Abstract
Multilocus sequence typing (MLST) is a sequence-based method used to characterize bacterial genomes. This method was used to examine the genetic structure of Medicago-nodulating rhizobia at the Amra site, which is located in an arid region of Tunisia. Here the annual medics Medicago laciniata and M. truncatula are part of the natural flora. The goal of this study was to identify whether distinct chromosomal groups of rhizobia nodulate M. laciniata because of its restricted requirement for specific rhizobia. The MLST analysis involved determination of sequence variation in 10 chromosomal loci of 74 isolates each of M. laciniata and M. truncatula. M. truncatula was used as a control trap host, because unlike M. laciniata, it has relatively unrestricted rhizobial requirements. Allelic diversity among the plasmid nodC alleles in the isolates was also determined. The 148 isolates were placed into 26 chromosomal sequence types (STs), only 3 of which had been identified previously. The rhizobia of M. laciniata were shown to be part of the general Medicago-nodulating population in the soil because 99.95% of the isolates had chromosomal genotypes similar to those recovered from M. truncatula. However, the isolates recovered from M. laciniata were less diverse than those recovered from M. truncatula, and they also harbored an unusual nodC allele. This could perhaps be best explained by horizontal transfer of the different nodC alleles among members of the Medicago-nodulating rhizobial population at the field site. Evidence indicating a history of lateral transfer of rhizobial symbiotic genes across distinct chromosomal backgrounds is provided.
Traditionally, legumes have been important in a practice referred to as low-input sustainable agriculture. The recognition of their agricultural importance led to the demonstration that legumes bearing nodules inhabited by bacteria assimilated dinitrogen in a process termed biological nitrogen fixation. The bacteria, commonly referred to as rhizobia, harbor the genetic information necessary for establishing the nitrogen fixation process when they reside symbiotically inside the nodules. Plants of the genus Medicago are legumes that benefit from such a symbiosis with rhizobia that are currently divided into two species, Sinorhizobium meliloti (11) and S. medicae (25).
The plant genus Medicago L., as currently defined, consists of about 85 species (28), including Medicago sativa (alfalfa), the world's most important cultivated forage crop. Heyn (16) reported that this genus is native to Western Asia and the Mediterranean countries, although many annual species have invaded wide areas in both the Old and New Worlds.
Among the 85 species, M. laciniata has symbiotic properties that are unusual. While most Medicago species nodulated and formed nitrogen-fixing symbioses when they were grown in Australian soils with naturalized rhizobial populations, Ballard and Charman (3) observed that there was a lower incidence of nodulation with M. laciniata. They attributed this observation to the specific rhizobial requirement of M. laciniata that had been reported previously by Brockwell and Hely (7). For nodulation of M. laciniata, Brockwell and Hely (7) suggested that its rhizobia have distinct nodule formation behavior that distinguishes them from other Medicago-nodulating rhizobia. Barran et al. (4) provided evidence from a complementation and site-directed mutagenesis analysis that the specific nodC allele carried on the pSymA plasmid of strain USDA 1170 was required for successful nodulation of M. laciniata. Subsequently, Villagas et al. (34) proposed subdivision of S. meliloti into two biovars, S. meliloti bv. meliloti and S. meliloti bv. medicaginis, to distinguish the rhizobia of M. laciniata from other Medicago-nodulating rhizobia. However, it is unclear whether these two biovars have distinctive chromosomal genotypes or whether they are members of the same Medicago-nodulating rhizobial population.
Of the techniques used to estimate the diversity among Medicago-nodulating rhizobia, DNA fingerprinting is the most popular approach (1, 5, 6, 10, 12, 23, 24, 26, 36). However, DNA fingerprinting is not sufficiently specific to estimate the genetic diversity within and among the three large replicons normally found in Medicago-nodulating rhizobia (17). Therefore, an alternative methodology is necessary to investigate the potential correspondence between chromosomal and plasmid genotypes in different rhizobial genomes.
Multilocus sequence typing (MLST) was originally developed by Maiden et al. (22) for identification of the virulent lineages of the bacterial pathogen Neisseria meningitidis. In their study Maiden et al. (22) determined the allelic variation of seven loci by direct nucleotide sequence comparison. A combination of the observed alleles for each locus was then used to derive an allelic profile or sequence type (ST) for each of their strains. This made it possible to compare large numbers of multilocus bacterial genotypes. MLST results have been reported for numerous pathogenic bacterial species (13). Because MLST has been used to determine patterns of chromosomal evolutionary descent among Medicago-nodulating rhizobia (33), this strategy also can be used to determine any potential correspondence between chromosomal background and the nodC allele carried on pSymA.
Therefore, the primary goal of this study was to identify distinct chromosomal groups of rhizobia that were isolated from M. truncatula and M. laciniata growing in a single Tunisian soil using the strategy developed by van Berkum et al. (33). The MLST results were subsequently used to link the chromosomal groups with the distribution of the nodC alleles present in the genomes of the rhizobial isolates.
MATERIALS AND METHODS
Bacterial isolation, counting, and genomic DNA extraction.
