Abstract
In Mycoplasma pneumoniae and several other mollicutes, the UGA opal codon specifies tryptophan rather than a translation stop. This often makes it difficult to express Mycoplasma proteins in heterologous hosts. In this work, we demonstrate that mollicute proteins can be fused to an affinity tag and be expressed directly in M. pneumoniae. The protein can then be purified by affinity chromatography and be used for biochemical or any other desired analysis.
Mycoplasma pneumoniae is a human pathogen that thrives on mucosal surfaces and causes diseases such as mild pneumonia, tracheobronchitis, pediatric encephalitis, and erythema multiforme (2, 11, 15, 18, 24). These bacteria possess one of the smallest genomes of any free-living organism known so far. This reduced genome has recently attracted much interest, as the analysis of M. pneumoniae and its close relatives may help to identify the minimal set of genes that is required for independent life (4, 9). This prompted the development of the new field of synthetic biology with projects aimed at the generation of artificial microbes based on Mycoplasma spp. (3, 14). Another interesting aspect of the small genome is the observation that several enzymes of Mycoplasma spp. are moonlighting: i.e., they have multiple unrelated functions (12). This was discovered for glycolytic kinases that are also active as nucleoside diphosphate kinases in M. pneumoniae and other Mycoplasma spp. (17).
The analysis of proteins from Mycoplasma spp. is hampered by a peculiarity of the genetic code of these bacteria: they use the UGA opal codon to incorporate tryptophan rather than as a stop codon as in the universal genetic code (10, 19). Thus, if cloned in Escherichia coli or other hosts, the genes from M. pneumoniae often contain many stop codons that prevent heterologous expression. Several strategies have been developed to solve this problem. Expression of mollicute genes in Spiroplasma spp. that read the UGA as a tryptophan codon has been reported; however, these bacteria are difficult to handle (21). E. coli suppressor strains expressing an opal suppressor tRNA have been developed; however, they fail if multiple opal codons are present (20). M. pneumoniae genes containing few UGA codons have been expressed in Bacillus subtilis with low efficiency (13). Recently, we developed a PCR-based strategy to exchange as many as nine opal codons in a single step. This approach, the multiple mutation reaction, allowed the expression of the M. pneumoniae glycerol kinase and has been used for several other proteins (7). However, there are cases in which it is desirable to purify a mollicute protein from the original host. This is of special importance for protein-protein interaction studies. To facilitate gene expression and protein purification in M. pneumoniae, we developed a system for the expression of proteins fused to an N-terminal streptavidin (Strep) tag in M. pneumoniae. These proteins can easily be purified by affinity chromatography using Strep-tactin columns due to the highly specific interaction between the Strep-tag peptide and the artificial protein Strep-tactin (23).
Outline of the expression system.
To allow efficient expression and subsequent purification of a protein of interest in M. pneumoniae, the respective gene has to be fused to an affinity tag and a promoter has to be provided. We chose to use the Strep tag because this peptide binds with high affinity to Strep-tactin materials and can be eluted in a single step using desthiobiotin. In our hands, the purification using the Strep tag is much more efficient than the popular His tag for purification from both E. coli and B. subtilis (8, 16). To drive the expression of the fusion proteins, we selected a promoter fragment of the M. pneumoniae ackA gene encoding acetate kinase. This promoter was recently shown to confer active transcription to a reporter gene in M. pneumoniae (6). Since there are no replicative plasmids for M. pneumoniae available, we introduced the artificial gene by transposition into the chromosome of M. pneumoniae and assayed several clones for expression of the recombinant protein using antibodies recognizing the Strep tag.
Cloning of M. pneumoniae hprK and expression of the protein.
