Abstract
We engineered a strain of the bacterium Caulobacter crescentus to fluoresce in the presence of micromolar levels of uranium at ambient temperatures when it is exposed to a hand-held UV lamp. Previous microarray experiments revealed that several Caulobacter genes are significantly upregulated in response to uranium but not in response to other heavy metals. We designated one of these genes urcA (for uranium response in caulobacter). We constructed a reporter that utilizes the urcA promoter to produce a UV-excitable green fluorescent protein in the presence of the uranyl cation, a soluble form of uranium. This reporter is specific for uranium and has little cross specificity for nitrate (<400 μM), lead (<150 μM), cadmium (<48 μM), or chromium (<41.6 μM). The uranium reporter construct was effective for discriminating contaminated groundwater samples (4.2 μM uranium) from uncontaminated groundwater samples (<0.1 μM uranium) collected at the Oak Ridge Field Research Center. In contrast to other uranium detection methodologies, the Caulobacter reporter strain can provide on-demand usability in the field; it requires minimal sample processing and no equipment other than a hand-held UV lamp, and it may be sprayed directly on soil, groundwater, or industrial surfaces.
The role of nuclear technology in our energy resource portfolio is likely to become increasingly important as the global demand for energy continues to rise. Uranium processing, both for energy and for nuclear weapons, has resulted in a multitude of contaminated sites worldwide. In the United States specifically, there are more than 120 uranium-contaminated sites containing approximately 6.4 trillion liters of waste (22). As a heavy metal as well as a radionuclide, enriched uranium is toxic to cellular functions. Remediation strategies have utilized plants to extract uranium from contaminated sites (15) and recently have focused on containment to minimize migration of uranium in groundwater and prevent infiltration into drinking water supplies (6). The uranyl ion (UO22+) is the most water-soluble and bioavailable form of uranium (38) and poses the greatest threat to human health. Due to the ease of uranyl ion spread through groundwater systems, most bioremediation strategies attempt to prevent contaminating uranium spread by utilizing microorganisms to reduce the oxidation state of uranium from U(VI), found in uranyl, to less soluble forms of uranium, including U(IV) (17, 36, 37).
Detecting the presence of low concentrations of uranium is necessary to identify contaminated areas and to assess the progress of remediation efforts. Methods employed to detect and quantify uranium concentrations often exploit the physical properties of uranium, including phosphorescence kinetics (5), atomic emission (16), and mass spectrometry characteristics (4). These methods are extremely sensitive and selective for uranium, but they allow low sample throughput and are not very portable. Additionally, these technologies measure the total amount of uranium present in a given sample, as opposed to the quantity of bioavailable uranium. Recently, several uranyl biosensors have been reported; these biosensors include a uranium immunosensor in which a fluorescently labeled monoclonal antibody selectively binds to chelated uranyl (3) and a catalytic beacon sensor consisting of a DNA enzyme that in the presence of uranyl catalyzes DNA cleavage, leading to an increase in fluorescence (19). The catalytic beacon biosensor rivals the most sensitive analytical instruments for uranium detection, with a detection limit of 45 pM, a linear detection range of up to 400 nM, and extremely good specificity for the uranyl ion. However, catalytic beacon sensor measurement must be performed with individually isolated and prepared samples.
In addition to biosensors based upon in vitro methods, such as the immunosensor and catalytic beacon sensor described above, there are precedents for whole-cell heavy metal biosensors (1, 7, 18, 28, 30, 33) utilizing either luciferase or fluorescent protein reporters. In contrast to other methodologies, whole-cell biosensors may be dispensed directly on the site of interest, detecting the presence of a bioavailable heavy metal in situ. This is possible because whole-cell biosensors that utilize a UV-excitable green fluorescent protein, GFPuv (8, 33), require little or no sample preparation. The heavy metals detectable thus far by whole-cell biosensors include mercury, chromate, arsenic, and copper, but to the best of our knowledge, no whole-cell biosensor for uranium has been reported previously.
In the work presented here, we bioengineered a strain of the bacterium Caulobacter crescentus to become fluorescently green in the presence of toxic levels of uranium. Caulobacter is a widely distributed bacterial genus that is able to survive in low-nutrient environments, including freshwater, seawater, soil (29), contaminated groundwater (21), wastewater (20), and habitats where contamination with uranium may be present (27). This great diversity of viable habitats, including habitats contaminated with uranium, suggests that a Caulobacter whole-cell in situ uranium biosensor could robustly function across a wide spectrum of environmental conditions and ambient temperatures. Caulobacter is particularly resistant to the lethal effects of uranium up to a uranyl nitrate concentration of 1 mM (13), and previous microarray experiments have demonstrated that several Caulobacter genes are significantly upregulated in response to uranium but not in response to other heavy metals (13). Building upon these results, we constructed a uranium reporter that places GFPuv under the control of the promoter that is most strongly upregulated under uranium stress conditions.
MATERIALS AND METHODS
Materials.
T4 DNA ligase, shrimp alkaline phosphatase (SAP), and endonucleases were purchased from Fermentas (Hanover, MD) and New England Biolobs (Ipswich, MA). DNA oligonucleotides were purchased from the Stanford Protein and Nucleic Acid Biotechnology Facility (Stanford, CA). OneShot Top10 chemically competent Escherichia coli and 0.1-cm electroporation cuvettes were purchased from Invitrogen (Carlsbad, CA). DNA sequencing was performed by Sequetech (Mountain View, CA). KOD Hot Start DNA polymerase was purchased from Novagen (Madison, WI). DNA miniprep and gel extraction kits were purchased from Qiagen (Valencia, CA). An ND-3300 fluorospectrometer was purchased from NanoDrop (Wilmington, DE). Cadmium sulfate (CdSO4), potassium chromate (K2CrO4), lead nitrate [Pb(NO3)2], and depleted uranyl nitrate [UO2(NO3)2 · 6H2O] were purchased from Sigma-Aldrich, and stock solutions were prepared as described previously (13).
