Abstract
Repeated dog deaths occurred in 2002, 2003, and 2005 after the animals drank water from the shoreline of the Tarn River in southern France. Signs of intoxication indicated acute poisoning due to a neurotoxin. Floating scum and biofilms covering pebbles were collected in the summers of 2005 and 2006 from six different sites along 30 km from the border of this river. The cyanobacterial neurotoxic alkaloid anatoxin-a and/or its methyl homolog, homoanatoxin-a, was detected in the extracts of most samples examined by gas chromatography-mass spectrometry. Fifteen filamentous cyanobacteria of the order Oscillatoriales were isolated and displayed four distinct phenotypes based on morphological characteristics and pigmentation. Three of the phenotypes can be assigned to the genus Oscillatoria or Phormidium, depending on the taxonomic treatises (bacteriological/botanical) employed for identification. The fourth phenotype is typical of the genus Geitlerinema Anagnostidis 1989. Eight strains rendered axenic were analyzed for production of anatoxin-a and homoanatoxin-a, and all strains of Oscillatoria/Phormidium proved to be neurotoxic. The genetic relatedness of the new isolates was evaluated by comparison of the intergenic transcribed spacer sequences with those of six oscillatorian strains from the Pasteur Culture Collection of Cyanobacteria. These analyses showed that the neurotoxic representatives are composed of five different genotypes, three of which correspond to phenotypes isolated in this study. Our findings prove that neurotoxic oscillatorian cyanobacteria exist in the Tarn River and thus were most likely implicated in the reported dog poisonings. Furthermore, they reemphasize the importance of monitoring benthic cyanobacteria in aquatic environments to fully assess the health risks associated with these organisms.
Neurotoxicosis and deaths of wild and domestic animals associated with cyanobacterial blooms in fresh and brackish waters have been reported for four continents: America, Eurasia, Africa, and Australia (3, 16, 24, 31). Anatoxin-a (ANTX), an alkaloid which acts as a potent agonist of the nicotinic acetylcholine receptor, and other neurotoxins, such as the sodium channel blocker saxitoxin and the organophosphorus cholinesterase inhibitor ANTX(s), are generally produced by planktonic freshwater cyanobacteria of the genera Anabaena, Planktothrix, and Aphanizomenon (21, 29, 32, 37, 38). These cyanobacteria may accumulate in massive quantities at the surface of water bodies depending on environmental factors favorable to their growth (temperature and abundant nutrient loads) or as a result of physical parameters, such as stratification and long retention times of the water (25). In recent decades, however, several studies reported animal intoxications by ingestion of ANTX and/or its homolog homoanatoxin-a (HANTX) produced by benthic cyanobacteria identified as species of Oscillatoria and Phormidium (7, 8, 32, 33). Furthermore, examination of the toxin production of axenic strains representative of 13 cyanobacterial genera confirmed the high incidence of neurotoxicity among benthic members of the genus Oscillatoria (2).
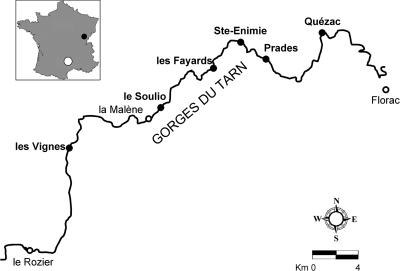
When 20 dogs died between August and September 2002 after drinking water from the Tarn River (see Fig. 1), a popular site for tourists in southwestern France, the French national sanitary service paid particular attention to the risks that animals may encounter by drinking or bathing in the river. One year later, several intoxications and deaths of dogs occurred again at the same location, and two other cases were reported for the shoreline of the La Loue River (Fig. 1) in eastern France (8). All deaths happened within 30 min of the start of clinical signs. Indications typical of neurotoxicosis (10) included convulsions, coma, rigors, and limb twitching of the animals.
FIG. 1.
Locations of the sampling sites along the Tarn River, France. The locations of the sampling sites are indicated by black circles. (Inset) Map of France showing the two regions, Tarn River (white circle) and the La Loue River (small black circle), where dog poisonings were reported.
In this study, we wished to provide answers to the following questions. (i) Are benthic neurotoxic cyanobacteria a likely cause for the dog deaths in the Tarn River? (ii) Do the six sampling sites differ in neurotoxin content and cyanobacterial representatives? (iii) Do morphologically similar cyanobacterial isolates represent the same genotype? (iv) Are ANTX- and/or HANTX-producing strains from the Tarn River genetically distinct from a selection of oscillatorian cyanobacteria of the Pasteur Culture Collection of Cyanobacteria (PCC) (three of which are known to produce this class of neurotoxins [2])?
MATERIALS AND METHODS
Site description and sample collection.
The sampling sites are located along the Tarn River in the south of France (Fig. 1). Ste-Enimie, Les Vignes, and Prades are characterized by large river banks that form lagoons. Les Fayards and Le Soulio are camping places along forested rock shores of the river. The village Quézac, known for its plant of bottled mineral water, is located on the river bank. During the sampling periods (summers 2005 and 2006), the river did not contain any blooms of planktonic cyanobacteria, but isolated patches of floating scum material were abundant and pebbles close to the edge of the river were covered by thin biofilms. Scum and biofilm samples were collected wearing disposable gloves, changed at each sampling, for the protection of the operator and to avoid cross-contamination. Scum material was captured directly with and stored in sterile plastic screw-cap vials (50 ml) (Falcon; BD Biosciences, Le Pont de Claix, France). Biofilms were detached from three pebbles (diameter, 10 to 12 cm) per sample, using a new toothbrush for each site, and washed into the Falcon vials with small amounts of river water. All samples were placed into a cooling box for transport and stored at 4°C upon arrival in the laboratory. Examinations by light microscopy for identification of cyanobacterial representatives and toxin analyses were made within a few days.
Strain isolation and culture conditions.
Single filament isolates of the field samples, spotted (10 to 30 μl) onto solid media immediately upon arrival in the laboratory, were made by micromanipulation by the method of Rippka (26). To optimize the chances of growth, two media were used, both containing 1 mM NaHCO3. One was modified medium Z8, described as medium 551 (12) but incorrectly cited as medium Z8o (which lacks nitrate), and the other was medium 552 (12), called medium 2N (2). The liquid media were solidified either with 0.9% (wt/vol) of washed agar (Sigma A 8678) or 0.8% (wt/vol) of agarose (Litex LSL 4000; FMC, Vallensbaek Strand, Denmark). Most plates were incubated at 23 to 25°C under continuous illumination with Osram white universal fluorescent tubes providing a photosynthetic photon flux density (PPFD) of 15 to 20 μmol quanta m−2 s−1 (measured with a Licor LI-185B quantum/radiometer/photometer equipped with a LI-190SB quantum sensor). For one strain (Oscillatoria PCC 10601), part of the isolation and purification procedure was performed at 22°C, and a PPFD of 5 to 10 μmol quanta m−2 s−1 was received over a light/dark cycle of 14 h/10 h. Purity to the axenic state was confirmed as described previously (2). Irrespective of the media employed for isolation, all axenic strains grew well in medium 2N (2), which was used for toxicity tests. The 100-ml flasks containing 40 ml of medium were incubated without shaking at 25°C under continuous white light as described above and a PPFD of 20 μmol quanta m−2 s−1. If necessary, the desired biomass (0.6 g [wet weight]) was obtained by pooling the pellets of several cultures.
