | −23.2 | |
| Podoviridae | Cyanophage | P-SSP7 | AY939843 | 45 | 543 | 8.4 | −43.5 |
| Cyanophage | P60 | AF338467 | 48 | 368 | 5.7 | −25.0 | |
| Roseophage | SIO1 | AF189021 | 40 | 118 | 1.8 | −16.7 | |
| Siphoviridae | Temperate phage | Lambda | J02459 | 49 | 39 | 0.6 | −11.7 |
| Phycodnaviridae | PbCV-1 | U42580 | 331 | 59 | 0.9 | −25.0 | |
| EsV-1 | AF204951 | 336 | 12 | 0.2 | −8.5 |
Each genome was compared separately, so numbers of homologous sequences and library percentages are neither mutually exclusive nor additive. Only homologs with E values of <10−6 were considered significant.
FIG. 5.
Positions of Chesapeake Bay virioplankton BLAST homologs on the Prochlorococcus phage P-SSM2 genome. Regions with high levels of coverage are indicated by brackets. Only translated BLAST homologs with E values below 10−6 are shown. psbA, core photosystem II reaction center protein; nrdA&B, alpha and beta subunits of ribonucleoside reductase.
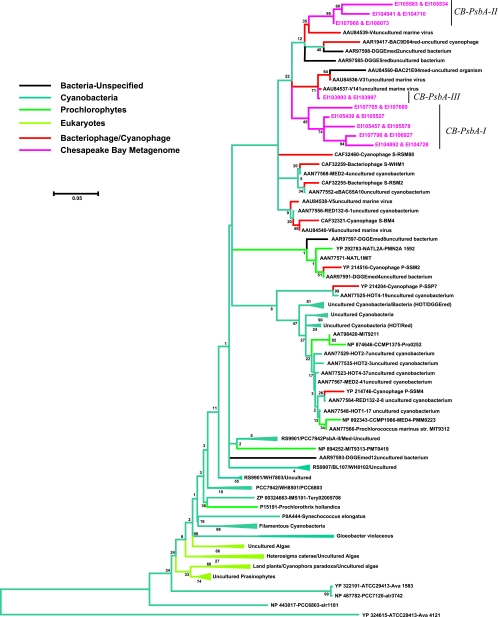
Energy metabolism functionality was found with unexpected frequency in metagenome homologs, and over 60 virioplankton sequences were classified in this category (Fig. 4). These homologs included genes encoding cytochrome c oxidase, high-light-inducible proteins, and ferredoxin, as well as other photosynthetic genes. Most notably, 26 high-confidence (E value, <10−45) homologs to the photosystem II core reaction center protein DI encoded by the psbA gene were detected. The inferred PsbA amino acid sequences from Chesapeake Bay virioplankton formed three distinct phylogenetic groups containing five (group CB-PsbA-I), three (group CB-PsbA-II), and one (group CB-PsbA-III) nonidentical PsbA sequences (Fig. 6). The CB-PsbA-I and CB-PsbA-II groups appear to be more closely related to one another than they are to sequences retrieved from GenBank. The two GenBank sequences (accession numbers AAU84537 and AAU84539) most similar to Chesapeake Bay sequences were recovered from uncultured marine viruses in coastal waters of the eastern Mediterranean Sea near Haifa, Israel, in 2004. The monophyletic nature of the Chesapeake Bay sequences was consistently found in all tree topologies, indicating a level of endemism among Chesapeake Bay cyanophages. This finding agreed with recent molecular phylogenetic data showing that seasonally dynamic endemic populations of Synechococcus and cyanophage are present in the Chesapeake Bay (24, 66).
FIG. 6.
Phylogenetic tree of PsbA amino acid sequences deduced by a comparison of viral metagenome sequences with PsbA amino acid sequences derived from public databases. The tree is based on alignment of 187 homologous positions. Scale bar = 0.05 substitution per position.