To test the anticipated expression system, we selected the M. pneumoniae HPr kinase/phosphorylase (HPrK; encoded by the hprK gene). We previously expressed HPrK with an N-terminal Strep tag and demonstrated biochemical activity of the fusion protein (16). Plasmid pGP1012 allowing the expression of HPrK carrying an N-terminal Strep tag under the control of the ackA promoter was constructed as follows. The DNA fragment corresponding to the hprK gene with a sequence specifying the Strep tag II at its 5′ end was amplified from plasmid pGP611 (16) using the oligonucleotides SS22 (5′ AAACTGCAGTGGAGCCACCCGCAGTTCG) and SS23 (5′ AAAGCGGCCGCGGTCTGCTACTAACACTAGGATTCATC). In addition, the ackA promoter fragment, which also contains the Shine-Dalgarno sequence and the ATG start codon of the acetate kinase open reading frame, was amplified from M. pneumoniae chromosomal DNA using the primer pair SH58 (6) and SS21 (5′ AAACTGCAGCATTTTTATCTAATAGGTAACAA). The promoter and the hprK gene fragments were digested with EcoRI/PstI and PstI/NotI, respectively, and ligated to pMT85 (25) linearized with EcoRI and NotI in a three-fragment ligation mixture. The correctness of the resulting plasmid, pGP1012, was verified by restriction and sequence analysis. Plasmid pMT85 (and thus, pGP1012) carries an aac-aphD gentamicin resistance determinant as well as a derivative of Tn4001 that lacks the transposase gene (mini-Tn4001) to avoid transposition after the initial integration into the M. pneumoniae chromosome. The tranposase gene is located on the plasmid backbone that is lost upon transposition (25). This allows the random integration of the cassette carrying the ackA promoter and the artificial strep-hprK gene in the chromosome of M. pneumoniae. For this purpose, we introduced pGP1012 (and as a control, pMT85) by electroporation into M. pneumoniae M129 as described earlier (1). Transformants were selected on plates containing gentamicin.
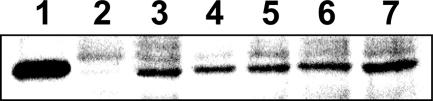
To check the expression of the tagged HPrK in the transformants, we cultivated five independent clones obtained with pGP1012 and one transformant carrying the empty mini-Tn4001 derived from pMT85 and prepared cell extracts. These cell extracts were separated by one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were transferred to a polyvinylidene difluoride membrane by electroblotting. The presence of Strep-HPrK was verified using polyclonal antibodies directed against Strep-glycerol kinase (GlpK) from M. pneumoniae (SeqLab, Göttingen, Germany). We observed that these antibodies were able to efficiently cross-react with the Strep tag in addition to M. pneumoniae GlpK (data not shown). As shown in Fig. 1, a Strep-tagged protein corresponding in size exactly to the Strep-HPrK purified from E. coli was present in all clones carrying the minitransposon derived from pGP1012 but not in those carrying the empty mini-Tn4001 (pMT85). Since the minitransposon can be inserted at a different position in each clone, we cannot exclude position effects on the expression of the construct. Based on the Western blot shown in Fig. 1, we selected clone 1 for the purification of Strep-HPrK. This strain was designated GPM78.
FIG. 1.
Evaluation of the expression of Strep-tagged HPrK in M. pneumoniae GPM78 by Western blot analysis. Crude extracts of different M. pneumoniae GPM78 clones grown in the presence of glucose (1% [wt/vol]) were separated by SDS-PAGE. Strep-HPrK was detected by using polyclonal rabbit antibodies raised against M. pneumoniae Strep-tagged GlpK due to cross-reaction to the Strep tag. Ten micrograms of cell extract was applied to each lane. A total of 500 ng recombinant Strep-tagged HPrK served as a control (lane 1). Lane 2, M. pneumoniae GPM79 (empty vector); lanes 3 to 7, M. pneumoniae GPM78 clones 1 to 5.
Purification of Strep-HPrK and activity assay.
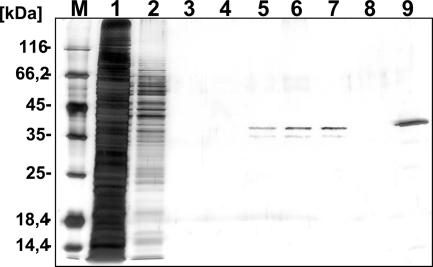
To purify the tagged HPrK, we cultivated M. pneumoniae GPM78 in 400 ml of modified Hayflick medium (5). The cells were disrupted by sonication, and the cell extracts were passed over a Strep-tactin Sepharose column (IBA GmbH, Göttingen, Germany). After five washing steps, the protein carrying a Strep tag was eluted by applying desthiobiotin (2.5 mM) to the columns as described previously (16). The purified protein was further analyzed by SDS-PAGE (Fig. 2). Using this procedure, we obtained 25 μg of virtually pure protein from a culture of 400 ml (concentration, 100 ng/μl).
FIG. 2.