Soil samples, taken at a depth of 0 to 20 cm, were collected in the summer of 2005 at the Amra site, which is located in the central region of Tunisia and is within the arid climatic zone. The Amra site does not have a history of agriculture. Both M. laciniata and M. truncatula are native to this location and served as sources of seeds. The seeds were sown and plants were cultivated in the Amra soil in 500-ml sterilized plastic pots. Plants were grown in an environmentally controlled greenhouse with a photoperiod consisting of 16 of light and 8 h of darkness at a constant temperature of 25°C and 60 to 80% relative humidity. The plants grown in the pots were used as a source of nodules for isolation of the rhizobia. Sixty-day-old plants were uprooted, and nodules were selected at random for isolation of the rhizobia from each host species using standard procedures and yeast extract mannitol agar as described by Vincent (35). Cultures were grown at 28°C and were subsequently stored in 25% glycerol at −80°C. Yeast extract mannitol agar slants for 78 isolates from each host species were prepared for shipment to Beltsville, MD. Upon arrival, each culture was plated to check for purity and was subsequently grown in 10 ml modified arabinose-gluconate broth (30) for 3 days to prepare DNA. PCR-quality template DNA was prepared from 1 ml of each culture with 200 μl Prepman Ultra (Applied Biosystems, Foster City, CA) by following the manufacturer's protocol. DNA samples were stored at −20°C, and the cultures were stored as glycerol suspensions at −70°C and as lyophilized samples.
The numbers of rhizobia present in the Amra soil able to nodulate M. truncatula and M. laciniata were estimated from most probable numbers using the methodology outlined by Vincent (35).
PCR primer design and PCR amplification of chromosomal loci and nodC.
The loci chosen for MLST analysis were described previously by van Berkum et al. (33). The primers for PCR and sequence analysis of loci encoding glyceraldehyde-3-phosphate dehydrogenase (gap), protein-PII uridylyltransferase (glnD), 6-phosphogluconate dehydrogenase (gnd), and the putative oxidoreductase protein (ordL2) were the primers described previously (33). Separate PCR and sequence primers were designed for the other six loci examined (Table 1) by using the primer design software package Oligo Primer Analysis, version 6.65 (Molecular Biology Insights, Inc., Cascade, CO). Similarly, PCR and sequence primers were designed to determine the nodC alleles present in each isolate and to sequence the entire 1,281-bp nodC gene present in representative isolates (Table 1). The oligonucleotides (Table 1) were synthesized by Sigma-Genosys (The Woodlands, TX) and were received as dried preparations. Upon receipt, the primers were dissolved in 10 mM Tris buffer (pH 8.0) to obtain a final concentration of 1,000 pmol/ml and were stored at −20°C. The PCRs for each locus were then optimized by using a FailSafe PCR premix selection kit (Epicenter, Madison, WI) and the thermal cycle protocol described by van Berkum and Fuhrmann (31) with a PTC-225 Peltier thermal cycler (MJ Research Inc., Waltham, MA), using genomic DNA of USDA 1002 and A321 as templates. The reaction mixtures were analyzed by horizontal agarose gel electrophoresis to select the FailSafe PCR system (Epicenter, Madison, WI) determined to be most suitable for PCR amplification of the DNA preparations of all 148 isolates used in this investigation. The presence of a single PCR product of the expected molecular size for each primer pair using each template was verified by horizontal gel electrophoresis. Each PCR product was then purified, especially to remove the PCR primers, by using the Ampure PCR purification system (Agincourt Bioscience Corporation, Beverly, MA).
TABLE 1.
Primer sequences used in MLST and nodC analysis of 148 Medicago-nodulating rhizobia originating from a single arid Tunisian soil collected at the Amra site
| Locusa | Directionb | Primer sequence | Product length (bp) or designation | Location on chromosome or primer location |
|---|---|---|---|---|
| PCR primers | ||||
| asd | F | 5′-TGTCGGCCGGGAGATGCTGAA-3′ | 799 | 3617313-3618347 |
| R | 5′-GCCGCCGTTCTCATGCTTGT-3′ | |||
| edD | F | 5′-CGTCCCCAATCTCGGCATCAT-3′ | 603 | 766634-768454 |
| R | 5′-GCCCGCTTCGTCGCTTCCCTG-3′ | |||
| nuoE1 | F | 5′-GGGCCGAGGCAACGATCAAGA-3′ | 537 | 1381334-1382161 |
| R | 5′-CCGGCGCCTGCAACGTCGTCA-3′ | |||
| recA | F | 5′-CGGAGCGAAGGACAGCGTAGT-3′ | 721 | 1948178-1949224 |
| R | 5′-AACGCCTTCGCCATACATGAT-3′ | |||
| sucA | F | 5′-GCGAGGCCGAGGTGCATCAGT-3′ | 886 | 3313082-3316078 |
| R | 5′-GCCGCGTCGCCGTGCAGGAT-3′ | |||
| zwf | F | 5′-TCCCGTCGAACCGTTTGACTA-3′ | 641 | 769253-770728 |
| R | 5′-CGGCTTCGGAAACGGTGATCT-3′ | |||
| nodC | F | 5′-CGCRATTGTTGATGCAGTGCT-3′ | 1,958 | 465-485 in nodB |
| R | 5′-CGCGCGTGCTCATCCKGAAGT-3′ | 487-467 in nodI | ||
| Sequence primers | ||||
| asd | F | 5′-TCCGGCCGACGAAGTGGTG-3′ | 81-99 | |
| R | 5′-ATGCGCTTGGTGAACTTCTTG-3′ | 575-555 | ||
| edd | F | 5′-TCGGCATCATCACCTCCTACA-3′ | 209-229 | |
| R | 5′-GGCGTGCCGGGATTGATGAAG-3′ | 761-741 | ||
| nuoE1 | F | 5′-TCGGCGGTCATTCCCCTGCTG-3′ | 118-138 | |
| R | 5′-CCTCGAACGTGTCCTTGAAGA-3′ | 475-455 | ||
| recA | F | 5′-GCGAAGGACAGCGTAGTTGAG-3′ | 82-102 | |
| R | 5′-GCGGTGCCATCTTGTTCTTGA-3′ | 754-734 | ||
| sucA | F | 5′-CGGCGTCGAATTCATGCATAT-3′ | 609-629 | |
| R | 5′-CGCGCGCCTTGCCCATGACGA-3′ | 1126-1106 | ||