The axenic strains Oscillatoria PCC 6304, PCC 6407, PCC 7112, PCC 7515, PCC 9240, and PCC 10111 were from the Pasteur Culture Collection of Cyanobacteria (PCC) and grown as described above, except that for four of them (strains PCC 6304, PCC 6407, PCC 7112, and PCC 7515) modified BG11 medium was used, which corresponds to medium 539 (12).
Photomicroscopy.
Aliquots of the filament aggregates were placed on a dry plate of solid growth medium. Once the liquid surrounding the deposit was absorbed, a small block (about 2 to 3 mm2) of this area was cut out with a sterile microspade and transferred to a slide. The agar block was then homogenized with the vertical edge of the coverslip prior to placing and pressing the coverslip in its final position. Photography was performed using a Plan-Neofluar objective (40×/0.75 Oil Ph) and an Axioskop 2 microscope (Carl Zeiss Jena). Images were taken with a charge-coupled-device camera (DEI-750; Optronics Engineering, Goleta, CA), transferred to a personal computer, and further processed with Paintshop Pro 6 or Photoshop 7.0 (Adobe Europe). Measurements of cell length and width were made on printed images (15 × 20 cm) by comparison to an image of a micrometer scale enlarged to the same dimensions. The data reported are the averages of measurements made on 50 cells of a minimum of three filaments, excluding apical cells.
Determination of phycobiliproteins.
Aliquots of liquid cultures equivalent to 300 mg (wet weight) were centrifuged at 12,000 × g for 10 min at 20°C. The pellets were resuspended in 500 μl of 20 mM sodium acetate buffer (pH 5.5) supplemented with 2 mM sodium azide and 10 mM disodium-EDTA. The suspension was transferred to Lysing Matrix A tubes (Qbiogene, Illkirch, France) and exposed to two 30-second runs at a speed setting of 5.0 in a FastPrep instrument (Qbiogene). After lysis of the cells, 500 μl of additional extraction buffer was added, and the crude cell extracts were precipitated with 1% (wt/vol) streptomycin sulfate for 30 min at 4°C and centrifuged at 10,000 × g for 10 min in order to eliminate membrane fragments containing chlorophyll. The amounts of phycoerythrin (PE) and phycocyanin (PC) in the 1-ml samples were calculated from the absorption spectra of the supernatant fractions using previously described equations (35). Given that the amount of cell material in the samples varied, the data are expressed as the ratios of PE/PC.
DNA extraction and PCR amplification.
Aliquots (2 ml) of liquid cultures were centrifuged at 12,000 × g for 10 min at 20°C and washed twice with sterile distilled water. The final pellets were resuspended in 200 μl of sterile distilled water and frozen in liquid N2. Lysates of the frozen samples were prepared by a minimum of five alternating cycles of thawing at 100°C for 5 min and refreezing in liquid N2. Aliquots (100 μl) of the lysis products were fixed onto FTA cards (Whatman, Dassel, Germany) and conserved at room temperature. Amplifications of the intergenic transcribed spacer (ITS) regions of the ribosomal operon were carried out by PCR using primers 322 and 340 as described previously (13). The PCR mixtures, in a final volume of 50 μl, contained the FTA sample disk cut with a Uni-Core punch (Whatman), 5 μl of 10× buffer, 4 μl of 25 mM MgCl2, 75 μM of each deoxynucleoside triphosphate, 250 ng of each primer, and 1 U Taq polymerase (Promega Corporation, Charbonnières, France). After an initial step consisting of 5 min at 95°C, 30 cycles of amplification were performed; each amplification cycle consisted of 30 s at 95°C, 30 s at 60°C, and 1 min at 72°C. A final elongation step was carried out for 7 min at 72°C. All PCRs were performed in a 9700 Perkin-Elmer thermocycler (Applied Biosystem Cycler, Foster City, CA), and oligonucleotides were purchased from MWG Biotech (Ebersberg, Germany). A total of 5 μl of each sample was analyzed by electrophoresis on 0.6% (wt/vol) agarose (Litex FMC) gels in 0.5× Tris-borate-EDTA buffer at pH 8.0 (28). For visualization of the products, the gels were stained with ethidium bromide (0.5 μg/ml). PCR products were purified using the Qiaquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced by Genome Express (Meylan, France).
Sequence alignment of the ITS regions.
The nucleotide sequences of ITS regions reported in this study were first aligned using ClustalX in Bioedit software version 1.83 (36). The alignment was then manually refined using GeneDoc version 2.6.002 (20) by reference to secondary structure models of the 16S rRNA-ITS-23S rRNA region proposed by Iteman et al. (13). Based on these alignments, and irrespective of large insertions/deletions (indels) in some of the ITS sequences, a distance matrix was created in ClustalX with gaps being counted as differences, and a dendrogram was drawn using TreeView program (Win32) (http://taxonomy.zoology.gla.ac.uK/rod/treeview.html).
The percentage of identity in pairwise comparisons of the ITS sequences was determined by counting manually the number of nonidentical nucleotides for a given set of sequences, aligned as described above, and expressing identical nucleotides as a fraction of the total.
Toxin extraction and gas chromatography (GC)-MS analysis of ANTX and HANTX.
Filament aggregates from the field material (10 g [wet weight]), drained with a spatula directly in the Falcon sampling tubes, or cell pellets of axenic cultures (0.6 g [wet weight]) were extracted with 15 ml of 50 mM acetic acid, sonicated twice for 3 min, and centrifuged at 12,000 × g for 30 min at 4°C. The supernatant was adjusted to pH 9 with NH4OH. Solid-phase extraction was performed using a C18 column (500 mg Bakerbond resin; Mallinckrodt Baker, Paris, France) previously conditioned with 2 ml methanol (MeOH) and 6 ml H2O. The sample was loaded by gravity, and the column was washed successively with 6 ml of H2O and 2 ml of 10% (vol/vol) MeOH-H2O. Fractions containing the toxin were eluted with 6 ml MeOH. After solvent evaporation, the residues were dissolved in 200 μl CH3CN, and samples of 1 μl were injected into the gas chromatograph-mass spectrometer. Mass spectrometry (MS) was performed with a Varian 3800 gas chromatograph coupled to a Saturn 2000 ion trap mass spectrometer (Varian, Les Ulis, France). Chromatographic separation was achieved on a Varian CP-Sil 8 CB low-bleed/MS capillary column (30 m × 0.25 mm × 0.25 μm), and the injection was operated in the splitless mode. Helium was used as a carrier gas at 1 ml/min in a constant flow rate. The injector temperature was 220°C. The oven temperature was maintained at 50°C for 1 min, programmed at 50 to 275°C (5°C/min) with a hold at 275°C for 5 min. Mass spectra in the electron impact ionization (EI) mode were recorded at 70 eV with an ionization current of 22 μA; the ion trap temperature was 140°C, and the transfer line temperature was set at 170°C. A thermal regeneration step was intercalated between each run to avoid contamination by carryover of residual analytes.