PurcA lacZ reporter strain.
The genomic region containing the urcA promoter was amplified from C. crescentus CB15N genomic DNA with KOD Hot Start DNA polymerase and oligonucleotides NJH144 and NJH121 (see Table S1 in the supplemental material). The 50-μl PCR mixture, containing 5% dimethyl sulfoxide, was made using the manufacturer's protocol. The PCR was initiated by 1.75 min of melting at 94°C, followed by 32 cycles of 15 s of melting at 94°C, 30 s of annealing at 58°C, and 70 s of extension at 68°C. The PCR product was then purified by electrophoresis through 1.2% agarose, followed by gel extraction, and then reamplified with oligonucleotides NJH120 (see Table S1 in the supplemental material) and NJH121 as described above. The second PCR product was then digested with BglII and KpnI (using the protocol recommended by the manufacturer), ligated overnight at 16°C with T4 DNA ligase (using the protocol recommended by the manufacturer) with the similarly digested pPR9TT vector (31) backbone, and then transformed into OneShot Top10 chemically competent E. coli cells (using the protocol recommended by manufacturer). The sequence of the resulting plasmid, pNJH123, was confirmed by primer extension sequencing using oligonucleotides NJH155 and NJH156 (see Table S1 in the supplemental material). C. crescentus CB15N ΔCC_1634 (strain LS4358) was transformed with plasmid pNJH123 by electroporation as previously described (10) to obtain the urcA promoter LacZ reporter strain NJH199 (Table 1. The in-frame ΔCC_1634 deletion reduces the background β-galactosidase activity of Caulobacter (J. C. Chen, unpublished results).
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Description | Reference |
|---|---|---|
| Plasmids | ||
| pPR9TT | Translational fusion LacZ reporter vector | 31 |
| pNJH123 | pPR9TT-PurcAlacZ reporter vector | This study |
| pBAD-GFP | Prokaryotic GFPuv expression vector | 8 |
| pRVYFP-2 | Low-copy-number expression vector | M. Thanbichler, unpublished |
| pNJH193 | pRVYFP-2-PurcAgfpuv reporter vector | This study |
| pNJH198 | pRVYFP-2-PurcA His6gfpuv reporter vector | This study |
| pNJH200 | pRVYFP-2-PurcA_RBS His6gfpuv reporter vector | This study |
| pBVMCS-2 | High-copy-number expression vector | M. Thanbichler, unpublished |
| pNJH201 | pBVMCS-2-PurcA_RBS His6gfpuv reporter vector | This study |
| pX31 | High-copy-number xylose-inducible expression vector | A. Iniesta, unpublished |
| pNJH153 | pX31-Pxylgfpuv expression vector | This study |
| pRSET-B-mCherry | mCherry expression vector | 32 |
| pMT383 | pPvan-ftsZ-eyfp expression integration vector | 35 |
| pNJH15 | pPvan-ftsZ-mcherry expression integration vector | This study |
| pMT397 | Low-copy-number pPvan-MCS-eYFP expression vector | M. Thanbichler, unpublished |
| pNJH156 | pX31-Pxylgfpuv (or eyfp) expression vector | This study |
| pNJH169 | pX31-PxylurcA-mcherry expression vector | This study |
| Strains | ||
| LS4358 | C. crescentus CB15N ΔCC_1634 | J. C. Chen, unpublished |
| NJH199 | C. crescentus CB15N ΔCC_1634(pNJH123) | This study |
| LS101 | C. crescentus CB15N | 11 |
| NJH371 | C. crescentus CB15N(pNJH201) | This study |
| NJH250 | C. crescentus CB15N(pNJH153) | This study |
| NJH300 | C. crescentus CB15N(pNJH169) | This study |
PurcA lacZ reporter activity assays.
Cultures of strain NJH199 were grown at 28°C in M2G medium (10). Overnight cultures were diluted to an optical density at 660 nm (OD660) of 0.1 with fresh M2G medium and then grown for an additional 2 h at 28°C to obtain exponential growth again before the cells were stressed with either a mock treatment or indicated concentrations of uranyl nitrate, sodium nitrate, lead nitrate, cadmium sulfate, or potassium chromate. The stressed cultures were then grown on an orbital shaker for 2 h (see Fig. 3B) or the amount of time indicated below (see Fig. 3A) before liquid culture β-galactosidase assays were conducted, as previously described (24).
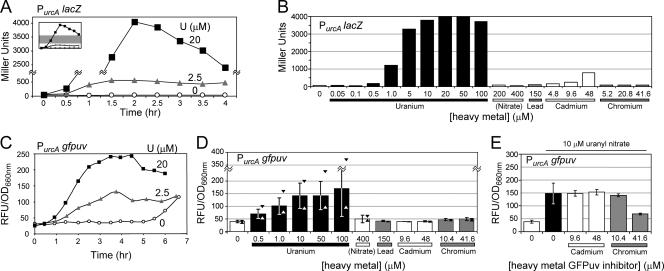
FIG. 3.