| zwf | F | 5′-GGGGGCACCGGCGATCTTG-3′ | 49-67 | |
| R | 5′-AGCGCAGTGCCATCAGATTCT-3′ | 583-563 | ||
| nodC | F | 5′-GGCCGCGGTCCGACACA-3′ | 333-349 in nodB | |
| F | 5′-CGGCCCGGTGCAATCGTG-3′ | 496-513 in nodB | ||
| F | 5′-CCGCCGCTATCTCAATCT-3′ | 26-43 in nodC | ||
| F | 5′-CGCGCGATCCGAGGTTCA-3′ | 1566U | 311-328 | |
| F | 5′-TGCCGTGGTGGACAATTTTGA-3′ | 1001-1021 | ||
| F | 5′-ACCCATCAACCTCTTTCT-3′ | 1107-1124 | ||
| R | 5′-CCGCTCTGCCTCTTCCATAAT-3′ | 756-736 in nodI | ||
| R | 5′-AGCCGTTCCCAAATCAAGTGA-3′ | 689-669 in nodI | ||
| R | 5′-CGCCCGTTTCCAGTCATGCTA-3′ | 17 to −4 in nodI | ||
| R | 5′-GAGGCCGCGCAACAGAG-3′ | 2136L | 897-881 in nodC | |
| R | 5′-GGCCGCAGCAACACATAA-3′ | 628-611 | ||
| R | 5′-TCCGCTTTCCGACGTTCT-3′ | 364-347 |
The genes encode the following proteins: asd, aspartate-semialdehyde dehydrogenase; edd, phosphogluconate dehydratase; gap, glyceraldehyde-3-phosphate dehydrogenase; glnD, protein-PII uridylyltransferase; gnd, 6-phosphogluconate dehydrogenase; nuoE1, NADH dehydrogenase I chain E protein; ordL2, putative oxidoreductase protein; recA, DNA strand exchange and recombination protein; sucA, 2-oxoglutarate dehydrogenase E1; zwf, glucose-6-phosphate 1-dehydrogenase; nodC, NodC N-acetylglucosaminyltransferase.
F, forward; R, reverse.
Sequence analysis.
For MLST the purified PCR products were used in two sequence reactions each, either with one of the original PCR primers (for the gap, glnD, gnd, and ordL2 loci) or with nested primers designed for the remaining six loci (Table 1). The nodC allele present in each isolate was determined by sequence analysis of the 570-bp nodC PCR products using primers 1566U and 2136L (Table 1). Subsequently, sequences were obtained for the entire nodC genes in strains USDA 1002 (S. meliloti type strain), A321 (S. medicae type strain), and USDA 1170, as well as 10 isolates that represented each of the alleles that had been detected. The 16S rRNA gene sequences of six Tunisian isolates representing chromosomal allelic profiles that were the most different from that of strain 1021 were determined by using the protocols described by van Berkum and Fuhrmann (31). An Applied Biosystems 3100 genetic analyzer in combination with a Dye Deoxy terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) was used to sequence the purified PCR products as described previously (32).
Data analysis.
A Microsoft Access database was created to compile the data collected for the 148 isolates. The sequence lengths entered for each locus were the same, and the same alleles were identified using the software Sequence Comparator (version 2.0.1; Keith Jolley). As additional alleles were identified, they were assigned different allele numbers in the database. In each case, allelic variation was verified by confirming the sequence disparity using Genedoc (version 2.6.001; K. B. Nicholas and H. B. Nicholas [http://www.nrbsc.org/gfx/genedoc/index.html]) and then checking the electropherograms produced by the sequencing analysis to substantiate differences. When there were ambiguities, the sequencing analysis was repeated.
When the database was complete, the relationship function in Microsoft Access was used to create a query for the allelic allocation for each of the 10 loci across the 148 isolates. The resulting tabulated data were then exported as a Microsoft Excel file to prepare data input files. For Fig. 1 and 2 the data input files for the 148 isolates and 231 strains of Medicago-nodulating rhizobia (33) were combined. Based on the allele frequencies among the STs, the genetic diversity of each locus was determined using a computer program by T. S. Whittam (27). Sequence Type Analysis and Recombinational Tests (START) version 2 (19) was used to determine both the allele and profile frequencies and to create a Nexus input file for SplitsTree version 4.6 (18). SplitsTree 4.6 was used to represent the genetic relationships among the Medicago-nodulating rhizobia in the form of a NeighborNet. NeighborNet is a linkage tree algorithm similar to neighbor joining or the unweighted-pair group method using average linkages (UPGMA), but the pairing and combining of nodes are different to take into consideration the fact that the phylogenetic signals may conflict or that there may be alternate evolutionary histories (8) (for example, as a result of recombination). In this case, the input file used was a treelike distance matrix resulting in a splits graph. For analysis with eBURST, STs were classified as single-locus variants (SLVs), double-locus variants, or singletons (STs differing at three or more loci) according to the convention reported by Feil et al. (14), and then the data were combined with the data published by van Berkum et al. (33). The number of groups, the clonal complex in each group, and a population snapshot of the chromosomal variation were then generated using eBURST. The null hypothesis of linkage equilibrium for the multilocus sequence data as defined by Maynard Smith et al. (22a) was evaluated for the 148 isolates from Tunisia using the program LIAN 3.0 (15). The nodC sequence alignment, created in Genedoc (http://www.nrbsc.org/gfx/genedoc/index.html), included the nodC genes of Rhizobium leguminosarum (accession number Y00548) and R. tropici (accession number X98514) as references. The alignment was used to obtain nucleotide and derived amino acid sequence similarity matrices for construction of genetic distance trees using the UPGMA algorithm (21).