Nucleotide sequence accession numbers.
The ITS sequences reported in this paper were deposited in the DDBJ-EMBL-GenBank database under the following accession numbers (the strains are shown in the parentheses): EF061067 (PCC 10601), EF061068 (PCC 10609), EF061069 (PCC 10610), EF061070 (PCC 10611), EF061071 (PCC 10612), EF061072 (Fil.10 PR), EF061073 (Fil.1 SO), EF061074 (Fil.2D1 FY), EF061075 (PCC 10608), EF061076 (Fil.1 SE), EF061077 (PCC 10702), EF061078 (PCC 10602), EF061079 (PCC 9240), EF061080 (PCC 10111), EF178272 (PCC 7515), EF178273 (PCC 6407), and EF178274 (PCC 7112).
RESULTS
Environmental samples, toxin content, and strain isolation.
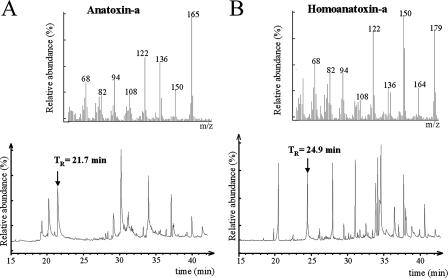
Fifteen samples were collected in July and August 2005 and July 2006 from six sites spread over a distance of 30 km between the villages of Quézac and Les Vignes in France (Fig. 1). ANTX and HANTX content of the samples was examined by GC-MS analyses. Two examples are shown in Fig. 2. Except for three samples from the same site (Les Fayards), all contained ANTX and/or HANTX (Table 1). Microscopic observation of the scum samples and pebble biofilms revealed that filamentous nonheterocystous cyanobacteria of the order Oscillatoriales (Pseudanabaena, Geitlerinema, and Oscillatoria/Phormidium) formed the dominant photosynthetic microbial population, but small amounts of diatoms, green algae, and unicellular cyanobacteria were also found. Given the high incidence of ANTX and/or HANTX production among members of the genera Oscillatoria and Phormidium (2, 3, 8), particular attention was paid to the isolation of such representatives, although for comparison one isolate of Geitlerinema, present in most samples, was also made. From each of the toxin-containing samples (12/15), at least one isolate of neurotoxic cyanobacteria was obtained with the exception of two samples from the Le Soulio site (Table 1), which for unknown reasons did not yield viable filaments. A total of 15 filamentous cyanobacteria, recognized by light microscopy as four different phenotypes, were isolated and eight were rendered axenic. As indicated by their respective PCC numbers (Table 1), the axenic strains were integrated in the PCC and are publicly available upon request.
FIG. 2.
GC-MS analysis of extracts from environmental samples. (A) GC chromatogram of a sample from Le Soulio, France, in July 2005. (Inset) EI mass spectrum at a retention time (TR) of 21.7 min, characteristic of anatoxin-a (molecular ion m/z 165 and fragments m/z 150, 136, and 122). (B) GC chromatogram of a sample from Ste Enimie, France, in July 2005. (Inset) EI mass spectrum at retention time of 24.9 min characteristic of homoanatoxin-a (molecular ion m/z 179 and fragments m/z 164, 150, 136, and 122).
TABLE 1.
Environmental samples collected from the Tarn River in France
| Sampling site | Sampling date | Type of sample | Neurotoxin(s) in samplea | Isolate designationb | Phenotypec |
|---|---|---|---|---|---|
| Quézac | July 2006 | Scum | HANTX | Fil.6 QZ | 2 |
| Prades | July 2006 | Scum | HANTX | Fil.10 PR | 1 |
| Ste Enimie | July 2005 | Scum | HANTX | PCC 10608 | 3 |
| PCC 10609 | 1 | ||||
| August 2005 | Scum | ANTX-HANTX | PCC 10610 | 1 | |
| July 2006 | Scum | HANTX | Fil.1 SE | 3 | |
| Fil.6 SE | 1 | ||||
| Pebble | HANTX | Fil.3.2 SE | 3 | ||
| Les Fayards | July 2005 | Scum | ND | Fil.2D1 FY | 1 |
| August 2005 | Scum | ND | None | ||
| July 2006 | Pebble | ND | None | ||
| Le Soulio | July 2005 | Scum | ANTX | PCC 10601 | 1 |
| PCC 10602 | 4 | ||||
| August 2005 | Scum | ANTX | None | ||
| July 2006 | Scum | ANTX-HANTX | Fil.1 SO | 1 | |
| Pebble | ANTX-HANTX | None | |||
| Les Vignes | August 2005 | Scum | ANTX-HANTX | PCC 10611 | 1 |
| PCC 10612 | 1 | ||||
| July 2006 | Pebble | HANTX | PCC 10702 | 2 |
Presence of ANTX and/or HANTX. ANTX-HANTX, both detected; ND, not detected.
Axenic isolates have been deposited in the PCC; all others are clonal but not exempt from bacterial contamination.
Arbitrary numbers distinguishing the different phenotypes.
Characterization of the Tarn River isolates.
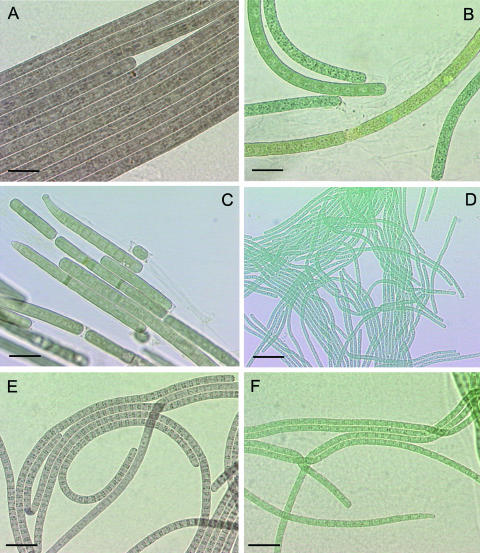
Morphological characters, such as cell dimensions, shape of terminal cells, and PE/PC ratios, allowed us to distinguish more precisely between representative strains of the four different phenotypes (Fig. 3 and Table 2). Properties common to all strains are rapid gliding motility by rotation, little or no sheath formation visible by light microscopy, and lack of pronounced constrictions between the cells composing the filaments. Phenotypes 1, 2, and 3 (phenotypes numbered arbitrarily) have cells of similar dimensions (diameter of 7.8 to 8.2 μm) that are shorter than wide (width/length ratio of >2), whereas those of phenotype 4 are more narrow (2.5 μm) and longer than wide (width/length ratio of <1). The presence of granular cell inclusions varies with the age of the cultures and was not taken into account for discrimination of the phenotypes.
