Abstract
Fluorescence in situ hybridization and real-time PCR analysis targeting the 16S rRNA gene of Akkermansia muciniphila were performed to determine its presence in the human intestinal tract. These techniques revealed that an A. muciniphila-like bacterium is a common member of the human intestinal tract and that its colonization starts in early life and develops within a year to a level close to that observed in adults (108 cells/g) but decreases (P < 0.05) in the elderly.
The human gastrointestinal (GI) tract is colonized by a complex and diverse microbiota that shows different compositions and activities depending on environmental and genetic factors (8, 14, 16, 18, 19). The intestinal microbiota starts to establish at birth and continues to develop and change during life and also provides a barrier against colonization of pathogens and harmful substances, protecting intestinal integrity, and stimulates the development of the immune system (10, 14).
A mucus layer covering the GI tract has been reported to serve as a source of nutrients for bacterial growth (6). Thus, its presence influences intestinal colonization by attracting bacteria that have the ability to survive and multiply within the mucus layer. The mucus layer also contributes to host defense by preventing bacterial adhesion or invasion and toxin binding to the mucosal surface (11). Nevertheless, the association of the microbiota with the mucus is not well understood and requires further investigation.
A novel mucin-degrading bacterium designated Akkermansia muciniphila has been isolated and characterized from a fecal sample from a healthy adult (6). Subsequent studies based on 16S rRNA gene cloning and sequencing have demonstrated that A. muciniphila is present in distinct parts of the human mucosa as well as in fecal samples (7, 9, 17). The relation between mucin and bacteria varies depending on the microbiota, and several studies have reported a potential involvement of mucin-degrading bacteria in pathogenesis of intestinal diseases (3). Mucosal surfaces, microbiota, and mucus secretion may be altered due to environmental factors, including age. As a consequence, the composition and thickness of the mucus layer may be modified (4). Such changes may alter the contact between intestinal microbiota and mucosal dendritic cells. As the mucus quality and quantity alter during ageing, we hypothesized that the levels of intestinal A. muciniphila could be different during human life, and detection of the presence of this bacterium requires detailed investigation.
Fecal samples from healthy infant subjects 1 month old (n = 50), 6 months old (n = 50), and 12 months old (n = 50); healthy adults from 25 to 35 years old (n = 54); and elderly subjects from 80 to 82 years old (n = 45) were collected and analyzed. Two grams of fresh fecal samples was used and suspended in 20 ml of phosphate-buffered saline. The suspension was vortexed thoroughly with 3-μm-diameter glass beads and centrifuged at 800 × g for 1 min to remove debris. Different aliquots from the pure culture and fecal samples were used to extract DNA and to fix cells for fluorescence in situ hybridization (FISH) analysis. One volume of the supernatant was transferred into 3 volumes of fresh 4% paraformaldehyde and fixed at 4°C overnight. The bacteria were stored in 50% ethanol-phosphate-buffered saline at −20°C until analysis. One volume was used for DNA extraction by using a QIAamp DNA stool mini kit (Qiagen, Hilden, Germany), following the manufacturer's instructions.
Two specific primers were designed from the variable regions of the 16S rRNA gene sequence of A. muciniphila. The GenBank program from NCBI (BLAST) was used to verify that both primers were specific to the target organism only. The primers selected for detection of A. muciniphila were named on the basis of nomenclature from Alm et al. (1). These primers were S-St-Muc-1129-a-a-20 (AM1), with the sequence 5′CAG CAC GTG AAG GTG GGG AC, and S-St-Muc-1437-a-A-20 (AM2), with the sequence 5′CCT TGC GGT TGG CTT CAG AT (Table 1). All primers were purchased from MWG (Ebersgerg, Germany). To check the specificity of the amplification, standard PCR amplification was performed, using as the template DNA from 20 intestinal isolates (Table 2); 96 cloned 16S rRNA genes from uncultured bacteria belonging to the most dominant groups found in the GI tract, including Bacteroidetes, Clostridium cluster XIVa, and Clostridium cluster IV; and other clones belonging to disparate clusters, including Bifidobacterium, Lactobacillus, and Atopobium (5, 15, 18). The PCR primers were specific for A. muciniphila at 60°C, with amplification of a product of the expected size (327 bp) (data not shown). All other nontargeted strains tested and the cloned 16S rRNA genes from uncultured GI tract bacteria showed no amplification during PCR (data not shown).