PsbA-encoding genes have previously been identified in marine cyanophages (39, 58, 59) and are known to be transcribed during lytic infection (37). The Chesapeake Bay metagenome contained a total of 17,124 bp of psbA homologous sequence, which is 5.0% of the 341 kb of cyanophage homologs identified in the library. Comparison of the metagenome library with recently sequenced cyanophage genomes (Fig. 5 and Table 3) suggested that the total number of cyanophage homologs was underestimated by the initial BLAST search. A revised estimate which incorporated the new genomes (namely, P-SSM2, P-SSM4, and P-SSP7) increased the size of the total cyanophage sequence in the library to 610 kb. Using this revised total, the psbA homologous sequence still accounted for 2.8% of all cyanophage homologous sequences observed. In cyanophages known to carry psbA, this gene has been observed to account for between ∼0.5% (43, 58) and 2.4% (58) of the total genome length, with the highest percentage seen in the cyanopodophage P-SSP7. Comparing these percentages to the 2.8% psbA length fraction in the Chesapeake Bay metagenome, it appears that the vast majority of cyanophages carried psbA at the time of sampling and that the strategy for maintaining host photosystem functionality during infection may be nearly universal among cyanophages in the Chesapeake Bay. Recently, examination of over 30 cyanophages showed that 88% of them carried psbA, and the propensity for these phages to carry both psbA and psbD appeared to coincide with the host specificity and/or genome size of a given strain (59). Broad-host-range cyanomyophages with larger genomes carried both genes, while narrow-host-range cyanopodo- and cyanosiphophages carried only psbA. Combining the 2.8% psbA length fraction of cyanophage homologous sequence in the library with the fact that psbD was rarely observed in the library (10-fold-fewer significant BLAST homologs than psbA) indicated that the Chesapeake Bay cyanophage assemblage was dominated by small-genome, narrow-host-range cyanopodophages at the time of sampling.
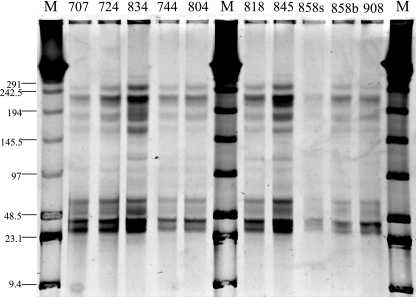
In contrast to the high degree of genetic homology to known cyanophages, contig spectra community analysis suggested that there was a virioplankton assemblage that was evenly distributed among thousands of genotypes, with the most abundant genotype accounting for <0.1% of the community (Table 4). For the assembly of all reads, the power law model estimated that there were 4,110 and 1,650 total genotypes for average genome sizes of 50 and 125 kb, respectively. PFGE of virioplankton assemblages indicated that the sizes of the viral genomes in samples used for library construction ranged from ∼30 to ∼250 kb and that there were two major subpopulations: viruses with moderate-size genomes (∼30 to ∼60 kb) and viruses with larger genomes (∼125 to ∼250 kb) (Fig. 7). Fifty-kilobase genomes were most abundant, while 125 kb was the mean size of all genomes observed (unweighted for abundance). In all assemblies, the estimates for the most abundant genotype ranged from 0.04 to 0.6% of the total community (Table 4), resulting in evenness estimates very close to 1 and Shannon diversity indices near the maximum value allowed for the estimated number of genotypes (55). Recent assessments of virioplankton richness based on short-read sequences resulted in similarly divergent conclusions, i.e., a high level of BLAST homology to cyanophage genomes and an extremely diverse global ocean community containing between 57,600 and 129,000 viral genotypes estimated by contig spectral analysis (6). These discrepancies likely reflect the relative sensitivities of the two types of analyses. While tBLASTx tolerates a greater range of sequence diversity because it relies on translated amino acid sequences, contig spectra rely on the outcome of a high-stringency nucleotide sequence assembly.
TABLE 4.