FIG. 3.
Photomicrographs of six cyanobacterial strains examined in this study. (A to D) Strains PCC 10601 (A), PCC 10702 (B), PCC 10608 (C), and PCC 10602 (D) correspond to phenotypes 1, 2, 3, and 4, respectively, described in Table 2. (E and F) Strains Oscillatoria sp. strain PCC 9240 (E) and Oscillatoria formosa PCC 10111 (F) studied for comparison. Bars, 20 μm.
TABLE 2.
Phenotypic and genotypic characteristics and neurotoxin content of representative isolates from the Tarn River (France) compared to six PCC strains of Oscillatoria
| Strain | Cell width (μm)a | Cell length (μm)a | Morphology of terminal cells | PE/PC ratiob | Neurotoxin(s)c | Length of ITS amplicons (bp) | Generic and specific assignments
|
|
|---|---|---|---|---|---|---|---|---|
| Bacteriological code | Botanical codef | |||||||
| Isolates from the Tarn River | ||||||||
| PCC 10601 (phenotype 1) | 7.4-9.0 (8.2) | 2.4-5.0 (3.4) | Rounded | 1.0 | ANTX | ∼800 | Oscillatoria (cluster 2.C)e | Phormidium sp. |
| PCC 10702 (phenotype 2) | 7.5-8.8 (8.1) | 2.0-3.7 (3.2) | Rounded | 0.5 | ANTX-HANTX | ∼600 | Oscillatoria (cluster unknown)e | Phormidium sp. |
| PCC 10608 (phenotype 3) | 6.9-8.5 (7.8) | 2.5-3.7 (3.2) | Conical/capitate | 0.4 | ANTX-HANTX | ∼800 | Oscillatoria (cluster 2.D)e | Phormidium sp. |
| PCC 10602 (phenotype 4) | 2.2-2.7 (2.5) | 3.8-4.2 (4.0) | Rounded | NA | ND | ∼600 | Geitlerinema (cluster unknown)e | Geitlerinema amphibiumg |
| Oscillatoria strains of the PCC used for comparison | ||||||||
| PCC 7515 | 15-16l | Rounded | 1.9 | ND | ∼800 | Oscillatoria (cluster 1)d | Oscillatoria sanctah | |
| PCC 7112 | 7-8l | Rounded | 1.3 | ND | ∼800 | Oscillatoria (cluster 2)d | Phormidium nigro-viridisi | |
| PCC 6304 | 4-5l | Hook-like curved | NA | ND | ∼340; ∼540 | Oscillatoria (cluster 3)d | Phormidium acuminatumj | |
| PCC 6407 | 4-5l | Conical/capitate | NA | HANTX | ∼800 | Oscillatoria (cluster 4)d | Phormidium formosumk | |
| PCC 9240 | 3.8-5.3 (4.6) | 1.9-4.5 (2.8) | Rounded | 1.1 | ANTX-HANTX | ∼800 | Oscillatoria (cluster 2.B)e | Phormidium sp. |
| PCC 10111 | 3.7-4.4 (4.1) | 2.2-3.3 (2.7) | Conical/capitate | NA | HANTX | ∼800 | Oscillatoria (cluster 4)e | Phormidium formosumk |
The range and mean value for cell width and cell length are shown. The mean value is shown in parentheses.
Phycoerythrin (PE)/phycocyanin (PC) ratios were determined as described in Materials and Methods. NA, not applicable.
Presence of ANTX and/or HANTX. ANTX-HANTX, both detected; ND, toxin not detected.
Assignment according to Castenholz et al. (6).
Assignment made in this study.
Sensu Komárek and Anagnostidis 2005 (15).
Based on Oscillatoria amphibia Agardh ex Gomont 1892. Authority for transfer to the genus Geitlerinema, Anagnostidis 1989 (1).
Oscillatoria sancta Kützing ex Gomont 1892 (see reference 15). This species remained in the genus Oscillatoria (see reference 15).
Based on Oscillatoria nigro-viridis Thwaites ex Gomont 1892. Authority for transfer to the genus Phormidium, Anagnostidis and Komárek 1988 (see reference 15).
Based on Oscillatoria acuminata Gomont 1892. Authority for transfer to the genus Phormidium, Anagnostidis and Komárek 1988 (see reference 15).
Based on Oscillatoria formosa Bory ex Gomont 1892. Authority for transfer to the genus Phormidium, Anagnostidis and Komárek 1988 (see reference 15).
The measurements were taken from Castenholz et al. (6).
The most frequently encountered (Table 1) filamentous cyanobacterium (phenotype 1 [Fig. 3A]) is brownish in color due to a high PE/PC ratio. Apical cells are generally rounded to dome-shaped and may occasionally carry a thickened colorless membrane (calyptra). Thylakoids can be observed as longitudinal patches, giving the filaments a striated pattern when viewed under bright-field illumination. This feature distinguishes phenotype 1 from phenotypes 2 and 3, in which a longitudinal thylakoid arrangement can be deduced only with difficulty by light microscopy.
Phenotype 2 (Fig. 3B) has dark-green filaments containing small amounts of PE. As for phenotype 1, the terminal cells are rounded to dome-shaped.
Phenotype 3 (Fig. 3C) exhibits filaments that, like phenotype 2, are dark green due to a low PE content. Its filaments can readily be distinguished, both from phenotypes 1 and 2, by their straight or curved apices formed by three to six tapering cells. The terminal cells are conical and often carry a capitate, dome-shaped extension.
Phenotype 4 (Fig. 3D), easily distinguished by its much more narrow blue-green trichomes, lacks PE. Terminal cells are rounded to slightly conical. Granular inclusions are often visible at both poles of the cells. Thylakoids seem to form a tight cortical layer, as judged by the lighter color in the center of the cells when viewed under bright-field illumination.
Comparison of the Tarn River isolates with six other PCC strains.
The reference strains of clusters 1, 2, 3, and 4 of the genus Oscillatoria (6), corresponding, respectively, to the species O. sancta, O. nigro-viridis, O. acuminata, and O. formosa (6), were included in this study for comparison (Table 2). In addition, two Scandinavian strains, Oscillatoria sp. strain PCC 9240 and Oscillatoria formosa PCC 10111, known to be neurotoxic (30, 33), but for which little other information is available, were also characterized. Strain PCC 9240 (Fig. 3E and Table 2) is an isolate from Lake Hormajärvi (Finland), first described as Oscillatoria sp. strain 193 (30). Except for the narrower cell diameter (∼5 μm), it resembles strains of phenotype 1 from the Tarn River with respect to color, thylakoid arrangement, and terminal cells. The pronounced granulation at the cross walls, visible in most of the filaments shown in Fig. 3E, is observed only in freshly transferred cultures. Strain PCC 10111 (Fig. 3F and Table 2) is an axenic subisolate (2) of Oscillatoria formosa NIVA-CYA-92 (33), which was isolated from Lake Levrasjön (Sweden). It lacks PE, and the blue-green trichomes (cell diameter of ∼4 μm) are composed of almost isodiametric cells (cell width/length ratio of ∼1.4). The trichomes taper slightly towards their termini and often carry curved conical or dome-shaped capitate terminal cells. This strain is most similar to Oscillatoria PCC 6407 (Table 2).