TABLE 1.
Primer sequences used in this study
TABLE 2.
Species and origins of reference strains used for validation of the Akkermansia muciniphila primers (AM1 and AM2) and probe (MUC-1437)
| Species | Originb |
|---|---|
| Akkermansia muciniphila | ATCC BAA-835 |
| Atopobium minutuma | DSM 20586 |
| Bacteroides distasonis | DSM 20701 |
| Bacteroides fragilisa | DSM 2151 |
| Bacteroides pyogenes | CCUG 15419 |
| Bacteroides tectus | CCUG 25929 |
| Bacteroides thetaiotaomicrona | RRI |
| Bacteroides uniformisa | DSM 6597 |
| Bifidobacterium lactisa | DSM 10140 |
| Bifidobacterium bifiduma | DSM 20082 |
| Bifidobacterium adolescentisa | DSM 20083 |
| Bifidobacterium brevea | DSM 20091 |
| Bifidobacterium longuma | DSM 20090 |
| Clostridium aminophilum | ATCC 49906 |
| Clostridium sticklandii | ATCC 12662 |
| Clostridium perfringensa | DSM 756 |
| Clostridium sporogenes | DSM 1664 |
| Colinsella aerofaciensa | DSM 3979 |
| Coprococcus sp.a | RRI |
| Enterococcus faecalis | RRI |
| Escherichia colia | NCTC 12900 |
| Eubacterium biforme | DSM 3989 |
| Eubacterium cylindroides | RRI |
| Eubacterium rectale | RRI |
| Eubacterium ruminantium | RRI |
| Faecalibacterium prausnitzii | RRI |
| Lachnospira multiparus | RRI |
| Lactobacillus acidophilusa | ATCC 4356 |
| Lactobacillus amylovorusa | DSM 20531 |
| Lactobacillus rhamnosusa | ATCC 53103 |
| Lactobacillus plantaruma | NCIMB 8826 |
| Lactobacillus reuteria | DSM 20016 |
| Megasphaera elsdenii | NCIMB 8927 |
| Mitsuokella multiacidus | RRI |
| Peptostreptococcus anaerobius | ATCC 27337 |
| Pepstostreptococcus micros | DSM 20468 |
| Prevotella albensis | DSM 11370 |
| Prevotella brevis | ATCC 19188 |
| Prevotella bryantiia | DSM 11371 |
| Prevotella ruminocolaa | ATCC 19189 |
| Roseburia intestinalis | RRI |
| Ruminococcus albus | RRI |
| Ruminococcus bromii | RRI |
| Ruminococcus flavefasciens | RRI |
| Ruminococcus hansenii | DSM 20583 |
| Ruminococcus productus | DSM 2950 |
| Streptococcus bovis | RRI |
| Veilonella parvula | RRI |
| Victivallis vadensisa | DSM 14823 |
The strains of these species were tested using the AM1 and AM2 primers.
RRI, Rowett Research Institute (Aberdeen, United Kingdom).
Quantitative PCR amplification and detection were performed with optical-grade 96-well plates with an ABI PRISM 7300-PCR sequence detection system (Applied Biosystems, United Kingdom). Each reaction mixture of 25 μl was composed of SYBR green PCR master mix (Applied Biosystems, United Kingdom), 1 μl of each of the specific primers at a final concentration of 0.25 μM, and 1 μl of template DNA. The PCR conditions were as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 15 s, 60°C for 40 s, and 72°C for 30 s, and a final extension at 72°C for 5 min. The fluorescent products were detected at the last step of each cycle. A melting curve analysis was made after amplification to distinguish the targeted PCR product from the nontargeted PCR product. Standard curves were created using serial 10-fold dilutions of A. muciniphila pure culture DNA corresponding to 102 to 1010 cells as determined by flow cytometry counts. The bacterial concentration of each sample was calculated by comparing the threshold cycle (CT) values obtained from the standard curve. All samples were analyzed in duplicate.