PurcA lacZ and PurcA gfpuv reporter kinetics, sensitivity, and specificity. (A) Time course of strain NJH199 β-galactosidase activity in liquid culture after induction with uranyl nitrate. The middle section of the plot, indicated by the gray region of the inset, has been removed. (B) Strain NJH199 β-galactosidase activity after 2 h of heavy metal exposure. (C) Time course of GFPuv fluorescence (expressed in relative fluorescence units [RFU] divided by the culture OD660) for strain NJH371 (PurcA gfpuv) after induction with uranyl nitrate. (D) Strain NJH371 GFPuv fluorescence after 4 h of heavy metal exposure. The error bars indicate one standard deviation from the mean; the triangles indicate the maximum and minimum observed fluorescence values. The data are aggregate results from uranyl and mock treatments (n = 7) or from replicate experiments (n = 3). (E) Inhibitory effect of high concentrations of chromium on GFPuv reporter function. Strain NJH371 was induced with 10 μM uranyl nitrate (indicated by the horizontal line at the top) with or without cadmium or chromium for 4 h before GFPuv fluorescence was assayed. The data are aggregate results obtained with 10 μM uranyl nitrate alone (n = 5) or in replicate experiments (n = 3).
PurcA gfpuv reporter strain.
An NcoI/NheI DNA fragment containing gfpuv was amplified from plasmid pBAD-GFP (8). An NcoI restriction site internal to gfpuv was silently mutated using splicing by overlap extension (SOE) (12). The pBAD-GFP template was amplified with KOD Hot Start DNA polymerase using oligonucleotides NJH122 and NJH237 (5′ SOE PCR) (see Table S1 in the supplemental material) or NJH236 and NJH123 (3′ SOE PCR) (see Table S1 in the supplemental material), as described above. The first-round SOE PCR products were mixed 1:1 and used as the template for the second-round SOE PCR and then amplified using oligonucleotides NJH122 and NJH123, as described above. The second-round SOE PCR product was then digested with NcoI and NheI.
An AscI/NcoI DNA fragment containing the urcA promoter was amplified from plasmid pNJH123 with KOD Hot Start DNA polymerase using oligonucleotides NJH144 and NJH238 (see Table S1 in the supplemental material). The PCR product was then digested with AscI and NcoI, ligated with the NcoI/NheI-digested gfpuv fragment (described above) and the AscI/NheI-digested low-copy-number pRVYFP-2 vector (M. Thanbichler, unpublished) backbone (triple ligation), and then transformed into OneShot Top10 chemically competent E. coli cells. The sequence of the resulting plasmid, pNJH193, was confirmed by primer extension sequencing using oligonucleotides NJH237 and NJH244 (see Table S1 in the supplemental material).
In an effort to enhance folding and stability, we added a His6 tag to the N terminus of GFPuv. To do this, oligonucleotides NJH246 and NJH247 (see Table S1 in the supplemental material) (100 pmol/μl each) were mixed 1:1 to obtain a 50-μl (total volume) mixture, heated at 94°C for 2 min, and then annealed at room temperature. The annealed mixture of NJH246 and NJH247 was diluted 1:400 and then mixed 1:1 with the NcoI-digested, SAP-treated pNJH193 vector backbone for ligation. This ligation mixture was then transformed into OneShot Top10 chemically competent E. coli cells. The sequence of the resulting plasmid, pNJH198, was confirmed by primer extension sequencing using oligonucleotide NJH241 (see Table S1 in the supplemental material).
In an attempt to increase the strength of the ribosome-binding site within pNJH198, we added the ribosome-binding site through the ATG start codon of pRKLac290 (M. R. K. Alley and J. Gober, unpublished) in frame with the His6-GFPuv protein sequence of pNJH198. To do this, oligonucleotides NJH248 and NJH249 (see Table S1 in the supplemental material) (100 pmol/μl each) were mixed 1:1 to obtain a 50-μl (total volume) mixture, heated at 94°C for 2 min, and then annealed at room temperature. The annealed mixture of NJH248 and NJH249 was diluted 1:400 and then mixed 1:1 with the NcoI-digested, SAP-treated pNJH198 vector backbone for ligation. The ligation mixture was then transformed into OneShot Top10 chemically competent E. coli cells. The sequence of the resulting plasmid, pNJH200, was confirmed by primer extension sequencing using oligonucleotide NJH241.
To place the urcA promoter GFPuv reporter into a higher-copy-number plasmid, we amplified an AscI/SpeI DNA fragment from plasmid pNJH200 with KOD Hot Start DNA polymerase using oligonucleotides NJH144 and NJH239 (see Table S1 in the supplemental material), as described above except that the extension time was 1.75 min. The PCR product was then digested with AscI and SpeI and ligated with the similarly digested pBVMCS-2 vector (M. Thanbichler, unpublished) backbone and then transformed into OneShot Top10 chemically competent E. coli cells. The sequence of the resulting plasmid, pNJH201, was confirmed by primer extension sequencing using oligonucleotides NJH240, NJH241, NJH242, and NJH243 (see Table S1 in the supplemental material). C. crescentus CB15N (strain LS101) was transformed with plasmid pNJH201 by electroporation as previously described to obtain the urcA promoter GFPuv reporter strain NJH371.