Taxonomic assignments.
Assignments at the generic/specific level of the Tarn River isolates and the PCC strains used for comparison are provided in Table 2, following either bacteriological (6) or botanical (15) systems of nomenclature. Strains of phenotypes 1, 2, and 3 can be assigned to the genera Oscillatoria (6) or Phormidium (15), whereas the single strain of phenotype 4 is a representative of genus Geitlerinema Anagnostidis 1989 (1). For the seemingly conflicting bacteriological and botanical generic assignments, see references 6, 15, and 18.
Confirmation of neurotoxin production.
The content of ANTX and/or HANTX was determined for all axenic strains isolated from the Tarn River (France), the clonal but impure strain Fil.2D1 FY (Table 1), and the PCC strains of Oscillatoria used for comparison (Table 2). Cultures of phenotypes 1, 2, and 3 produced either ANTX or HANTX or both of these toxins. In strain PCC 6407 (Oscillatoria cluster 4) and Oscillatoria formosa strain PCC 10111, only HANTX was detected, whereas strain Oscillatoria PCC 9240 produced both ANTX and HANTX (Table 2), confirming the presence of this class of toxins (2, 30, 33). However, two of these strains (PCC 6407 and PCC 9240) were previously thought to produce exclusively ANTX (2, 30). The single isolate of phenotype 4 produced neither ANTX nor HANTX. The lack of this class of neurotoxins observed for strains Oscillatoria PCC 6304, PCC 7112, and PCC 7515 (Table 2) is in agreement with a previous study by Aráoz et al. (2).
ITS sequence analysis.
The ITS regions of the 15 isolates from the Tarn River (France) and the PCC strains of Oscillatoria were amplified and sequenced. The PCR products examined by gel electrophoresis gave a single band of ∼600 bp or ∼800 bp for each strain (Table 2 and data not shown), except for strain PCC 6304, which exhibited two bands of ∼340 and ∼540 bp. Sequencing of the PCR products, except for those of strain PCC 6304 and the impure isolates Fil.3.2 SE (phenotype 3) and Fil.6 QZ (phenotype 2) that were not further analyzed, showed that the ITS lengths varied between 392 and 632 bp. This length range is in overall agreement with the bands observed by gel electrophoresis for the respective amplicons that, with the primers used, should be about 200 bp longer (∼150 bp being generated by the 3′ end of the 16S rRNA gene and ∼50 bp by the 5′ end of the 23S rRNA gene).
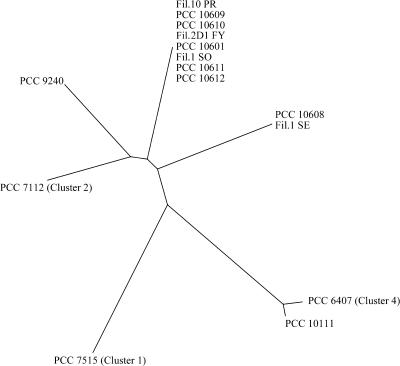
A preliminary alignment of all ITS sequences (data not shown) showed that the multiple isolates of the neurotoxic phenotypes 1 and 3 (eight isolates of phenotype 1 and two isolates of phenotype 3) have almost identical nucleotide sequence (99 to 100% sequence identity), which differ, however, between the two phenotypes. Furthermore, the alignment revealed that the largest conserved block of the sequences corresponds to the tRNAIle gene and that the short ITS sequences (392 and 401 bp, respectively) in strains PCC 10702 (phenotype 2) and PCC 10602 (phenotype 4) lack the tRNAAla gene. Consequently, a second alignment (Fig. 4) was created using exclusively the ITS sequences of 450 to 632 bp and only one sequence each of phenotypes 1 and 3, respectively, represented by those of strains PCC 10601 and Fil.1 SE (see the legend to Fig. 4). The seven ITSs can be distinguished based on their lengths, those of strains PCC 9240, Fil.1 SE, PCC 10601, and PCC 7112 being in the range of 534 to 574 bp. Due to insertions/deletions (indels) with respect to the former four ITSs, the sequence of strain PCC 7515 is significantly shorter (450 bp), whereas those of strains PCC 6407 and PCC 10111 are longer (630 and 632 bp). In order to visualize the overall relatedness of the ITSs, without attempting to infer precise phylogenetic relationships, a dendrogram (Fig. 5) was constructed from all sequences, irrespective of their different lengths.
FIG. 4.
Alignment of the nucleotide sequences of the ITSs. Percentage identity is represented by three colors: blue for 100%, green for values above 80%, and red for those above 60%. Gaps (dashes) are treated as differences. The conserved domains (D1, D1′, D2, D3, and D4), the tRNA genes, and the antiterminator (box B and box A) are indicated. The ITS sequences of strains PCC 10609, PCC 10610, PCC 10611, and PCC 10612 and the impure isolates Fil.10 PR and Fil.2D1 FY, corresponding to phenotype 1 are 99 to 100% identical to that of strain PCC 10601 and are not included in the alignment. The full-length ITS of strain Fil.1 SE is representative of phenotype 3. It was preferred over the identical but slightly incomplete sequence of strain PCC 10608, which was excluded from the alignment.
FIG. 5.
Dendrogram depicting the overall nucleotide differences in the ITS sequences (see Fig. 4 and legend) of 10 isolates from the Tarn River (France) in comparison to five other PCC strains, including the reference strains of Oscillatoria clusters 1, 2, and 4 (Table 2). Given that accurate phylogenetic inference cannot be drawn from this representation (see Materials and Methods), no scale bar is given for the nucleotide substitution per site.
This representation (Fig. 5) clearly shows that the neurotoxic isolates of phenotypes 1 and 3 (strains PCC 10601 and Fil.1 SE, respectively) from the Tarn River (France) represent two different genotypes. Both are closest to, but distinct from, the Finnish neurotoxic strain PCC 9420 and the nontoxic strain PCC 7112 (reference strain of Oscillatoria cluster 2), the latter two of which can also be recognized as two different genotypes. In contrast, the HANTX-producing strains Oscillatoria formosa PCC 10111 and strain PCC 6407 (Oscillatoria cluster 4), which have the longest ITS (630 to 632 bp [Fig. 4]), group closely together but are very distant from the former four genotypes. The nontoxic strain PCC 7515 (Oscillatoria cluster 1), characterized by the shorter ITS (450 bp [Fig. 4]), is well separated from all other strains (Fig. 5).