In addition, FISH coupled with flow cytometry (FCM-FISH) was carried out by following the procedure described in reference 5, using a specific oligonucleotide probe, MUC-1437 (Table 3), targeting one region of the 16S rRNA gene of A. muciniphila, to compare and validate the method. Total bacterial cell numbers were enumerated, using fluorescein-labeled probe EUB-338 (2, 13) as a positive control and probe Non-EUB (2, 13) as a negative control, to monitor the nonspecific hybridizations (Table 3). Absolute bacterial cell counts were determined by addition of Flow-Count fluorospheres, following the supplier's instructions (Beckman Coulter). Flow-cytometric analyses were performed using a BD LSR II flow cytometer (Becton Dickinson) equipped with a 488-nm laser at 15 mW. Data were stored as list mode files and analyzed offline using BD FACSDiva version 4.1.1 software (Becton Dickinson).
TABLE 3.
Probe sequences and hybridization conditions used in this study
A linear relationship was observed between the cell counts and CT values (r2 = 0.9906) when the number of cells per reaction mixture was between 102 and 109. Reliable quantification of cells was not possible at concentrations below 10 cells per reaction mixture. This corresponds to a detection limit of 5 × 102 cells/g feces. Melting curve analysis of the amplicons obtained by real-time PCR generated a specific peak at 85°C in the 16S rRNA gene of A. muciniphila. The inter-PCR reproducibility of the quantification was found to be very high based on the CT values obtained for two DNA extracts (a pure culture and a fecal specimen harboring approximately 108 to 109 A. muciniphila-like bacteria/g) in five replicate runs. The average CT values obtained for those samples were 11.0 (range, 10.9 to 11.1) and 11.2 (range, 11.0 to 11.4). Similar results were observed with other samples and in other dilutions (average and standard deviation, 11.1 ± 0.5). By analysis of duplicate DNA extracts from fecal samples, the interextract variability was found to be less than 10% (data not shown).
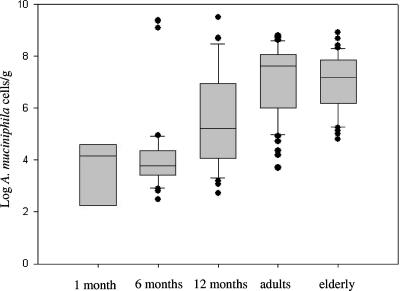
Application of the real-time-PCR approach to fecal samples of infants (up to 1 year old), young adults (25 to 35 years old), and elderly subjects (80 to 82 years old) showed that A. muciniphila-like bacteria appeared in early life and increased significantly in number from early life to adult age (Fig. 1 and Table 4). The presence of A. muciniphila-like bacteria was detected in 8 of the 50 fecal samples of 1-month-old infants (16% of the samples) at levels from 2.05 to 4.36 log cells/g. Bacteria related to A. muciniphila were detected in 36 out of 50 children aged 6 months (72% of the samples) and 45 of 50 children at 12 months of age (90% of the samples). The values ranged from 2.50 to 7.30 log cells/g and 2.80 to 9.50 log cells/g in samples from 6- and 12-month-old infants, respectively. The number of bacteria related to A. muciniphila increased significantly (P < 0.05) from early life to adult age and reached levels ranging from 5.00 to 8.80 log cells/g in all samples from adults. These data indicate that A. muciniphila-like bacteria are colonizing the intestinal tract in early life and develop within a year to a level close to that observed in adults. Remarkably, the concentrations of A. muciniphila cells in fecal samples from elderly subjects were significantly decreased (>1 logarithmic unit, to 6.00 log cells/g; P < 0.05) compared to those in samples from adults.
FIG. 1.
A. muciniphila levels in human fecal samples as determined by real-time PCR over a life span. Data represent the positive samples. The line in the box represents the median (50th percentile), with the lower line the 25% border (25th percentile) and the upper line the 75% border (75th percentile). The end of the upper vertical line represents the maximum data value, outliers not considered. The end of the lower vertical line represents the lowest value, outliers not considered. The separate dots indicate outliers.
TABLE 4.