Overproduction and purification of recombinant M. pneumoniae HPrK. Cells of M. pneumoniae GPM78 were grown in modified Hayflick medium containing 1% (wt/vol) glucose. After 72 h, cells were harvested and disrupted by sonication. The crude extract was passed over a Strep-tactin column (400-μl bed volume; IBA, Göttingen, Germany). The recombinant protein was eluted with desthiobiotin (IBA [final concentration, 2.5 mM]) after extensive washing with a buffer containing 100 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 1 mM EDTA. Aliquots of the individual fractions were separated on 12% SDS-polyacrylamide gels followed by silver staining. A protein mass marker (Fermentas) served as a standard (lane M). Lane 1, washing step 1; lane 2, washing step 2; lane 3, washing step 5; lanes 4 to 8, elution fractions 1 to 5; lane 9, Strep-HPrK purified from E. coli DH5α (500 ng).
The purified Strep-HPrK was used for activity assays: i.e., the phosphorylation of M. pneumoniae HPr protein at the expense of ATP. For this purpose, we purified HPr carrying an N-terminal His tag using E. coli DH5α/pGP217 (22). The activity assays were performed as described previously (22). As shown in Fig. 3, the Strep-HPrK purified from M. pneumoniae was active in the phosphorylation of HPr.
FIG. 3.
In vitro phosphorylation assay to test the activity of purified Strep-HPrK in M. pneumoniae GPM78 elution fractions. M. pneumoniae His6-HPr (20 μM) was incubated with 12 μl of each elution fraction and 0.5 mM ATP in assay buffer (25 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 1 mM dithiothreitol) in a final volume of 20 μl at 37°C for 120 min. Subsequently, the HPrK was heat inactivated for 10 min at 95°C. The assay mixtures were analyzed by 10% native PAGE. The first lanes are controls with M. pneumoniae His6-HPr (first lane) and His6-HPr(Ser-P) prepared in vitro (second lane). In addition, 5 μg of M. pneumoniae GPM78 crude extract served as a control (third lane). Lanes 1 to 5, elution fractions 1 to 5; lane 6, elution fraction 3 without addition of His6-HPr.
This study demonstrates that the purification of Strep-tagged proteins directly from M. pneumoniae is an alternative to their expression in heterologous hosts such as E. coli, which often requires the laborious replacement of opal codons to prevent premature translation termination. We are confident that other Mycoplasma species such as M. genitalium might also be used as expression hosts. Moreover, the application of novel, potentially stronger promoters and translation signals may increase the efficiency of protein expression. The proteins isolated in this way can be used for biochemical characterization or for the generation of antibodies. Moreover, the expression in M. pneumoniae also allows the isolation of functional protein complexes (8; our unpublished results).
Acknowledgments
We are grateful to Richard Herrmann for the generous gift of plasmid pMT85 and to Julia Busse for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
Footnotes
Published ahead of print on 12 October 2007.
REFERENCES
- 1.Catrein, I., R. Dumke, J. Weiner III, E. Jacobs, and R. Herrmann. 2004. Cross-complementation between the products of the genes P1 and ORF6 of Mycoplasma pneumoniae subtypes 1 and 2. Microbiology 150:3989-4000. [DOI] [PubMed] [Google Scholar]
- 2.Christie, L. J., S. Honarmand, D. F. Talkington, S. S. Gavali, C. Preas, C. Y. Pan, S. Yagi, and C. A. Glaser. 2007. Pediatric encephalitis: what is the role of Mycoplasma pneumoniae? Pediatrics 120:305-313. [DOI] [PubMed] [Google Scholar]
- 3.Drubin, D. A., J. C. Way, and P. A. Silver. 2007. Designing biological systems. Genes Dev. 21:242-254. [DOI] [PubMed] [Google Scholar]
- 4.Glass, J. I., N. Assad-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchison III, H. O. Smith, and J. C. Venter. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA 103:425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halbedel, S., C. Hames, and J. Stülke. 2004. In vivo activity of enzymatic and regulatory components of the phosphoenolpyruvate:sugar phosphotransferase system in Mycoplasma pneumoniae. J. Bacteriol. 186:7936-7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halbedel, S., H. Eilers, B. Jonas, J. Busse, M. Hecker, S. Engelmann, and J. Stülke. 2007. Transcription in Mycoplasma pneumoniae: analysis of the promoters of the ackA and ldh genes. J. Mol. Biol. 371:596-607. [DOI] [PubMed] [Google Scholar]