Pairwise comparison of the ITS sequences (see Table S1 in the supplemental material) revealed that strains of phenotypes 1 and 3 share only 77% sequence identity, and both phenotypes are distinct from Finnish neurotoxic strain PCC 9240 (79% and 77% sequence identity, respectively). With respect to strain PCC 7112, which is most closely related to strain PCC 9240 (82% sequence identity), phenotypes 1 and 3 show 74% and 69% sequence identity, respectively. Based on these relationships, strain PCC 9240 and strains of phenotypes 1 and 3 are distinguished from the reference strain, PCC 7112, of Oscillatoria cluster 2 (6), by assigning them to three new subclusters called 2.B, 2.C, and 2.D, respectively (Table 2).
The high relatedness of strains PCC 6407 and PCC 10111 (97% ITS sequence identity) is in full agreement with their common phenotypic characters (Table 2). These data provide the first molecular evidence that the previous assignment to the same nomenspecies, Oscillatoria formosa (6, 33) synonymous with Phormidium formosum (Bory ex Gomont) Anagnostidis and Komárek 1988 (see reference 15), is justified for strains PCC 6407 and PCC 10111, in spite of their different geographical origin (strain PCC 6407 was isolated in California, and strain PCC 10111 was isolated in Sweden).
The very short ITS sequences (392 and 401 bp, respectively) of strains PCC 10702 (phenotype 2) and PCC 10602 (phenotype 4), which both lack the tRNAAla gene, are substantially different from one another, in agreement with the very different morphologies of these two strains (Table 2).
DISCUSSION
Benthic neurotoxic cyanobacteria are the likely cause for the dog deaths at the Tarn River in France.
The clinical signs of animal poisoning, death within a short time, and the absence of pesticides (Direction Départementale des Affaires Sanitaires et Sociales de la Lozère [DDASS], 2005, personal communication) suggested that the dog deaths along the Tarn River were due to ingestion of water containing neurotoxins, potentially produced by cyanobacteria.
With the exception of samples from one site (Les Fayards), all cyanobacterial samples collected in the summers of 2005 and 2006 from five different sites along the Tarn River in France contained ANTX and/or HANTX. From toxin-containing samples, 15 filamentous cyanobacteria was isolated, and all but one (phenotype 4) were neurotoxin-producing strains. Cyanobacteria of phenotype 1 were most frequently encountered and were exclusively isolated from scum samples. This is in contrast to members of phenotypes 2 and 3 that were isolated either from scum material or pebbles, which may indicate a lower degree of adherence to solid substrate (i.e., pebbles) for the representatives of phenotype 1. For two of the sampling sites (Ste Enimie and Le Soulio [Table 1]), the persistence of cyanobacteria of phenotype 1 over two consecutive years could be confirmed, demonstrating their successful establishment and survival in the river. This also emphasizes the need to monitor benthic neurotoxin-producing cyanobacteria with particular care once they have been detected for the first time. Strain Fil.2D1 FY (Table 1), corresponding to phenotype 1, was shown to produce ANTX (data not shown), although it was isolated from an apparently nontoxic site (Les Fayards). This surprising result may be due to subdetectable concentrations of the toxins in the corresponding samples, possibly due to photolysis and nonphotochemical degradation at high light intensities or alkaline pH (14, 34). Alternatively, the possibility that both toxic and nontoxic representatives of phenotype 1 may occur at this site and that, fortuitously, the single filament isolate Fil.2D1 FY proved to be a toxin producer cannot be entirely excluded.
Neurotoxin-producing strains from the Tarn River (France) differ phenotypically.
Characterization of phenotypic properties and neurotoxin analyses on cultures of representative axenic strains clearly demonstrated the coexistence of three different ANTX- and HANTX-producing phenotypes in the Tarn River (Fig. 2 and Table 2). The relative amounts of the two homologs differed between these strains but were not quantified. The simultaneous presence of ANTX and/or HANTX in strains of phenotypes 2 and 3 may, however, be only a minor phenotypic difference. Both toxins were previously reported for Raphidiopsis mediterranea (19), a filamentous heterocystous cyanobacterium, and for one strain of Oscillatoria (2). Furthermore, in another strain of Oscillatoria (PCC 6506), the presence of one or the other of the two homologs varied with the growth conditions of the organism (2). Thus, it seems possible that many, if not all, ANTX-producing strains may also be capable of producing HANTX. This conclusion would also explain the discrepancy in the toxin content observed for strains PCC 9240 and PCC 6407 (see Results and Table 2) compared to previous data (2, 30). Biosynthetic studies on ANTX and HANTX suggest that these homologs share glutamic semialdehyde as a common precursor (11). However, more research on the regulation of synthesis and activities of the enzyme(s) that convert this central intermediate to either ANTX or HANTX needs to be performed in order to elucidate the factors responsible for the preferential production of one of the homologs or their different ratios.
Neurotoxin-producing strains from the Tarn River (France) represent three different genotypes.
Sequence comparisons of the ITSs, known to provide a better resolution of discrimination than the 16S rRNA genes (4, 5, 9, 17, 22, 23, 27), showed that phenotypes 1, 2, and 3 from the Tarn River are three different genotypes. Multiple isolates of the same phenotype, for both phenotypes 1 and 3, share almost identical nucleotide sequences. This confirms that our phenotypic identifications were reliable and that isolates of the same phenotype, even if originating from different sampling sites, are also genetically identical as judged by the ITS as a molecular marker.
Genetic relationships of the French Tarn River isolates to other oscillatorian cyanobacteria.
Comparison of the full-length ITS sequences (see Table S1 in the supplemental material) with those available in the NCBI database showed that the ITSs of strains of phenotype 1 share 94% identity with the sequence (accession number EF222209.1) of a HANTX-producing Phormidium strain isolated from the Hutt River in New Zealand (39), demonstrating the widespread geographical distribution of these closely related toxic oscillatorian cyanobacteria. Strains of both phenotypes 1 and 3 are also relatively closely related to Phormidium autumnale CCAP 1462/10 (toxicity unknown), a strain from Antarctic soil (18), and form an ITS sequence cluster with the Finnish strain PCC 9240, a third distinct neurotoxin producer, and the nontoxic strain PCC 7112. The fourth group of neurotoxic oscillatorian strains is composed of strains PCC 6407 and PCC 10111. The toxin-producing strain PCC 10702 (phenotype 2) has no close relatives in the NCBI database and seems to represent a fifth distinct neurotoxic genotype.
The ITS sequence of strain PCC 6407 determined in this study shows 100% identity with that published previously (18), and both strains Oscillatoria formosa PCC 6407 and PCC 10111 are most closely related (92 and 93% ITS sequence identity, respectively) to Phormidium inundatum strain SAG 79.79, isolated from thermal water in France (18), which consequently should be checked for neurotoxin content. The relatedness of the ANTX-producing strains described as Phormidium formosum and Phormidium amoenum from an Australian water reservoir (3), and the representative of Phormidium favosum (8) in the La Loue River, France, to the five neurotoxic genotypes discovered in this study, cannot, unfortunately, be assessed, for lack of the corresponding ITS sequences.