Community analysis of Chesapeake Bay viral metagenome based on contig spectraa
| Reads | Richness (total no. of genotypes)
|
Evenness
|
Most abundant genotype (% of community)
|
Shannon diversity index (H′ in nats)
|
||||
|---|---|---|---|---|---|---|---|---|
| 50-kb sequencesb | 125-kb sequencesb | 50-kb sequences | 125-kb sequences | 50-kb sequences | 125-kb sequences | 50-kb sequences | 125-kb sequences | |
| Forward only | 2,380 | 953 | 1 | 1 | 0.042 | 0.105 | 7.77 | 6.86 |
| Reverse only | 3,180 | 1,300 | 0.99 | 0.987 | 0.297 | 0.606 | 7.98 | 7.08 |
| All | 4,110 | 1,650 | 0.999 | 0.998 | 0.065 | 0.153 | 8.31 | 7.4 |
For all analyses, the contig spectrum vector included values for the first 12 contig types (up to 12 sequences in one contig), and the power law model equation was used to calculate the rank-abundance distribution.
Average viral genome size used to calculate the parameters shown.
FIG. 7.
PFGE gel of virioplankton concentrates used to construct the Chesapeake Bay metagenome library. The numbers above the lanes indicate the stations in the bay (see Materials and Methods for the location of each station). For station 858, surface and bottom samples are indicated by the suffixes “s” and “b,” respectively. The numbers on the left indicate marker band sizes (in kilobases). Marker lanes M contained concatemers of phage λ genomes (with resolvable bands at positions ranging from 291 to 48.5 kb) mixed with a HindIII digest of λ genomic DNA (23.1 and 9.4 kb). The viral concentrate from the bottom water sample at station 858 was not used in construction of the metagenome library in this study.
Assuming that there were 4,110 viral genotypes (Table 4) with an average genome size of 50 kb, we estimated a theoretical total virioplankton metagenome size of 205.5 Mb in our sample. If our estimate that 30% of the sequences are novel is correct, complete sequencing of all Chesapeake Bay viral genomes would yield up to 61.6 Mb of novel sequence. Clearly, these data demonstrate that environmental dsDNA viruses contain a sizeable proportion of unexplored genetic diversity and that viral metagenomics is a critical step towards identifying the expanse of novel and uncharacterized genes in the biosphere.
This report describes the first detailed examination of an estuarine viral metagenome. An extensive effort was made to place observed genetic diversity in a functional and taxonomic context. Overall, the results demonstrate the unique capabilities of long-read metagenomic sequence data for characterization of natural viral communities. The large amount of unknown and novel DNA sequence observed in this study dramatically underscores the fact that extant gene diversity among dsDNA viruses is poorly constrained and illustrates the need to develop additional approaches that move beyond cataloging and on to determining the ecological significance and evolutionary advantages conferred by the genes to the viruses that carry them.
Supplementary Material
Acknowledgments
We gratefully acknowledge the support of a USDA National Needs Fellowship to S.R.B. and NSF Microbial Observatories grant MCB-0132070 awarded to K.E.W.
We thank Mya Breitbart and Forest Rohwer for useful suggestions on the contig spectrum analyses. We are also indebted to Larry Tindell, Leo Genyuk, Jaysheel Bhavsar, and Sowmya Vijayaraghavan for computational assistance and to the captains and crew of the R/V Cape Henlopen for assistance during research cruises.