Pxyl gfpuv strain.
pBAD-GFPuv was digested with EcoRI, followed by a 1-h limited digestion with NdeI (gfpuv contains an internal NdeI site), to obtain the desired 900-bp band containing full-length gfpuv, which was isolated by gel electrophoresis. The NdeI/EcoRI gfpuv fragment was ligated with similarly digested pX31 (A. Iniesta, unpublished), a pBBR1MCS-based vector containing 500 bp of the xylose promoter inserted in front of the unique NdeI site, and then transformed into OneShot Top10 chemically competent E. coli cells. C. crescentus CB15N was transformed with the resulting plasmid, pNJH153, by electroporation as previously described to obtain the Pxyl gfpuv strain NJH250.
PurcA gfpuv reporter activity assays.
Cultures of strain NJH371 (PurcA gfpuv) were grown overnight at 28°C in M2G medium to an OD660 of about 0.4. These cultures were then stressed either by mock treatment or by addition of the indicated concentrations of uranyl nitrate, sodium nitrate, lead nitrate, cadmium sulfate, and/or potassium chromate (see Fig. 3C, 3D, and 3E), or they were stressed by addition (1:1, by volume) of M2G medium or uranium-contaminated (4.2 μM uranium) or uncontaminated (<0.1 μM uranium) variants of Oak Ridge Field Research Center groundwater sample FW231-17 (see Fig. 4) that was supplemented or not supplemented with 50 μM uranyl nitrate. The stressed cultures were then grown on an orbital shaker for 4 h (see Fig. 3D, 3E, and 4) or the amount of time indicated below (see Fig. 3C) before the GFPuv fluorescence intensity was measured with an ND-3300 fluorospectrometer, the cultures were excited with a UV light-emitting diode, and emission was monitored at 509 nm, as directed by the manufacturer. Replicate experiments were performed on separate days. Digital photographs of strain NJH371 (see Fig. 4B) were acquired with a tripod-mounted Canon Powershot A630 automatic camera, using either daylight or a hand-held UV lamp (366 nm) as the light source. Adobe Photoshop CS2 was utilized to isolate the green channel of the UV-illuminated RGB (red, green, blue) image, as well as to scale the green channel intensity, maintaining a gamma of one, to maximize the dynamic display range.
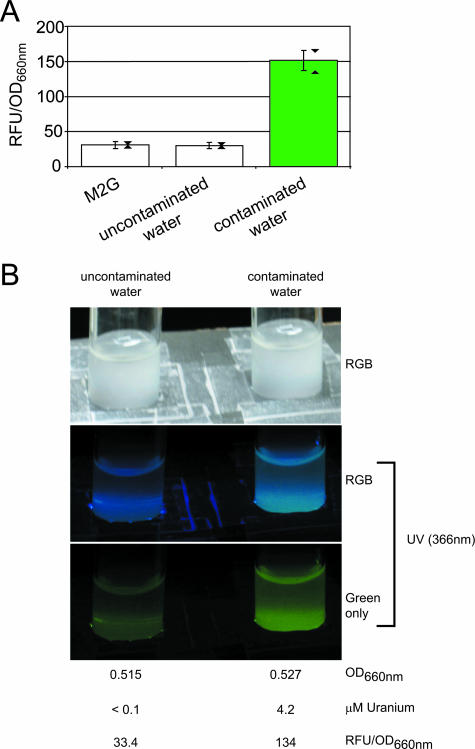
FIG. 4.
PurcA gfpuv reporter detection of uranium-contaminated groundwater. (A) Strain NJH371 GFPuv fluorescence was assayed after 4 h of exposure to uranium-contaminated (4.2 μM uranium) or uncontaminated (<0.1 μM uranium) Oak Ridge Field Research Center water samples. M2G minimal medium was used as a negative control. For an explanation of the error bars and triangles, see the legend to Fig. 3D. The data are aggregate results from four experiments. (B) Cultures of strain NJH371, illuminated with either daylight or a hand-held UV lamp, were photographed after 4 h of exposure to uranium-contaminated or uncontaminated water samples. The isolated green channel of the UV-illuminated RGB image (red, green, and blue channels), as well as culture OD660 and GFPuv fluorescence values for the cultures photographed, are shown. RFU, relative fluorescence units.
Pxyl urcA-mcherry strain.
The genomic region containing urcA was amplified from C. crescentus CB15N genomic DNA with KOD Hot Start DNA polymerase using oligonucleotides NJH204 and NJH205 (see Table S1 in the supplemental material), as described above except that the extension time was 2.5 min. The PCR product was then reamplified using oligonucleotides NJH200 and NJH213 (see Table S1 in the supplemental material), as described above except that the extension time was 30 s. The second PCR product containing urcA was then digested with NdeI and EcoRI.
An AgeI/BsrGI fragment containing mcherry was amplified from pRSET-B mcherry (32) using oligonucleotides NJH25 and NJH26 (see Table S1 in the supplemental material) as described above except that the extension time was 1 min, digested with AgeI and BsrGI, and ligated into similarly digested pMT383 (35), and the sequence of the resulting plasmid, pNJH15, was confirmed by primer extension sequencing using oligonucleotides NJH25 and NJH26. Plasmid pNJH15 was digested with EcoRI and BsrGI to obtain a 750-bp EcoRI/BsrGI fragment containing mcherry, which was isolated by gel electrophoresis.