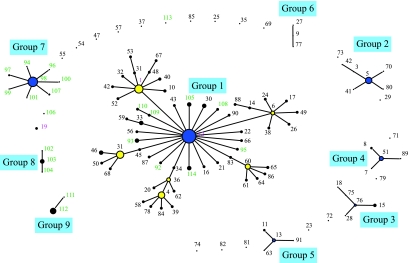
FIG. 1.
Snapshot of a population of 378 Medicago-nodulating rhizobia derived from allelic variation at 10 chromosomal loci. A matrix of the STs followed by the allele labels for each ST was used in eBURST (14) to generate a diagram of the evolutionary patterns among the strains and isolates. The snapshot was produced by setting the group definition to 0/10 genes. The sizes of the circles are related to the numbers of strains and isolates in each ST. The founder and cofounder genotypes are blue and yellow, respectively. The lengths of connecting lines between STs are arbitrary.
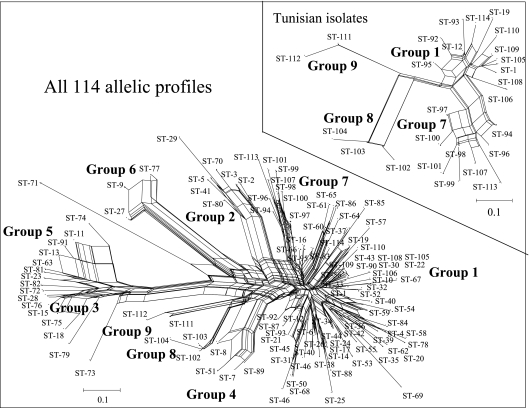
FIG. 2.
NeighborNet for 378 Medicago-nodulating rhizobia derived from allelic variation at 10 chromosomal loci. The inset shows the NeighborNet for the 148 Tunisian isolates. Matrices of the strain or isolate identification and the ST followed by the allele designations for each taxon was used in the START program (version 1.05) (19) to generate a Nexus file for input in Splitstree (18) to generate the NeighborNet (8).
Nucleotide sequence accession numbers.
The nucleotide sequences of alleles for each locus have been deposited in the GenBank database under accession numbers EF428921 to EF428961.
RESULTS
The most-probable-number estimates obtained for the Amra soil indicated that there were more rhizobia capable of nodulating M. truncatula than rhizobia capable of nodulating M. laciniata. The average concentration was 103 cells per g when M. truncatula was used as the trap host for counting, compared with 3 × 102 cells per g when M. laciniata was used.
As a result of this study, the documented number of alleles present in the chromosomes of Medicago-nodulating rhizobia (33) was increased. In addition to the 91 STs reported previously by van Berkum et al. (33), another 23 STs were discovered. Specifically, additional alleles were obtained for the following loci: asd, edd, zwf, gap, glnD, gnd, nuoE, ordL, recA, and sucA (one, five, one, one, one, two, two, one, three, and one alleles, respectively). This increased the mean number of alleles from 11.3 as reported by van Berkum et al. (33) to 13.1. However, the genetic diversity among the loci decreased with the exception of gnd and recA, for which increases were recorded (Table 2). The mean number of alleles for all the isolates from the Amra site in Tunisia was 3.7, and the mean genetic diversity across the chromosomes was 0.396. The genetic diversity and the number of alleles for the isolates originating from nodules of M. truncatula were higher than the genetic diversity and the number of alleles for the isolates from M. laciniata (Table 2).
TABLE 2.
Number of alleles and genetic diversity across chromosomes of Medicago-nodulating rhizobia
| Locus | All cultures
|
All Tunisian isolates
|
Isolates from M. truncatula
|
Isolates from M. laciniata
|
||||
|---|---|---|---|---|---|---|---|---|
| No. of alleles | Genetic diversity | No. of alleles | Genetic diversity | No. of alleles | Genetic diversity | No. of alleles | Genetic diversity | |
| asd | 17 | 0.526 | 4 | 0.345 | 4 | 0.333 | 2 | 0.389 |
| edd | 23 | 0.812 | 9 | 0.766 | 9 | 0.814 | 2 | 0.389 |
| zwf | 11 | 0.614 | 2 | 0.077 | 2 | 0.091 | 1 | 0.000 |
| gap | 10 | 0.532 | 3 | 0.280 | 3 | 0.325 | 1 | 0.000 |
| glnD | 11 | 0.747 | 4 | 0.686 | 4 | 0.658 | 3 | 0.722 |
| gnd | 10 | 0.410 | 3 | 0.495 | 3 | 0.515 | 2 | 0.500 |
| nuoE | 13 | 0.399 | 3 | 0.151 | 2 | 0.091 | 2 | 0.222 |
| ordL | 10 | 0.553 | 2 | 0.077 | 2 | 0.091 | 2 | 0.222 |
| recA | 13 | 0.591 | 4 | 0.689 | 4 | 0.697 | 3 | 0.722 |
| sucA | 13 | 0.577 | 3 | 0.394 | 3 | 0.385 | 2 | 0.389 |
| Total | 13.1 | 0.576 | 3.7 | 0.396 | 3.6 | 0.400 | 2 | 0.356 |
The likely patterns of chromosomal evolutionary descent and the identities of the probable founding genotypes were inferred from an eBURST analysis (14) of the MLST data. In this analysis the number of resamplings used for bootstrapping was 1,000 and the number of identical loci used for group definition was nine. Using these parameters, the number of groups that were identified increased from the six previously reported (33) to nine. The largest of these groups (group 1) contained 58 STs, which represented 234 of the 378 rhizobial cultures analyzed (Fig. 1). Within group 1 the predicted founding type was ST-12, with a bootstrap confidence value of 98%, while the second highest value was the value for ST-1 (39%). The analysis of the 148 Tunisian isolates increased the size of group 1 by eight STs. In addition to these newly identified STs, 53 of the Tunisian isolates were placed with the founding genotype (ST-12), increasing the number of isolates from 24 (33) to 77. Also, two isolates were placed in ST-1, increasing the number of isolates from 25 to 27. None of the Tunisian isolates were single SLV or double-locus variants of the cofounding types ST-1, ST-4, ST-6, ST-31, and ST-60. The only other previously recorded ST that was recognized among the 148 Tunisian isolates was the ST for two isolates from M. truncatula that were placed in ST-19.