Footnotes
Published ahead of print on 5 October 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abascal, F., R. Zardoya, and D. Posada. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104-2105. [DOI] [PubMed] [Google Scholar]
- 2.Adolf, J. E., C. L. Yeager, W. D. Miller, M. E. Mallonee, and L. W. Harding. 2006. Environmental forcing of phytoplankton floral composition, biomass, and primary productivity in Chesapeake Bay, USA. Estuar. Coast. Shelf Sci. 67:108-122. [Google Scholar]
- 3.Altschul, S., T. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Angly, F., B. Rodriguez-Brito, D. Bangor, P. McNairnie, M. Breitbart, P. Salamon, B. Felts, J. Nulton, J. Mahaffy, and F. Rohwer. 2005. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics 6:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angly, F. E., B. Felts, M. Breitbart, P. Salamon, R. A. Edwards, C. Carlson, A. M. Chan, M. Haynes, S. Kelley, H. Liu, J. M. Mahaffy, J. E. Mueller, J. Nulton, R. Olson, R. Parsons, S. Rayhawk, C. A. Suttle, and F. Rohwer. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azam, F., T. Fenchel, J. G. Field, J. S. Gray, L. A. Meyerreil, and F. Thingstad. 1983. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10:257-263. [Google Scholar]
- 8.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2003. GenBank. Nucleic Acids Res. 31:23-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 10.Binnewies, T. T., Y. Motro, P. F. Hallin, O. Lund, D. Dunn, T. La, D. J. Hampson, M. Bellgard, T. M. Wassenaar, and D. W. Ussery. 2006. Ten years of bacterial genome sequencing: comparative-genomics-based discoveries. Funct. Integr. Genomics 6:165-185. [DOI] [PubMed] [Google Scholar]
- 11.Bouvier, T. C., and P. A. del Giorgio. 2002. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol. Oceanogr. 47:453-470. [Google Scholar]
- 12.Bratbak, G., F. Thingstad, and M. Heldal. 1994. Viruses and the microbial loop. Microb. Ecol. 28:209-221. [DOI] [PubMed] [Google Scholar]
- 13.Breitbart, M., B. Felts, S. Kelley, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2004. Diversity and population structure of a near-shore marine-sediment viral community. Proc. R. Soc. London Ser. B 271:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breitbart, M., I. Hewson, B. Felts, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185:6220-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breitbart, M., J. H. Miyake, and F. Rohwer. 2004. Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol. Lett. 236:249-256. [DOI] [PubMed] [Google Scholar]
- 16.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brussaard, C. P. D. 2004. Viral control of phytoplankton populations—a review. J. Eukaryot. Microbiol. 51:125-138. [DOI] [PubMed] [Google Scholar]
- 18.Buchen-Osmond, C. 2003. The universal virus database ICTVdB. Comput. Sci. Eng. 5:16-25. [Google Scholar]
- 19.Calendar, R. (ed.). 2005. The bacteriophages, 2nd ed., vol. 1. Oxford University Press, New York, NY.
- 20.Cann, A. J., S. E. Fandrich, and S. Heaphy. 2005. Analysis of the virus population present in equine faeces indicates the presence of hundreds of uncharacterized virus genomes. Virus Genes 30:151-156. [DOI] [PubMed] [Google Scholar]
- 21.Chen, F., and J. Lu. 2002. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl. Environ. Microbiol. 68:2589-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, F., J. R. Lu, B. J. Binder, Y. C. Liu, and R. E. Hodson. 2001. Application of digital image analysis and flow cytometry to enumerate marine viruses stained with SYBR gold. Appl. Environ. Microbiol. 67:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, F., and C. A. Suttle. 1996. Evolutionary relationships among large double-stranded DNA viruses that infect microalgae and other organisms as inferred from DNA polymerase genes. Virology 219:170-178. [DOI] [PubMed] [Google Scholar]
- 24.Chen, F., K. Wang, J. J. Kan, M. T. Suzuki, and K. E. Wommack. 2006. Diverse and unique picocyanobacteria in Chesapeake Bay, revealed by 16S-23S rRNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 72:2239-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daughney, C. J., X. Chatellier, A. Chan, P. Kenward, D. Fortin, C. A. Suttle, and D. A. Fowle. 2004. Adsorption and precipitation of iron from seawater on a marine bacteriophage (PWH3A-P1). Mar. Chem. 91:101-115. [Google Scholar]