Plasmid pMT397 (M. Thanbichler, unpublished) was sequentially digested with SmaI and then EcoRI to obtain an 800-bp EcoRI/SmaI fragment containing eyfp, which was isolated by gel electrophoresis and then ligated into the vector backbone of similarly digested pNJH153 to obtain pNJH156. Plasmid pNJH156 was digested with NdeI/BsrGI, the vector backbone was ligated with the NdeI/EcoRI fragment containing urcA (see above) and the EcoRI/BsrGI fragment containing mcherry (see above) (triple ligation), and the sequence of the resulting plasmid, pNJH169, was confirmed by primer extension sequencing using oligonucleotide NJH210. C. crescentus CB15N was transformed with plasmid pNJH169 by electroporation as previously described to obtain the Pxyl UrcA-mCherry strain NJH300. Strain NJH300 was grown overnight at 28°C in M2G medium to an OD660 of about 0.3, induced with 0.3% xylose for 3 h at 28°C, and immobilized onto 1.0% agar in M2G medium before images were obtained by phase-contrast and epifluorescence deconvolution microscopy with a Leica DM6000 microscope using ImagePro Plus v6.0 (with embedded SharpStack Plus) software.
RESULTS
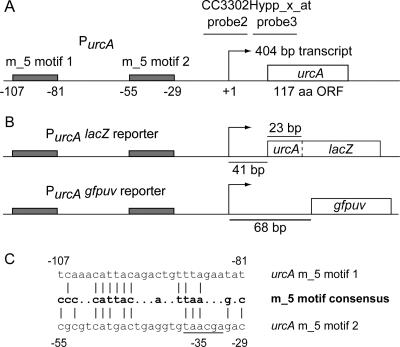
A custom-designed Affymetrix array of the C. crescentus genome, CauloHI1 (23), used to quantitate transcript levels upon exposure of Caulobacter to heavy metals, revealed that several genes were specifically upregulated upon exposure to uranyl nitrate (13). One of these genes was induced 27.5-fold under uranium stress but was not upregulated in response to other heavy metals in the test screen (13). We designated this gene urcA (for uranium response in caulobacter A) and selected the urcA promoter as a candidate to drive uranium reporter constructs. The results of the microarray experiments localized the urcA +1 transcriptional start site to a 10-bp window (13) (Fig. 1A). CC3302Hypp_x_at probe 3 is the most upstream probe in the Affymetrix array that matches the urcA transcript, placing its +1 site approximately 5 to 15 bp upstream from the end of the immediately adjacent probe 2 (chromosomal position 3552896). A uranium-inducible promoter sequence motif, present in the promoter regions of 11 Caulobacter genes, has been identified (23). The urcA promoter contains two matches to this uranium-specific m_5 motif, located 107 and 55 bp upstream of the putative +1 site (Fig. 1A and C). The urcA transcript overlaps the opposing strand of the CC_3302 gene in the original annotation of the Caulobacter genome (26), but the revised Glimmer (9) and GeneMark (2) annotations of the Caulobacter genome have identified urcA as the true open reading frame (spanning from chromosomal position 3552927 to position 3553280 on the positive strand), dispensing with the originally annotated CC_3302 gene. The urcA gene is not essential for viability and is not conserved among other α-proteobacteria.
FIG. 1.
urcA promoter region and PurcA reporter schematic diagrams. (A) CC3302Hypp_x_at probe 3 is the most upstream probe in the Affymetrix array that matches the 404-bp urcA transcript (13), placing the +1 transcriptional start site approximately 5 to 15 bp (10 bp as shown) from the end of the immediately adjacent probe 2. The locations of the tandem uranium-inducible m_5 motifs within the urcA promoter (23) are indicated by gray boxes with base pair numbering relative to the putative +1 site. (B) PurcA lacZ and PurcA gfpuv reporters utilize the urcA promoter, replacing urcA with a urcA/lacZ translational fusion and gfpuv, respectively. (C) Two sequence matches within the urcA promoter to the m_5 motif consensus (23). The vertical lines indicate identity. The underlined −35 region of the promoter overlaps the second urcA m_5 motif. aa, amino acids; ORF, open reading frame.
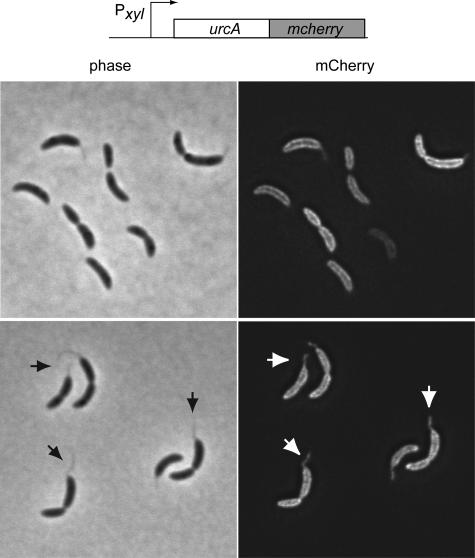
The urcA gene encodes a predicted 12.7-kDa protein. The signal sequence prediction tool SignalP (25) predicted with nearly 100% certainty that UrcA contains an N-terminal signal sequence whose most likely cleavage site is located between residues 30 and 31, and the prokaryotic subcellular protein localization tool SubLoc (14) predicted with 96% expected accuracy that UrcA is a periplasmic protein. To experimentally test the bioinformatic prediction of UrcA's periplasmic localization, we constructed a xylose-inducible UrcA-mCherry fusion (32). After xylose induction, the Pxyl urcA-mcherry strain was imaged by deconvolution microscopy (Fig. 2). The fluorescence images are consistent with the hypothesis that UrcA localizes to the cell periphery and to the cell stalk.
FIG. 2.