Numbers of A. muciphila-like bacteria per gram of feces in fecal samples analyzed by real-time PCR and FCM-FISH
| Sample group | No. of infected samples/total no. of samples | Log no. of cells/g fecesa as determined by:
|
|
|---|---|---|---|
| Quantitative PCR | FCM-FISH | ||
| Infant (1 mo old) | 8/50 | 3.90 (2.25-4.32)c | 4.56 (5.06-4.03)c |
| Infant (6 mo old) | 36/50 | 3.90 (3.52-4.36) | 6.50 (5.90-7.40)b |
| Infant (12 mo old) | 45/50 | 5.20 (4.08-7.00) | 7.33 (7.05-7.94)b |
| Adult | 54/54 | 7.61 (6.02-8.05) | 8.10 (7.90-8.32)b |
| Elderly | 43/45 | 6.00 (4.22-7.66) | 7.28 (6.63-7.96)b |
Total bacteria in samples ranged from 9.0 to 10 log cells/g of fecal samples. Due to nonnormal distribution, microbial data are expressed as medians, with interquartile ranges in parentheses. The Mann-Whitney U test was applied in comparisons among groups.
Significant differences (P < 0.05) were determined by a Mann-Whitney test comparing qPCR and FCM-FISH data.
All data from different age groups were significantly different (P < 0.05), with only one exception: data from 1-month-old and 6-month-old infants.
In addition, all fecal samples tested were analyzed also by FCM-FISH for comparison to the results obtained by quantitative real-time PCR. A good correlation between the results obtained with the two methods was found (Table 4). Nevertheless, in fecal samples harboring small numbers of A. muciniphila-like bacteria, the FCM-FISH approach estimated their numbers to be about 1 logarithmic unit higher than the real-time PCR method, probably due to background and/or unspecific signals (Table 4).
The results of the quantitative real-time PCR and FISH-FCM results showed a good correlation, but differences were found between these methods in samples with low numbers (<104 cells/g) of bacteria related to A. muciniphila. Similarly, FCM-FISH and real-time PCR results showed different levels of A. muciniphila-like bacteria when suspensions of pure cultures with concentrations ranging from 4 to 2 log cells/g were tested. FCM-FISH tended to detect more A. muciniphila-like bacteria than real-time PCR. In some cases, FCM-FISH could lead to an overestimation of the number of A. muciniphila-like bacteria in fecal samples with low content, due to the formation of unspecific hybridizations in complex matrices of fecal samples or to undescribed Akkermansia species that do not provide an amplification product with the developed primers targeted to A. muciniphila. In addition, cell hybridizations with fluorescent probes provide background levels that a flow cytometer may detect as cells, increasing the percentage of cells detected compared to that of cells obtained by PCR. Thus, the advantage of this specific PCR technique is that the method is approximately 10 to 100 times more sensitive and specific than the culture and FISH methods.
The presence of A. muciniphila-like bacteria was detected in fecal samples from infants 1 month old, but the number increased rapidly with the age (6- and 12-month-old infants and adults). The numbers of bacteria related to A. muciniphila almost doubled in the age period ranging from 6 to 12 months. These data are in agreement with earlier observations (12) which reported the establishment of mucin-degrading microbiota in children from birth to the age of 2 years, based on agar gel electrophoresis of their fecal samples. The establishment of mucin-degrading bacteria was reported during the first months of life and is completed when the children are around 2 years old (12). These data indicate that A. muciniphila is colonizing the intestinal tract in early life and develops within a year to a level close to that observed in adults (Fig. 1). Akkermansia was detected in all samples from adults at levels ranging from 5.00 to 8.80 log cells/g, and this is in agreement with early data (5). Our results suggest that the possible rise in concentrations of bacteria is related to A. muciniphila along with normal mucosa development. This rise may be associated with the integrity of the developing healthy intestinal tract with normal mucus production. However, the numbers of A. muciniphila-like bacteria in elderly subjects (80 to 82 years old) were significantly decreased (1 logarithmic unit; P < 0.05) compared to those in young and middle-aged adults. Although it cannot be ruled out that these bacteria do not show a dependency on mucin as a carbon and nitrogen source similar to that of A. muciniphila, it is likely that this reflects the coevolution of specific mucin-degrading bacteria and mucus production.