Conclusions.
Repeated public alert campaigns undertaken by public health services led to a decrease in the number of dog deaths at the Tarn River in France in the years 2002 to 2006. However, the most likely involvement of neurotoxic cyanobacteria in the health risks for animals drinking water from this river was shown only in this study. Our results prove that three different phenotypes of ANTX- and/or HANTX-producing cyanobacteria of the genera Oscillatoria and Phormidium coexist in the Tarn River. Furthermore, we demonstrated the usefulness of the ITS as a molecular marker by showing that each neurotoxic phenotype corresponds to a distinct genotype, all three of which differ from the neurotoxin-producing strains previously available in the PCC. Once a more extensive database of ITS sequences is available, it will be easy to recognize related neurotoxic, or potentially neurotoxic, cyanobacteria at the genetic level, without having access to the organisms. This is particularly important given the present taxonomic confusion, where generic and specific names are often contradictory and do not necessarily reflect the true relatedness of the cyanobacteria under study. As shown for the example of the neurotoxic Phormidium strain from the Hutt River (New Zealand), ITS sequence comparisons will also provide information concerning the geographic distribution of related toxin-producing strains. Finally, until genes encoding the enzyme(s) involved in ANTX/and or HANTX biosynthesis have been discovered, the different ITS sequences determined in this study may also prove useful for the design of genotype-specific primers for the detection and monitoring of potentially toxic Oscillatora/Phormidium representatives directly in the environment.
Supplementary Material
Acknowledgments
We are very grateful to Michael Herdman for valuable suggestions and help in aligning the ITS sequences.
This work was supported by the Institut Pasteur, the Centre National de la Recherche Scientifique (URA 2172), and Programme “Multiorganismes” from Aventis Pharma (Groupe Sanofi-Aventis) and Bayer Pharma. C. Peyraud-Thomas was a recipient of a fellowship from Programme “Multiorganismes” from Aventis Pharma (Groupe Sanofi-Aventis) and Bayer Pharma. S. Cadel-Six was a recipient of a Ph.D. fellowship funded by the Agence Française de Sécurité Sanitaire de l'Environnement et du Travail, project “Environnement et Santé”.
Footnotes
Published ahead of print on 12 October 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anagnostidis, K. 1989. Geitlerinema, a new genus of oscillatorialean cyanophytes. Plant Syst. Evol. 164:33-46. [Google Scholar]
- 2.Aráoz, R., H. O. Nghiêm, R. Rippka, N. Palibroda, N. Tandeau de Marsac, and M. Herdman. 2005. Neurotoxins in axenic oscillatorian cyanobacteria: coexistence of anatoxin-a and homoanatoxin-a determined by ligand-binding assay and GC/MS. Microbiology 151:1263-1273. [DOI] [PubMed] [Google Scholar]
- 3.Baker, P. D., D. A. Steffensen, A. R. Humpage, B. C. Nicholson, I. R. Falconer, B. Lanthois, K. M. Fergusson, and C. P. Saint. 2001. Preliminary evidence of toxicity associated with the benthic cyanobacterium Phormidium in South Australia. Environ. Toxicol. 16:506-511. [DOI] [PubMed] [Google Scholar]
- 4.Baurain, D., L. Renquin, S. Grubisic, P. Scheldeman, A. Belay, and A. Wilmotte. 2002. Remarkable conservation of internally transcribed spacer sequences of Arthrospira (“Spirulina”) (Cyanophyceae, Cyanobacteria) strains from four continents and of recent and 30-year-old dried samples from Africa. J. Phycol. 38:384-393. [Google Scholar]
- 5.Boyer, S. L., J. R. Johansen, V. R. Flechtner, and G. L. Howard. 2002. Phylogeny and genetic variance in terrestrial Microcoleus (Cyanophyceae) species based on sequence analysis of the 16S rRNA gene and associated 16S-23S ITS region. J. Phycol. 38:1222-1235. [Google Scholar]
- 6.Castenholz, R. W., R. Rippka, M. Herdman, and A. Wilmotte. 2001. Subsection III, order Oscillatoriales (Elenkin 1934), p. 539-562. In D. R. Boone, R. W. Castenholz, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. The Archaea and the deeply branching and phototrophic bacteria. Springer-Verlag, New York, NY. [Google Scholar]
- 7.Edwards, C., K. Beattie, C. Scrimgeour, and G. Codd. 1992. Identification of anatoxin-a in benthic cyanobacteria (blue-green algae) and in associated dog poisoning at Loch Insh, Scotland. Toxicon 30:1165-1175. [DOI] [PubMed] [Google Scholar]
- 8.Gugger, M., S. Lenoir, C. Berger, A. Ledreux, J. C. Druart, J. F. Humbert, C. Guette, and C. Bernard. 2005. First report in a river in France of the benthic cyanobacterium Phormidium favosum producing anatoxin-a associated with dog neurotoxicosis. Toxicon 45:919-928. [DOI] [PubMed] [Google Scholar]
- 9.Gugger, M., C. Lyra, P. Henriksen, A. Couté, J. F. Humbert, and K. Sivonen. 2002. Phylogenetic comparison of the cyanobacterial genera Anabaena and Aphanizomenon. Int. J. Syst. Evol. Microbiol. 52:1867-1880. [DOI] [PubMed] [Google Scholar]
- 10.Gunn, G. J., A. G. Rafferty, G. C. Rafferty, N. Cockburn, C. Edwards, K. A. Beattie, and G. A. Codd. 1992. Fatal canine neurotoxicosis attributed to blue-green algae (cyanobacteria). Vet. Rec. 130:301-302. [DOI] [PubMed] [Google Scholar]
- 11.Hemscheidt, T., J. Rapala, K. Sivonen, and O. M. Skulberg. 1995. Biosynthesis of anatoxin-a in Anabaena flos-aquae and homoanatoxin-a in Oscillatoria formosa. J. Chem. Soc. Chem. Commun. 13:1361-1362. [Google Scholar]
- 12.Herdman, M., I. Iteman, and R. Rippka. 2005. Catalogue of cyanobacterial strains, 2nd ed. Institut Pasteur, Paris, France.
- 13.Iteman, I., R. Rippka, N. Tandeau de Marsac, and M. Herdman. 2000. Comparison of conserved structural and regulatory domains within divergent 16S rRNA-23S rRNA spacer sequences of cyanobacteria. Microbiology 146:1275-1286. [DOI] [PubMed] [Google Scholar]
- 14.James, K. J., A. Furey, I. R. Sherlock, M. A. Stack, M. Twohig, F. B. Caudwell, and O. M. Skulberg. 1998. Sensitive determination of anatoxin-a, homoanatoxin-a and their degradation products by liquid chromatography with fluorimetric detection. J. Chromatogr. A 798:147-157. [DOI] [PubMed] [Google Scholar]
- 15.Komárek, K., and K. Anagnostidis. 2005. Cyanoprokaryota. Teil 2. Oscillatoriales 19/2, p. 1-655. In B. Büdel, L. Krienitz, G. Gärtner, and M. Schagerl (ed.), Süsswasserflora von Mitteleuropa. Elsevier GmbH, München, Germany.