UrcA-mCherry localization. Strain NJH300 was induced with 0.3% xylose for 3 h, and images were obtained by deconvolution microscopy. Phase-contrast and epifluorescence images are shown. The arrows indicate representative cell stalks visible in both phase-contrast and epifluorescence images.
To determine if the urcA promoter could be a candidate uranium biosensor, we constructed a plasmid-borne LacZ fusion reporter containing 1 kb of the promoter region upstream of the urcA start ATG codon through the first 8 amino acids of UrcA fused to LacZ to create strain NJH199 (Fig. 1B). The resulting PurcA lacZ reporter strain was exposed to 0, 2.5, or 20 μM uranyl, and the resulting kinetics of β-galactosidase activity was assayed in liquid culture (Fig. 3A). The PurcA lacZ strain was able to detect the presence of 2.5 μM uranyl, and maximal β-galactosidase activity occurred by 2 h after exposure to either 2.5 or 20 μM uranyl nitrate. To test the sensitivity and specificity of the reporter, the PurcA lacZ strain was exposed to a panel of heavy metals for 2 h and then assayed for β-galactosidase activity (Fig. 3B). The PurcA lacZ reporter's detection limit for uranyl after 2 h of exposure was about 1.0 μM. The maximum increase in the signal of the PurcA lacZ reporter was 65-fold over the background with 20 μM uranyl. The PurcA lacZ reporter was not stimulated by the presence of lead (150 μM) or chromium (41.6 μM), but it showed cross sensitivity to cadmium at a concentration of 48 μM. In addition to the analysis of the panel of heavy metals, the PurcA lacZ reporter response to nitrate was assayed as a negative control to ensure that the nitrate component of uranyl nitrate salt does not contribute to PurcA lacZ reporter activity.
After our success with the PurcA lacZ reporter, we constructed a reporter strain (NJH371) in which plasmid-borne PurcA drives the expression of UV-excitable green fluorescent protein fluorescence. The PurcA gfpuv reporter construct is shown in Fig. 1B. The time course of GFPuv signal kinetics for PurcA gfpuv after induction with uranyl nitrate is shown in Fig. 3C. The fluorescence activity of the PurcA gfpuv reporter strain reached a maximum between 3 and 4 h after exposure to uranyl, but one-half the maximum activity was observed after about 2 h. It should be noted that as the PurcA gfpuv reporter strain reached a high cell density at 6.5 h (OD660, >0.9), entering stationary phase in the absence of uranium, the basal activity level of PurcA gfpuv began to increase. This result indicates that high-density cultures of the PurcA gfpuv reporter strain should not be used, because they could lead to false positives when samples are probed for the presence of uranium.
The PurcA gfpuv reporter strain was exposed to a panel of heavy metals for 4 h and then assayed for fluorescence activity (Fig. 3D). The PurcA gfpuv reporter exhibited specificity for uranium, and there was little cross specificity for nitrate (<400 μM), lead (<150 μM), cadmium (<48 μM), or chromium (<41.6 μM). The PurcA gfpuv reporter's detection limit for uranyl after 4 h of exposure was around 0.5 μM. The mean signal increase for the PurcA gfpuv reporter was 4.2-fold over the background with 100 μM uranyl. Despite sizeable standard deviations in reporter fluorescence activity, it should be pointed out that the minimum measured activities of the reporter for uranyl concentrations above 0.5 μM (n = 7) were all greater than the maximum activities measured for nitrate, lead, cadmium, or chromium (n = 3). Interestingly, we did not observe low-level stimulation of the GFPuv reporter by cadmium, in contrast to the LacZ reporter results (Fig. 3B). An inhibitory effect of 41.6 μM chromium on GFPuv activity was observed for the PurcA gfpuv reporter (Fig. 3E), but cadmium levels less than 48 μM did not appear to significantly affect PurcA gfpuv reporter activity in the presence of 10 μM uranyl.
Figure 4 demonstrates that the PurcA gfpuv strain distinguished uranium-contaminated groundwater samples (4.2 μM uranium) from uncontaminated groundwater samples (<0.1 μM uranium) collected at the Oak Ridge Field Research Center. Adding 50 μM uranyl nitrate to the uncontaminated water sample yielded comparable photoemission. Using a hand-held UV lamp as the light source, the naked eye alone was sufficient to distinguish PurcA gfpuv reporter strain cultures exposed to the contaminated water (4.2 μM uranium) from cultures exposed to the uncontaminated water (Fig. 4B), although filtering out the blue region of the spectrum (as shown by isolating the green channel of the RGB image) facilitated discrimination. This key result provides proof of principle that the PurcA gfpuv reporter strain may be used to detect the presence of uranium contamination in real-world water samples, that the reporter's output may be successfully monitored with the naked eye without resorting to a fluorimeter, and that the background chemical composition of the water samples tested does not appear to induce false-positive or -negative results.
DISCUSSION
In the work presented here, we constructed a whole-cell uranium biosensor that can report the presence of micromolar amounts of the uranyl cation in situ with nothing other than a hand-held UV lamp. To accomplish this, we utilized a reporter construct that placed GFPuv under the control of the promoter of the Caulobacter gene urcA, which is strongly upregulated upon exposure to uranium. A promoter motif was identified in 11 Caulobacter genes that were induced in response to uranyl nitrate. The urcA promoter contains a tandem repeat of this uranium response motif, m_5, which may explain why urcA is upregulated under uranium stress so much more strongly than any other Caulobacter gene. The m_5 uranium response motif is similar to the cell cycle regulation promoter motif cc_1, which appears to be stress induced (23). As the OD660 of a PurcA gfpuv reporter strain culture begins to exceed about 0.92 at 6.5 h, the fluorescence activity becomes quantitatively comparable to the reporter's activity output after 4 h of exposure to 2.5 μM uranyl (Fig. 3C). The increase in the basal activity level of the PurcA GFPuv reporter upon entry into high cell density is consistent with UrcA's role in the stress response.