None of the Tunisian isolates were placed in groups 2 through 6 which were previously identified in the analysis of 231 strains (33). Instead, three new groups (groups 7 through 9) were identified. The largest of these (group 7) consisted of eight STs (Fig. 1). The predicted founding type was ST-98, with a bootstrap confidence value of 100%. ST-98 was distinguished for 35 of the 148 isolates, 22 and 13 of which originated from M. truncatula and M. laciniata, respectively. Within group 7 three and two isolates were placed in the SLVs ST-97 and ST-107, respectively, while only single isolates were associated with the remaining five SLVs of group 7. Group 7 (45 isolates) was larger than group 2, which had previously been identified as the second largest group of STs, with 18 isolates (33).
The other two newly identified groups (groups 8 and 9) were much smaller than group 7. Group 8 included three STs (ST-102, ST-103, and ST-104) associated with 10 of the isolates, while group 9 included two STs (ST-111 and ST-112) associated with 19 isolates. Two singletons also were identified, increasing the total number from 19 to 21.
The NeighborNet for the entire collection of 114 STs represents a summary of multiple trees (18). The extensive cross-linking within the network indicates that it would be inappropriate to use a single, nonreticulate tree to represent the genetic relationships among the STs (Fig. 2). Some, but not necessarily all, of the ambiguity may have been caused by recombination among the different chromosomes. Regardless, it was evident from the NeighborNet analysis that groups 3 and 5 continued to be the groups most distant from group 1, as previously reported by van Berkum et al. (33), even after the 26 Tunisian STs were included in the analysis.
Among the Tunisian STs, group 9 was the group most distant from group 1 (Fig. 2, inset). However, the group 9 STs were less distant from the group 1 STs than the group 6 STs and the singleton ST-71 (Fig. 2). The 16S rRNA gene sequences of ST-12 genome strain 1021 (9) and the rhizobia placed in groups 3, 5, and 6 and the singleton ST-71 were different (33). However, both the isolates with allelic profiles of ST-111 and two isolates each from M. laciniata and M. truncatula representing ST-112 had 16S rRNA gene sequences that were identical to that of strain 1021.
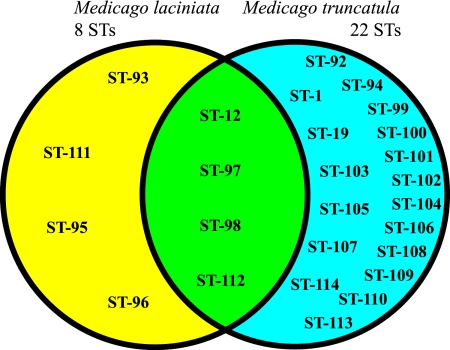
The 26 STs distinguished among the 148 Tunisian isolates were further subdivided based on the trap host into 22 and 8 STs that were isolated from nodules of M. truncatula and M. laciniata, respectively (Fig. 3). The four STs found in both species include the majority of the 148 isolates (73% or 108 isolates). Two of these four STs (ST-12 with 53 isolates and ST-98 with 35 isolates) were founders of groups 1 and 7, respectively. Three and 17 of the remaining isolates were placed in ST-97 (of group 7) and ST-112 (of group 9), respectively. The majority of the remaining 40 isolates originated from M. truncatula (32 isolates) and were placed into 18 STs that were not identified in the isolates from M. laciniata. Only eight isolates obtained from M. laciniata were allocated to four STs that were not found in the isolates recovered from M. truncatula (Fig. 3). However, these four STs were closely related to the four STs found in both host plant species, since ST-93 and ST-95 were both SLVs of ST-12, while ST-96 and ST-111 were SLVs of ST-98 and ST-112, respectively.
FIG. 3.
Diagram illustrating the host origins of rhizobia that belong to each of the STs. STs in the yellow, blue, and green areas originated from M. laciniata, from M. truncatula, and from both host species, respectively.
In a previous analysis of 231 strains, van Berkum et al. (33) reported strong linkage disequilibrium between the group 1 STs (S. meliloti) and the group 5 and 3 STs (S. medicae) shown in Fig. 1. One of the objectives of this study was to determine if there was also significant linkage disequilibrium among the four groups of STs identified among the Tunisian field isolates (Fig. 2, inset). Strong linkage disequilibrium was observed among the 26 STs identified among the 148 Tunisian isolates (Table 3). The STs of the major groups were partitioned to determine which, if any, of the four groups was responsible for the observed disequilibrium. There was no strong evidence of linkage disequilibrium within (or between) the two largest groups, group 1 and group 7. Because the two other groups (group 8 and group 9) had a very small number of STs (a total of five STs), it was not possible to complete meaningful linkage analyses for these groups. Consequently, the strong linkage disequilibrium that was evident among the entire collection of 26 STs probably was due to the five highly distinct STs of the two smallest groups (group 8 and group 9).