Taken together, the techniques described here are accurate, rapid, and easy methods for quantification of A. muciniphila in human feces. These methods will facilitate rapid and reliable counting of large numbers of samples, contributing to the efficient use of intestinal bacterial assays in research as well as in the assessment of dietary management of diseases. We also demonstrate for the first time the validation and development of a quantitative real-time PCR for detection and quantification of bacteria related to the mucin-degrading bacterium A. muciniphila. Our results demonstrate that A. muciniphila is present and colonizes the intestinal tract in early life and develops within a year to a level close to that observed in healthy adults. Further studies are needed to clarify the role of A. muciniphila in microbiota development and immune development in early childhood and old age.
Acknowledgments
M. C. Collado is the recipient of an Excellence Postdoctoral grant from Conselleria Empresa, Universidad y Ciencia de la Generalitat Valenciana, Spain (BPOSTDOC 06/016). M. Derrien was financed by the European Community, specific RTD program Quality of Life and Management of Living Resources, research project EU & Microfunction (QKL1-2001-00135).
Footnotes
Published ahead of print on 12 October 2007.
REFERENCES
- 1.Alm, E., D. Oerther, N. Larsen, D. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campieri, M., and P. Gionchetti. 2001. Bacteria as the cause of ulcerative colitis. Gut 48:132-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corfield, A. P., D. Carroll, N. Myerscough, and C. S. Probert. 2001. Mucins in the gastrointestinal tract in health and disease. Front. Biosci. 6:D1321-D1327. [DOI] [PubMed] [Google Scholar]
- 5.Derrien, M. 2007. Mucin utilisation and host interactions of the novel intestinal microbe Akkermansia muciniphila. Ph.D. thesis (ISBN 978-90-8504-644-8). Wageningen University, Wageningen, The Netherlands.
- 6.Derrien, M., E. E. Vaughan, C. M. Plugge, and W. M. de Vos. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54:1469-1476. [DOI] [PubMed] [Google Scholar]
- 7.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gueimonde, M., S. Tolkko, T. Korpimaki, and S. Salminen. 2004. New real-time quantitative PCR procedure for quantification of bifidobacteria in human fecal samples. Appl. Environ. Microbiol. 70:4165-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi, H., M. Sakamoto, and Y. Benno. 2002. Fecal microbial diversity in a strict vegetarian as determined by molecular analysis and cultivation. Microbiol. Immunol. 46:819-831. [DOI] [PubMed] [Google Scholar]
- 10.Hooper, L. V., and J. I. Gordon. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115-1118. [DOI] [PubMed] [Google Scholar]
- 11.Liévin-Le Moal, V., and A. L. Servin. 2006. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin. Microbiol. Rev. 19:315-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Midtvedt, A., B. Carlstedt-Duke, and T. Midtvedt. 1994. Establishment of a mucin-degrading intestinal microflora during the first two years of human life. J. Pediatr. Gastroenterol. Nutr. 18:321-326. [DOI] [PubMed] [Google Scholar]
- 13.Rigottier-Gois, L., A.-G. Le Bourhis, G. Gramet, V. Rochet, and J. Doré. 2003. Fluorescent hybridization combined with flow cytometry and hybridization of total RNA to analyse the composition of microbial communities in human faeces using 16S rRNA probes. FEMS Microbiol. Ecol. 43:237-245. [DOI] [PubMed] [Google Scholar]
- 14.Salminen, S., and M. Gueimonde. 2005. Gut microbiota in infants between 6 and 24 months of age. Nestle Nutr. Workshop Ser. Pediatr. Program 56:43-51. [DOI] [PubMed] [Google Scholar]
- 15.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaughan, E. E., H. G. Heilig, K. Ben-Amor, and W. M. de Vos. 2005. Diversity, vitality and activities of intestinal lactic acid bacteria and bifidobacteria assessed by molecular approaches. FEMS Microbiol. Rev. 29:477-490. [DOI] [PubMed] [Google Scholar]
- 17.Wang, M., S. Ahrné, B. Jeppsson, and G. Molin. 2005. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol. Ecol. 54:219-231. [DOI] [PubMed] [Google Scholar]
- 18.Zoetendal, E. G., A. D. Akkermans, and W. M. de Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zoetendal, E. G., E. E. Vaughan, and W. M. de Vos. 2006. A microbial world within us. Mol. Microbiol. 59:1639-1650. [DOI] [PubMed] [Google Scholar]