The PurcA gfpuv reporter strain was able to discriminate groundwater samples contaminated with micromolar levels of uranium from uncontaminated groundwater samples acquired from the Oak Ridge Field Research Center, demonstrating that this reporter may be successfully applied to real-world samples. High levels of contaminating chromium (41.6 μM), but not cadmium, decreased the uranyl-induced GFPuv fluorescence activity of the PurcA gfpuv reporter. The maximum signal of the PurcA gfpuv reporter was observed after 3 to 4 h of exposure to uranium, but the assay time could confidently be reduced to 2 h at the expense of increasing the detection limit from about 0.5 to 1.0 μM uranyl. Other uranium detection methods have shorter measurement times (about 8 min for the catalytic DNA beacon biosensor [19]), but the PurcA gfpuv reporter strain does not require any preliminary sample processing. It is worth pointing out that the reported catalytic DNA beacon detection of uranium in soil samples required 20 h of carbonate and biocarbonate soil extraction before the actual assay was conducted (19). The 0.5 μM uranyl detection limit of the PurcA gfpuv reporter corresponds well with the Environmental Protection Agency maximum contaminant level guideline, which is 0.13 μM uranium (13). The PurcA gfpuv reporter strain differs from more sensitive uranium detection methodologies in that it provides a signal only for toxic levels of bioavailable uranium contamination. Presumably, the detection limit of the urcA promoter has been tuned to coincide with the uranyl concentration above which uranium stress is toxic to Caulobacter. The PurcA gfpuv reporter strain method additionally differs from other uranium detection methods in that it requires minimal equipment and sample processing and operates at ambient temperatures. Future development of the PurcA gfpuv reporter will focus on field-ready application and spraying the strain directly on soil, groundwater, or industrial surfaces. Freeze-drying whole-cell bacterial cadmium biosensors has been shown to only moderately affect performance (34), and reconstituting the PurcA gfpuv reporter strain from a lyophilized powder could greatly enhance its on-demand usability in the field. In conjunction with bioremediation efforts, the PurcA gfpuv reporter could complement analytical uranium detection methodologies by rapid screening of many locations in parallel for toxic levels of bioavailable uranium contamination.
Supplementary Material
Acknowledgments
We thank Kevin Phillips for providing pBAD-GFP and Antonio Iniesta for providing pX31. We thank David Watson and T. C. Hazen for obtaining the groundwater samples from Oakridge Field Research Center.
This work was supported by Department of Energy Genomes to Life grant DE-FG02-05ER64136 (to L.S.) and by Damon Runyon Cancer Research Foundation fellowship DRG-1880-05 (to N.J.H.). G.L.A. and P.H. were funded by the Department of Energy Genomes to Life program, and the work was performed under the auspices of the U.S. Department of Energy at the University of California Lawrence Berkeley National Laboratory under contract DE-AC02-05CH11231.
Footnotes
Published ahead of print on 28 September 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alkorta, I., L. Epelde, I. Mijangos, I. Amezaga, and C. Garbisu. 2006. Bioluminescent bacterial biosensors for the assessment of metal toxicity and bioavailability in soils. Rev. Environ. Health 21:139-152. [DOI] [PubMed] [Google Scholar]
- 2.Besemer, J., and M. Borodovsky. 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33:W451—W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blake, R. C., A. R. Pavlov, M. Khosraviani, H. E. Ensley, G. E. Kiefer, H. Yu, X. Li, and D. A. Blake. 2004. Novel monoclonal antibodies with specificity for chelated uranium(VI): isolation and binding properties. Bioconjug. Chem. 15:1125-1136. [DOI] [PubMed] [Google Scholar]
- 4.Boomer, D. W., and M. J. Powell. 1987. Determination of uranium in environmental samples using inductively coupled plasma mass spectrometry. Anal. Chem. 59:2810-2813. [DOI] [PubMed] [Google Scholar]
- 5.Brina, R., and A. G. Miller. 1992. Direct detection of trace levels of uranium by laser-induced kinetic phosphorimetry. Anal. Chem. 64:1413-1418. [Google Scholar]
- 6.Brodie, E. L., T. Z. Desantis, D. C. Joyner, S. M. Baek, J. T. Larsen, G. L. Andersen, T. C. Hazen, P. M. Richardson, D. J. Herman, T. K. Tokunaga, J. M. Wan, and M. K. Firestone. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72:6288-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbisier, P., D. van der Lelie, B. Borremans, A. Provoost, V. de Lorenzo, N. L. Brown, J. R. Lloyd, J. L. Hobman, E. Csoregi, G. Johansson, and B. Mattiasson. 1999. Whole cell- and protein-based biosensors for the detection of bioavailable heavy metals in environmental samples. Anal. Chim. Acta 387:235-244. [Google Scholar]
- 8.Crameri, A., E. A. Whitehorn, E. Tate, and W. P. Stemmer. 1996. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14:315-319. [DOI] [PubMed] [Google Scholar]