TABLE 3.
Test of the null hypothesis of linkage equilibrium among 26 multilocus STs in a collection of 148 Tunisian Medicago isolates from a single field site
| STs includeda | No. of STs | Genetic diversity | Mismatch variation
|
Standardized ab | Monte Carlo P value | Reject null hypothesis | |
|---|---|---|---|---|---|---|---|
| Observed | Expected | ||||||
| All | 26 | 0.39 ± 0.08 | 3.25 | 1.80 | 0.09 | 0.001 | Yes |
| Groups 1 and 7 | 21 | 0.30 ± 0.08 | 1.73 | 1.50 | 0.02 | 0.162 | No |
| Group 1 | 12 | 0.19 ± 0.07 | 0.45 | 1.05 | −0.06 | 0.982 | No |
| Group 7 | 9 | 0.19 ± 0.06 | 0.42 | 1.25 | −0.07 | 0.959 | No |
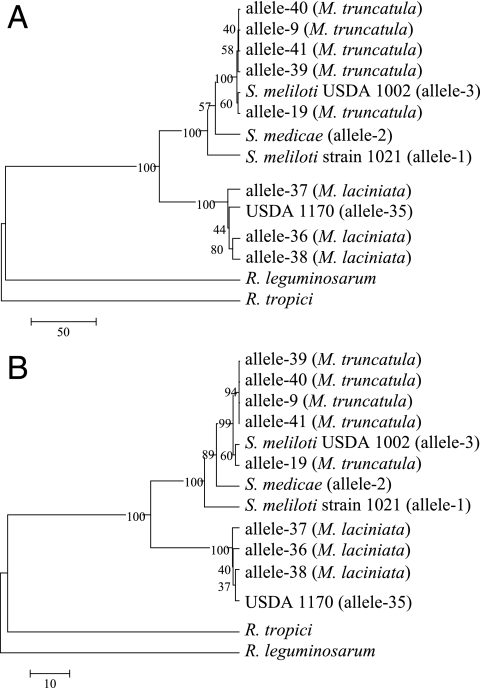
The 148 Tunisian isolates were examined for allelic variation in their plasmid-encoded nodC genes, since Barran et al. (4) reported evidence that the nodulation specificity of USDA 1170 for M. laciniata was mediated by the nodC allele carried on pSymA. Based on a partial sequence analysis (349 bp), 10 alleles were identified. The entire nodC locus present in the isolates representing the 10 different alleles was sequenced and was compared to the alleles present in strain 1021 and the type strains of S. meliloti (USDA 1002) and S. medicae (A321). The nodC genes of R. leguminosarum (accession number Y00548) and R. tropici (accession number X98514) were used as outgroups in the phylogenetic analysis (Fig. 4). Alleles 3, 9, 19, 39, 40, and 41 found in the isolates from M. truncatula were either identical to the nodC allele of the type strain of S. meliloti (allele 3) or were very similar. The maximum number of nucleotide sequence differences between the alleles was six (Fig. 4). The majority of the isolates from M. truncatula (59 isolates) harbored nodC allele 9, while alleles 3, 19, 39, 40, and 41 were present in six, three, one, one, and four of these isolates, respectively. The nodC allele present in USDA 1170 (allele 35) was identified in 47 of the isolates from M. laciniata. Alleles 36, 37, and 38 were present in 19, 5, and 3 of the remaining isolates, respectively. The most diverse nodC alleles in the isolates from M. laciniata were dissimilar at 19 nucleotide sequence positions. nodC allele 19, present in the isolates from M. truncatula, and allele 37, present in the isolates from M. laciniata, were the most divergent alleles in the 148 isolates and were only 89.7% similar (Fig. 4). By comparison, the similarity between the nodC alleles of the type strains of S. meliloti (USDA 1002) and S. medicae (A321) was 97.2%.
FIG. 4.
Phylogeny of nodC in rhizobia that form symbioses with the legume genus Medicago. The sequence alignment, created in Genedoc (http://www.nrbsc.org/gfx/genedoc/index.html) with the nodC genes of R. leguminosarum (accession number Y00548) and R. tropici (accession number X98514) used as outgroups, was 1,285 bp long. The msf file was converted to Mega format with Mega version 2.1 (21), which was also used to obtain nucleotide (A) and derived amino acid (B) sequence similarities for construction of trees using the UPGMA algorithm.
DISCUSSION
In a previous MLST analysis of Medicago-nodulating rhizobia, van Berkum et al. (33) identified 91 STs in 231 strains that had been obtained from many geographic locations. Another 23 STs were identified in the analysis of an additional 148 Tunisian isolates, and only three previously described STs (ST-1, ST-12, and ST-19) were recognized in this study. However, there are significant differences between this study and the previous study of van Berkum et al. (33). For instance, none of the 231 strains examined in the study of van Berkum et al. (33) were recovered from M. laciniata, and only 17 strains were obtained from M. truncatula, while 74 isolates from each Medicago species were analyzed in this study. Similarly, none of the 231 strains examined by van Berkum et al. (33) were from the African continent; all 17 strains from M. truncatula were from Jordan, Syria, and Turkey. Therefore, it is possible that African soils harbor genotypes of Medicago-nodulating rhizobia that are unique to this continent. Obviously, additional in-depth MLST investigations examining populations from other locations are needed to more fully evaluate this possibility.