- 16.Krienitz, L., A. Ballot, K. Kotut, C. Wiegand, S. Pütz, J. Metcalf, G. Codd, and S. Pflugmacher. 2003. Contribution of hot spring cyanobacteria to the mysterious deaths of lesser flamingos at Lake Bogoria, Kenya. FEMS Microbiol. Ecol. 43:141-148. [DOI] [PubMed] [Google Scholar]
- 17.Laloui, W., K. A. Palinska, R. Rippka, F. Partensky, N. Tandeau de Marsac, M. Herdman, and I. Iteman. 2002. Genotyping of axenic and non-axenic isolates of the genus Prochlorococcus and the OMF-‘Synechococcus’ clade by size, sequence analysis or RFLP of the internal transcribed spacer of the ribosomal operon. Microbiology 148:453-465. [DOI] [PubMed] [Google Scholar]
- 18.Marquardt, J., and K. A. Palinska. 2007. Genotypic and phenotypic diversity of cyanobacteria assigned to the genus Phormidium (Oscillatoriales) from different habitats and geographical sites. Arch. Microbiol. 187:397-413. [DOI] [PubMed] [Google Scholar]
- 19.Namikoshi, M., T. Murakami, M. F. Watanabe, T. Oda, J. Yamada, S. Tsujimura, H. Nagai, and S. Oishi. 2003. Simultaneous production of homoanatoxin-a, anatoxin-a, and a new non-toxic 4-hydroxyhomoanatoxin-a by the cyanobacterium Raphidiopsis mediterranea Skuja. Toxicon 42:533-538. [DOI] [PubMed] [Google Scholar]
- 20.Nicholas, K. B., and H. B. Nicholas, Jr. 1997. GeneDoc: a tool for editing and annotating multiple sequence alignments. Pittsburgh Supercomputing Center, Pittsburgh, PA.
- 21.Onodera, H., Y. Oshima, P. Henriksen, and T. Yasumoto. 1997. Confirmation of anatoxin-a(s), in the cyanobacterium Anabaena lemmermannii, as the cause of bird kills in Danish lakes. Toxicon 35:1645-1648. [DOI] [PubMed] [Google Scholar]
- 22.Otsuka, S., S. Suda, R. Li, M. Watanabe, H. Oyaizu, S. Matsumoto, and M. M. Watanabe. 1999. Phylogenetic relationships between toxic and non-toxic strains of the genus Microcystis based on 16S to 23S internal transcribed spacer sequence. FEMS Microbiol. Lett. 172:15-21. [DOI] [PubMed] [Google Scholar]
- 23.Otsuka, S., S. Suda, S. Shibata, H. Oyaizu, S. Matsumoto, and M. M. Watanabe. 2001. A proposal for the unification of five species of the cyanobacterial genus Microcystis Kützing ex Lemmermann 1907 under the Rules of the Bacteriological Code. Int. J. Syst. Evol. Microbiol. 51:873-879. [DOI] [PubMed] [Google Scholar]
- 24.Park, H. D., K. Bomchul, K. Enkyong, and O. Tokio. 1998. Hepatotoxic microcystins, and neurotoxic anatoxin-a in cyanobacteria blooms from Korean lakes. Environ. Toxicol. Water Qual. 13:225-234. [Google Scholar]
- 25.Reynolds, C. S. 1991. Ecology and control of cyanobacteria (blue-green algae). Public Health Lab. Ser. Microbiol. Digest 8:87-90. [Google Scholar]
- 26.Rippka, R. 1988. Isolation and purification of cyanobacteria. Methods Enzymol. 167:3-27. [DOI] [PubMed] [Google Scholar]
- 27.Rocap, G., D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68:1180-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1, p. 5.8. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 29.Selwood, A. I., P. T. Holland, S. A. Wood, K. F. Smith, and P. S. McNabb. 2007. Production of anatoxin-a and a novel biosynthetic precursor by the cyanobacterium Aphanizomenon issatschenkoi. Environ. Sci. Technol. 41:506-510. [DOI] [PubMed] [Google Scholar]
- 30.Sivonen, K., K. Himberg, R. Luukkainen, S. I. Niemelä, G. K. Poon, and G. A. Codd. 1989. Preliminary characterization of neurotoxic cyanobacteria blooms and strains from Finland. Toxic. Assess. 4:339-352. [Google Scholar]
- 31.Sivonen, K., and G. Jones. 1999. Cyanobacterial toxins, p. 41-111. In I. Chorus and J. Bartram (ed.), Toxic cyanobacteria in water: a guide line to public health significance, monitoring and management. E&FN Spon, London, United Kingdom.
- 32.Sivonen, K., S. I. Niemelä, R. M. Niemi, L. Lepistö, T. H. Luoma, and L. A. Räsänen. 1990. Toxic cyanobacteria (blue-green algae) in Finnish fresh and coastal waters. Hydrobiologia 190:267-275. [Google Scholar]
- 33.Skulberg, O. M., W. W. Carmichael, R. A. Andersen, S. Matsunaga, R. E. Moore, and R. Skulberg. 1992. Investigations of a neurotoxic oscillatorialean strain (Cyanophyceae) and its toxin. Isolation and characterization of homoanatoxin-a. Environ. Toxicol. Chem. 11:321-329. [Google Scholar]
- 34.Stevens, D. K., and R. I. Krieger. 1991. Stability studies on the cyanobacterial nicotinic alkaloid anatoxin-a. Toxicon 29:167-179. [DOI] [PubMed] [Google Scholar]
- 35.Tandeau de Marsac, N., and J. Houmard. 1988. Complementary chromatic adaptation: physiological conditions and action spectra. Methods Enzymol. 167:318-328. [Google Scholar]
- 36.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viaggiu, E., S. Melchiorre, F. Volpi, A. Di Corcia, R. Mancini, L. Garibaldi, G. Crichigno, and M. Bruno. 2004. Anatoxin-a toxin in the cyanobacterium Planktothrix rubescens from a fishing pond in northern Italy. Environ. Toxicol. 19:191-197. [DOI] [PubMed] [Google Scholar]
- 38.Wood, S. A., J. P. Rasmussen, P. T. Holland, R. Campbell, and A. L. M. Crowe. 2007. First report of cyanotoxin anatoxin-a from Aphanizomenon issatschenkoi (cyanobacteria). J. Phycol. 43:356-365. [Google Scholar]
- 39.Wood, S. A., A. I. Selwood, A. Rueckert, P. T. Holland, J. R. Milne, K. F. Smith, B. Smits, L. F. Watts, and C. S. Cary. 2007. First report of homoanatoxin-a and associated dog neurotoxicosis in New Zealand. Toxicon 50:292-301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.