- 27.Edwards, R. A., and F. Rohwer. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3:504-510. [DOI] [PubMed] [Google Scholar]
- 28.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 29.Fogel, G. B., C. R. Collins, J. Li, and C. F. Brunk. 1999. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb. Ecol. 38:93-113. [DOI] [PubMed] [Google Scholar]
- 30.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 31.Heidelberg, J. F., K. B. Heidelberg, and R. R. Colwell. 2002. Seasonality of Chesapeake Bay bacterioplankton species. Appl. Environ. Microbiol. 68:5488-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendrix, R. W., M. C. M. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hewson, I., D. M. Winget, K. E. Williamson, J. A. Fuhrman, and K. E. Wommack. 2006. Viral and bacterial assemblage covariance in oligotrophic waters of the West Florida Shelf (Gulf of Mexico). J. Mar. Biol. Assoc. U. K. 86:591-603. [Google Scholar]
- 34.Kan, J. J., B. C. Crump, K. Wang, and F. Chen. 2006. Bacterioplankton community in Chesapeake Bay: predictable or random assemblages. Limnol. Oceanogr. 51:2157-2169. [Google Scholar]
- 35.Karner, M. B., E. F. DeLong, and D. M. Karl. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507-510. [DOI] [PubMed] [Google Scholar]
- 36.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformat. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 37.Lindell, D., J. D. Jaffe, Z. I. Johnson, G. M. Church, and S. W. Chisholm. 2005. Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438:86-89. [DOI] [PubMed] [Google Scholar]
- 38.Mann, N. H. 2003. Phages of the marine cyanobacterial picophytoplankton. FEMS Microbiol. Rev. 27:17-34. [DOI] [PubMed] [Google Scholar]
- 39.Mann, N. H., A. Cook, A. Millard, S. Bailey, and M. Clokie. 2003. Marine ecosystems: bacterial photosynthesis genes in a virus. Nature 424:741. [DOI] [PubMed] [Google Scholar]
- 40.McDaniel, L., L. A. Houchin, S. J. Williamson, and J. H. Paul. 2002. Plankton blooms: lysogeny in marine Synechococcus. Nature 415:496. [DOI] [PubMed] [Google Scholar]
- 41.Middelboe, M., and P. G. Lyck. 2002. Regeneration of dissolved organic matter by viral lysis in marine microbial communities. Aquat. Microb. Ecol. 27:187-194. [Google Scholar]
- 42.Middelboe, M., L. Riemann, G. F. Steward, V. Hansen, and O. Nybroe. 2003. Virus-induced transfer of organic carbon between marine bacteria in a model community. Aquat. Microb. Ecol. 33:1-10. [Google Scholar]
- 43.Millard, A., M. R. J. Clokie, D. A. Shub, and N. H. Mann. 2004. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc. Natl. Acad. Sci. USA 101:11007-11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, R. V. 2001. Environmental bacteriophage-host interactions: factors contributing to natural transduction. Antonie Leeuwenhoek 79:141-147. [DOI] [PubMed] [Google Scholar]
- 45.Muhling, M., N. J. Fuller, A. Millard, P. J. Somerfield, D. Marie, W. H. Wilson, D. J. Scanlan, A. F. Post, I. Joint, and N. H. Mann. 2005. Genetic diversity of marine Synechococcus and co-occurring cyanophage communities: evidence for viral control of phytoplankton. Environ. Microbiol. 7:499-508. [DOI] [PubMed] [Google Scholar]
- 46.Peterson, J. D., L. A. Umayam, T. Dickinson, E. K. Hickey, and O. White. 2001. The Comprehensive Microbial Resource. Nucleic Acids Res. 29:123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poorvin, L., J. M. Rinta-Kanto, D. A. Hutchins, and S. W. Wilhelm. 2004. Viral release of iron and its bioavailability to marine plankton. Limnol. Oceanogr. 49:1734-1741. [Google Scholar]
- 48.Pride, D. T., T. M. Wassenaar, C. Ghose, and M. J. Blaser. 2006. Evidence of host-virus co-evolution in tetranucleotide usage patterns of bacteriophages and eukaryotic viruses. BMC Genomics 7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Proctor, L. M., and J. A. Fuhrman. 1990. Viral mortality of marine-bacteria and cyanobacteria. Nature 343:60-62. [Google Scholar]
- 50.Ptashne, M. 1992. A genetic switch: phage lambda and higher organisms, 2nd ed. Cell Press, Blackwell Scientific Publications, Cambridge, MA.