This study also differed from the study of van Berkum et al. (33) in terms of the strategy used to obtain the rhizobial isolates. In the current study all 148 isolates originated from a single soil at the Amra site in Tunisia. In contrast, only 1 or 2 of the 231 strains in the study of van Berkum et al. (33) originated from the same field site. Considering the range of the diversity present in the Amra soil, it appears that an intensive sampling strategy would be necessary to obtain more realistic local estimates of rhizobial diversity.
More chromosomal diversity was identified among the 74 isolates originating from M. truncatula (22 STs) than among the 74 isolates originating from M. laciniata (8 STs). Although both Medicago species are part of the natural flora of Tunisia (16), the lower rhizobial diversity in isolates nodulating M. laciniata at the Amra site may be related to the lower numbers of these isolates in the soil. Differential estimates of the most probable numbers for rhizobia nodulating different species of Medicago growing in soil from the Amra site were reported previously. Zribi et al. (36) indicated that the most probable number of rhizobia counted with M. sativa as the trap host was 6 × 102, while no rhizobia were found when M. polymorpha was used, possibly because this species is not part of the natural flora at the Amra site. Clearly, the sizes of the native populations of rhizobia that are adapted to specific Medicago host species vary widely at this location.
The majority of the rhizobia isolated from M. laciniata were placed in four STs that were also identified among the isolates from M. truncatula. Only eight isolates from M. laciniata were assigned to the four STs that were not detected in isolates from M. truncatula. However, the Amra soil probably does harbor rhizobia with these four chromosomal STs that nodulate M. truncatula, since these STs are SLVs of STs that originated from both host plant species. One reason why these rhizobia were not identified among the 74 isolates from M. truncatula may be their low numbers in the soil and the higher chromosomal diversity among the M. truncatula rhizobia. Despite the observation that these four STs were recovered only from M. laciniata, the soil at the Amra site is colonized by the same population of rhizobia that includes members that either have the capability to specifically nodulate M. laciniata or form symbioses with M. truncatula. Since the diversity among the isolates recovered from M. laciniata was more limited, it seems that the rhizobia with specificity for M. laciniata probably represent a subpopulation of the rhizobia that form symbioses with M. truncatula.
Even though most of the rhizobia of M. truncatula and M. laciniata from the Amra site share chromosomal identity, they differed in their plasmid-encoded nodC alleles. Barran et al. (4) previously implicated the nodC allele as an allele that is important in the host specificity of USDA 1170 for nodulation of M. laciniata. Whether the variation in nodC is associated with differences in entire nodulation operons or even dissimilar pSymA plasmids in the rhizobial genomes is not clear. This hypothesis could be evaluated by complete sequence analysis of the nod operon associated with each of the nodC alleles that were revealed in this study and by development and application of MLST using loci located on pSymA.
Finally, there also is uncertainty about the manner in which rhizobia with identical chromosomes could have acquired dissimilar nodC alleles, irrespective of whether it is due to a variation in the entire operon or pSymA itself. Using a phylogenetic approach, Bailly et al. (2) obtained evidence for the spread of nod genes among nitrogen-fixing symbionts of Medicago species. Kinkle and Schmidt (20) reported transfer of the pea symbiotic plasmid in nonsterile soil. Therefore, the most plausible explanation is that pSymA or distinct regions within pSymA were shared among members of the rhizobial population by lateral transfer and recombination.
Acknowledgments
This work was supported in part by an ASM MIRCEN Award.
We thank K. Lee Nash for excellent technical assistance.
Footnotes
Published ahead of print on 5 October 2007.
REFERENCES
- 1.Badri, Y., K. Zribi, M. Badri, T. Huguet, and M. E. Aouani. 2003. Sinorhizobium meliloti nodulates Medicago laciniata in Tunisian soils. Czech J. Genet. Plant Breed. 39(Special Issue):178-183. [Google Scholar]
- 2.Bailly, X., I. Olivieri, B. Brunel, J.-C. Cleyet-Marel, and G. Bena. 2007. Horizontal gene transfer and homologous recombination drive the evolution of the nitrogen-fixing symbionts of Medicago species. J. Bacteriol. 189:5223-5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballard, R. A., and N. Charman. 2000. Nodulation and growth of pasture legumes with naturalised soil rhizobia. 1. Annual Medicago spp. Aust. J. Exp. Agric. 40:939-948. [Google Scholar]
- 4.Barran, L. R., E. S. P. Bromfield, and D. C. W. Brown. 2002. Identification and cloning of the bacterial nodulation specificity gene in the Sinorhizobium meliloti-Medicago laciniata symbiosis. Can. J. Microbiol. 48:765-771. [DOI] [PubMed] [Google Scholar]
- 5.Biondi, E. G., E. Pilli, E. Giuntini, M. L. Roumiantseva, E. E. Andronov, O. P. Onichtchouk, O. N. Kurchak, B. V. Simarov, N. I. Dzyubenko, A. Mengoni, and M. Bazzicalupo. 2003. Genetic relationship of Sinorhizobium meliloti and Sinorhizobium medicae strains isolated from Caucasian region. FEMS Microbiol. 220:207-213. [DOI] [PubMed] [Google Scholar]