- 9.Delcher, A. L., K. A. Bratke, E. C. Powers, and S. L. Salzberg. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 11.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 13.Hu, P., E. L. Brodie, Y. Suzuki, H. H. McAdams, and G. L. Andersen. 2005. Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J. Bacteriol. 187:8437-8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hua, S., and Z. Sun. 2001. Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17:721-728. [DOI] [PubMed] [Google Scholar]
- 15.Huang, J. W., M. J. Blaylock, Y. Kapulnik, and B. D. Ensley. 1998. Phytoremediation of uranium-contaminated soils: role of organic acids in triggering uranium hyperaccumulation in plants. Environ. Sci. Technol. 32:2004-2008. [Google Scholar]
- 16.Huff, E. A., and D. L. Bowers. 1990. Icp Aes actinide detection limits. Appl. Spectrosc. 44:728-729. [Google Scholar]
- 17.Istok, J. D., J. M. Senko, L. R. Krumholz, D. Watson, M. A. Bogle, A. Peacock, Y. J. Chang, and D. C. White. 2004. In situ bioreduction of technetium and uranium in a nitrate-contaminated aquifer. Environ. Sci. Technol. 38:468-475. [DOI] [PubMed] [Google Scholar]
- 18.Larrainzar, E., F. O'Gara, and J. P. Morrissey. 2005. Applications of autofluorescent proteins for in situ studies in microbial ecology. Annu. Rev. Microbiol. 59:257-277. [DOI] [PubMed] [Google Scholar]
- 19.Liu, J., A. K. Brown, X. Meng, D. M. Cropek, J. D. Istok, D. B. Watson, and Y. Lu. 2007. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc. Natl. Acad. Sci. USA 104:2056-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacRae, J. D., and J. Smit. 1991. Characterization of caulobacters isolated from wastewater treatment systems. Appl. Environ. Microbiol. 57:751-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mannisto, M. K., M. A. Tiirola, M. S. Salkinoja-Salonen, M. S. Kulomaa, and J. A. Puhakka. 1999. Diversity of chlorophenol-degrading bacteria isolated from contaminated boreal groundwater. Arch. Microbiol. 171:189-197. [DOI] [PubMed] [Google Scholar]
- 22.McCullough, J., T. C. Hazen, S. M. Benson, F. B. Metting, and A. C. Palmisano. 2004. Bioremediation of metals and radionuclides: what it is and how it works, 2nd ed. Lawrence Berkeley National Laboratory, Berkeley, CA.
- 23.McGrath, P. T., H. Lee, L. Zhang, A. A. Iniesta, A. K. Hottes, M. H. Tan, N. J. Hillson, P. Hu, L. Shapiro, and H. H. McAdams. 2007. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat. Biotechnol. 25:584-592. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 25.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 26.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.North, N. N., S. L. Dollhopf, L. Petrie, J. D. Istok, D. L. Balkwill, and J. E. Kostka. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peitzsch, N., G. Eberz, and D. H. Nies. 1998. Alcaligenes eutrophus as a bacterial chromate sensor. Appl. Environ. Microbiol. 64:453-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poindexter, J. S. 1981. The caulobacters: ubiquitous unusual bacteria. Microbiol. Rev. 45:123-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberto, F. F., J. M. Barnes, and D. F. Bruhn. 2002. Evaluation of a GFP reporter gene construct for environmental arsenic detection. Talanta 58:181-188. [DOI] [PubMed] [Google Scholar]
- 31.Santos, P. M., I. Di Bartolo, J. M. Blatny, E. Zennaro, and S. Valla. 2001. New broad-host-range promoter probe vectors based on the plasmid RK2 replicon. FEMS Microbiol. Lett. 195:91-96. [DOI] [PubMed] [Google Scholar]
- 32.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567-1572. [DOI] [PubMed] [Google Scholar]
- 33.Shetty, R. S., S. K. Deo, Y. Liu, and S. Daunert. 2004. Fluorescence-based sensing system for copper using genetically engineered living yeast cells. Biotechnol. Bioeng. 88:664-670. [DOI] [PubMed] [Google Scholar]
- 34.Tauriainen, S., M. Karp, W. Chang, and M. Virta. 1998. Luminescent bacterial sensor for cadmium and lead. Biosens. Bioelectron. 13:931-938. [DOI] [PubMed] [Google Scholar]
- 35.Thanbichler, M., and L. Shapiro. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126:147-162. [DOI] [PubMed] [Google Scholar]
- 36.Wan, J., T. K. Tokunaga, E. Brodie, Z. Wang, Z. Zheng, D. Herman, T. C. Hazen, M. K. Firestone, and S. R. Sutton. 2005. Reoxidation of bioreduced uranium under reducing conditions. Environ. Sci. Technol. 39:6162-6169. [DOI] [PubMed] [Google Scholar]
- 37.Wu, W. M., J. Carley, T. Gentry, M. A. Ginder-Vogel, M. Fienen, T. Mehlhorn, H. Yan, S. Caroll, M. N. Pace, J. Nyman, J. Luo, M. E. Gentile, M. W. Fields, R. F. Hickey, B. Gu, D. Watson, O. A. Cirpka, J. Zhou, S. Fendorf, P. K. Kitanidis, P. M. Jardine, and C. S. Criddle. 2006. Pilot-scale in situ bioremedation of uranium in a highly contaminated aquifer. 2. Reduction of U(VI) and geochemical control of U(VI) bioavailability. Environ. Sci. Technol. 40:3986-3995. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, P., and B. Gu. 2005. Extraction of oxidized and reduced forms of uranium from contaminated soils: effects of carbonate concentration and pH. Environ. Sci. Technol 39:4435-4440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.