- 51.Riley, M. 1993. Functions of the gene products of Escherichia coli. Microbiol. Rev. 57:862-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rohwer, F., A. Segall, G. Steward, V. Seguritan, M. Breitbart, F. Wolven, and F. Azam. 2000. The complete genomic sequence of the marine phage Roseophage SIO1 shares homology with nonmarine phages. Limnol. Oceanogr. 45:408-418. [Google Scholar]
- 54.Rohwer, F., V. Seguritan, D. H. Choi, A. M. Segall, and F. Azam. 2001. Production of shotgun libraries using random amplification. BioTechniques 31:108-119. [DOI] [PubMed] [Google Scholar]
- 55.Shannon, C. E., and W. Weaver. 1998. The mathematical theory of communication, 3rd ed. University of Illinois Press, Urbana.
- 56.Short, C. M., and C. A. Suttle. 2005. Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Appl. Environ. Microbiol. 71:480-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Short, S. M., and C. A. Suttle. 2002. Sequence analysis of marine virus communities reveals that groups of related algal viruses are widely distributed in nature. Appl. Environ. Microbiol. 68:1290-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sullivan, M. B., M. L. Coleman, P. Weigele, F. Rohwer, and S. W. Chisholm. 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sullivan, M. B., D. Lindell, J. A. Lee, L. R. Thompson, J. P. Bielawski, and S. W. Chisholm. 2006. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 4:e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sullivan, M. B., J. B. Waterbury, and S. W. Chisholm. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424:1047-1051. [DOI] [PubMed] [Google Scholar]
- 61.Suttle, C. A. 2005. Viruses in the sea. Nature 437:356-361. [DOI] [PubMed] [Google Scholar]
- 62.Suttle, C. A., A. M. Chan, and M. T. Cottrell. 1990. Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347:467-469. [Google Scholar]
- 63.Thingstad, T. F. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45:1320-1328. [Google Scholar]
- 64.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfiel. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 65.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y.-H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 66.Wang, K., and F. Chen. 2004. Genetic diversity and population dynamics of cyanophage communities in the Chesapeake Bay. Aquat. Microb. Ecol. 34:105-116. [Google Scholar]
- 67.Weinbauer, M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127-181. [DOI] [PubMed] [Google Scholar]
- 68.Wheeler, D. L., C. Chappey, A. E. Lash, D. D. Leipe, T. L. Madden, G. D. Schuler, T. A. Tatusova, and B. A. Rapp. 2000. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 28: 10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilhelm, S. W., and C. A. Suttle. 1999. Viruses and nutrient cycles in the sea—viruses play critical roles in the structure and function of aquatic food webs. Bioscience 49:781-788. [Google Scholar]
- 70.Williamson, S. J., L. A. Houchin, L. McDaniel, and J. H. Paul. 2002. Seasonal variation in lysogeny as depicted by prophage induction in Tampa Bay, Florida. Appl. Environ. Microbiol. 68:4307-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson, W. H., D. C. Schroeder, M. J. Allen, M. T. G. Holden, J. Parkhill, B. G. Barrell, C. Churcher, N. Harnlin, K. Mungall, H. Norbertczak, M. A. Quail, C. Price, E. Rabbinowitsch, D. Walker, M. Craigon, D. Roy, and P. Ghazal. 2005. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science 309:1090-1092. [DOI] [PubMed] [Google Scholar]
- 72.Wommack, K., R. Hill, M. Kessel, E. Russek-Cohen, and R. Colwell. 1992. Distribution of viruses in the Chesapeake Bay. Appl. Environ. Microbiol. 58:2965-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wommack, K. E., J. Ravel, R. T. Hill, J. Chun, and R. R. Colwell. 1999. Population dynamics of Chesapeake Bay virioplankton: total-community analysis by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wommack, K. E., J. Ravel, R. T. Hill, and R. R. Colwell. 1999. Hybridization analysis of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 65:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, T., M. Breitbart, W. H. Lee, J. Q. Run, C. L. Wei, S. W. L. Soh, M. L. Hibberd, E. T. Liu, F. Rohwer, and Y. J. Ruan. 2006. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhong, Y., F. Chen, S. W. Wilhelm, L. Poorvin, and R. E. Hodson. 2002. Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20. Appl. Environ. Microbiol. 68:1576-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.