- 6.Bradić, M., S. Sikora, S. Redžepović, and Z. Štafa. 2003. Genetic identification and symbiotic efficiency of an indigenous Sinorhizobium meliloti field population. Food Technol. Biotechnol. 41:69-75. [Google Scholar]
- 7.Brockwell, J., and F. W. Hely. 1966. Symbiotic characteristics of Rhizobium meliloti: an appraisal of the systematic treatment of nodulation and nitrogen fixation interactions between hosts and rhizobia of diverse origins. Aust. J. Agric. Res. 17:885-899. [Google Scholar]
- 8.Bryant, D., and V. Moulton. 2004. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21:255-265. [DOI] [PubMed] [Google Scholar]
- 9.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Pühler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carelli, M., S. Gnocchi, S. Fancelli, A. Mengoni, D. Paffetti, C. Scotti, and M. Bazzicalupo. 2000. Genetic diversity and dynamics of Sinorhizobium meliloti populations nodulating different alfalfa cultivars in Italian soils. Appl. Environ. Microbiol. 66:4785-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dangeard, P. A. 1926. Recherches sur les tubercles radicaux des Légumineuses. Botaniste 16:1-275. [Google Scholar]
- 12.Del Papa, M. F., L. J. Balagué, S. C. Sowinski, C. Wegener, E. Segundo, F. M. Abarca, N. Toro, K. Niehaus, A. Pühler, O. M. Aguilar, G. Martínez-Drets, and A. Lagares. 1999. Isolation and characterization of alfalfa-nodulating rhizobia present in acidic soils of central Argentina and Uruguay. Appl. Environ. Microbiol. 65:1420-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feil, E. J. 2004. How stable are the core genes of bacterial pathogens? ASM News 6:234-238. [Google Scholar]
- 14.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequencing typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haubold, B., and R. R. Hudson. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847-848. [DOI] [PubMed] [Google Scholar]
- 16.Heyn, C. C. 1963. The annual species of Medicago. Scropta Hierosolymitana 12:1-154. [Google Scholar]
- 17.Honeycutt, R. J., M. McClelland, and B. W. S. Sobral. 1993. Physical map of the genome of Rhizobium meliloti 1021. J. Bacteriol. 175:6945-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 19.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 20.Kinkle, B. K., and E. L. Schmidt. 1991. Transfer of the pea symbiotic plasmid pJB5JI in nonsterile soil. Appl. Environ. Microbiol. 57:3264-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetic analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 22.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Maynard Smith, J., N. H. Smith, M. O' Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paffetti, D., C. Scotti, S. Gnocchi, S. Fancelli, and M. Bazzicalupo. 1996. Genetic diversity of an Italian Rhizobium meliloti population from different Medicago sativa varieties. Appl. Environ. Microbiol. 62:2279-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paffetti, D., F. Daguin, S. Fancelli, C. Scotti, S. Gnocchi, F. Lippi, and M. Bazzicalupo. 1998. Influence of plant genotype on the selection of nodulating Sinorhizobium meliloti strains by Medicago sativa. Antonie Leeuwenhoek 73:3-8. [DOI] [PubMed] [Google Scholar]
- 25.Rome, S., M. P. Fernandez, B. Brunel, P. Normand, and J.-C. Cleyet-Marel. 1996. Sinorhizobium medicae sp. nov., isolated from annual Medicago spp. Int. J. Syst. Microbiol. 46:972-980. [DOI] [PubMed] [Google Scholar]
- 26.Roumiantseva, M. L., E. E. Andronov, L. A. Sharypova, T. Dammann-Kalinowski, M. Keller, J. P. W. Young, and B. V. Simarov. 2002. Diversity of Sinorhizobium meliloti from the Central Asian alfalfa gene center. Appl. Environ. Microbiol. 68:4694-4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Small, E., and M. J. Jomphe. 1989. A synopsis of the genus Medicago (Leguminosae). Can. J. Bot. 67:3260-3294. [Google Scholar]
- 29.Reference deleted.
- 30.van Berkum, P. 1990. Evidence for a third uptake hydrogenase phenotype among the soybean bradyrhizobia. Appl. Environ. Microbiol. 56:3835-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Berkum, P., and J. J. Fuhrmann. 2000. Evolutionary relationships among the soybean bradyrhizobia reconstructed from 16S rRNA gene and internally transcribed spacer region sequence divergence. Int. J. Syst. Evol. Microbiol. 50:2165-2172. [DOI] [PubMed] [Google Scholar]
- 32.van Berkum, P., D. Beyene, and B. D. Eardly. 1996. Phylogenetic relationships among Rhizobium species nodulating the common bean Phaseolus vulgaris L. Int. J. Syst. Bacteriol. 46:240-244. [DOI] [PubMed] [Google Scholar]
- 33.van Berkum, P., P. Elia, and B. D. Eardly. 2006. Multilocus sequence typing as an approach for population analysis of Medicago-nodulating rhizobia. J. Bacteriol. 188:5570-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villegas, M. D. C., S. Rome, L. Mauré, L. Domerguea, O. Gardan, X. Bailly, J.-C. Cleyet-Marel, and B. Brunel. 2006. Nitrogen-fixing sinorhizobia with Medicago laciniata constitute a novel biovar (bv. medicaginis) of S. meliloti. Syst. Appl. Microbiol. 29:526-538. [DOI] [PubMed] [Google Scholar]
- 35.Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria. IBP handbook no. 15. Blackwell Scientific Publications Ltd., Oxford, United Kingdom.
- 36.Zribi, K., R. Mhamdi, T. Huguet, and M. E. Aouani. 2004. Distribution and genetic diversity of rhizobia nodulating natural populations of Medicago truncatula in Tunisian soils. Soil Biol. Biochem. 36:903-908. [Google Scholar]