- 7.Hames, C., S. Halbedel, O. Schilling, and J. Stülke. 2005. Multiple-mutation reaction: a method for the simultaneous introduction of multiple mutations into the glpK gene of Mycoplasma pneumoniae. Appl. Environ. Microbiol. 71:4097-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herzberg, C., L. A. Flórez Weidinger, B. Dörrbecker, S. Hübner, J. Stülke, and F. M. Commichau. SPINE: a method for the rapid detection and analysis of protein-protein interactions in vivo. Proteomics, in press. [DOI] [PubMed]
- 9.Hutchison, C. A., III, S. C. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 10.Inamine, J. M., K.-C. Ho, S. Loechel, and P.-C. Hu. 1990. Evidence that UGA is read as a tryptophan codon rather than as a stop codon by Mycoplasma pneumoniae, Mycoplasma genitalium, and Mycoplasma gallisepticum. J. Bacteriol. 172:504-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs, E. 1997. Mycoplasma infections of the human respiratory tract. Wien. Klin. Wochenschr. 109:574-577. [PubMed] [Google Scholar]
- 12.Jeffery, C. J. 2003. Moonlighting proteins: old proteins learning new tricks. Trends Genet. 19:415-417. [DOI] [PubMed] [Google Scholar]
- 13.Kannan, T. R., and J. B. Baseman. 2000. Expression of UGA-containing Mycoplasma genes in Bacillus subtilis. J. Bacteriol. 182:2664-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lartigue, C., J. I. Glass, N. Alperovich, R. Pieper, P. P. Parmar, C. A. Hutchison III, H. O. Smith, and J. C. Venter. 2007. Genome transplantation in bacteria: changing one species to another. Science 317:632-638. [DOI] [PubMed] [Google Scholar]
- 15.Lind, K. 1983. Manifestations and complications of Mycoplasma pneumoniae disease: a review. Yale J. Biol. Med. 56:461-468. [PMC free article] [PubMed] [Google Scholar]
- 16.Merzbacher, M., C. Detsch, W. Hillen, and J. Stülke. 2004. Mycoplasma pneumoniae HPr kinase/phosphorylase. Assigning functional roles to the P-loop and the HPr kinase/phosphorylase signature sequence motif. Eur. J. Biochem. 271:367-374. [DOI] [PubMed] [Google Scholar]
- 17.Pollack, J. D., M. A. Myers, T. Dandekar, and R. Herrmann. 2002. Suspected utility of enzymes with multiple activities in the small genome Mycoplasma species: the replacement of the missing “household” nucleoside diphosphate kinase gene and activity by glycolytic kinases. OMICS J. Integr. Biol. 6:247-257. [DOI] [PubMed] [Google Scholar]
- 18.Schalock, P. C., J. G. Dinulos, N. Pace, K. Schwarzenberger, and J. K. Wenger. 2006. Erythema multiforme due to Mycoplasma pneumoniae infection in two children. Pediatr. Dermatol. 23:546-555. [DOI] [PubMed] [Google Scholar]
- 19.Simoneau, P., C.-M. Li, S. Loechel, R. Wenzel, R. Herrmann, and P.-C. Hu. 1993. Codon reading scheme in Mycoplasma pneumoniae revealed by the analysis of the complete set of tRNA genes. Nucleic Acids Res. 21:4967-4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smiley, B. K., and F. C. Minion. 1993. Enhanced readthrough of opal (UGA) stop codons and production of Mycoplasma pneumoniae P1 epitopes in Escherichia coli. Gene 134:33-40. [DOI] [PubMed] [Google Scholar]
- 21.Stamburski, C., J. Renaudin, and J. M. Bove. 1991. First steps toward a virus-derived vector for gene cloning and expression in spiroplasmas, organisms which read UGA as a tryptophan codon: synthesis of chloramphenicol acetyltransferase in Spiroplasma citri. J. Bacteriol. 173:2225-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinhauer, K., T. Jepp, W. Hillen, and J. Stülke. 2002. A novel mode of control of Mycoplasma pneumoniae HPr kinase/phosphatase activity reflects its parasitic life style. Microbiology 148:3277-3284. [DOI] [PubMed] [Google Scholar]
- 23.Terpe, K. 2006. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Technol. 72:211-222. [DOI] [PubMed] [Google Scholar]
- 24.Waites, K. B., and D. F. Talkington. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17:697-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmerman, C.-U., and R. Herrmann. 2005. Synthesis of a small, cysteine-rich, 29 amino acids long peptide in Mycoplasma pneumoniae. FEMS Microbiol. Lett. 253:315-321. [DOI] [PubMed] [Google Scholar]