Abstract
We found that a robust energy taxis response mediated by the Aer receptor can sometimes mask chemotaxis mediated by other methyl-accepting chemotaxis proteins (MCPs) in Pseudomonas aeruginosa. We identified PA2652 as a chemoreceptor for malate by screening aer mcp double mutants by using swarm plate assays.
Flagellated bacteria swim toward chemicals in the environment by a directed movement known as chemotaxis (24). As defined for Escherichia coli, chemotaxis refers to a response that does not require metabolism of the chemoattractant. Motile bacteria also use energy taxis, also known as taxis to metabolizable organic compounds, aerotaxis, and electron acceptor taxis to migrate to environments that support the optimal generation of cellular energy (27). E. coli has served as the model organism for extensive genetic and structural studies of chemotactic signal transduction. E. coli has four different transmembrane chemoreceptors called methyl-accepting chemotaxis proteins (MCPs) (24), each of which binds a discrete and limited set of organic and inorganic attractants and repellents. It also has a fifth MCP called Aer that mediates energy taxis by sensing a perturbation in protonmotive force through an FAD moiety present in its sensory PAS domain, rather than by binding a particular compound (6, 27, 29). Many of the compounds that are detected by E. coli MCPs, were identified by screening for mutants defective in chemotaxis ring formation in soft agar swarm plates containing various amino acids and sugars. The principle of this method, which is rapid and easy to carry out, is that bacteria inoculated at the center of a petri plate containing growth medium solidified with a low concentration of agar (11, 18) swim through the soft agar and up the concentration gradient of the attractant that they generate as they metabolize compounds present in the growth medium. Chemotaxis is visualized as a sharp ring of growth that gradually spreads to the edge of the petri dish. The E. coli mcp tsr, trg, and tar mutants form defective swarm rings in soft agar medium containing their cognate chemoattractants (8, 11, 23). The E. coli aer mutant forms defective swarm rings in soft agar medium containing metabolizable organic compounds, such as glycerol or succinate, that are not recognized by any of its four other MCPs (6, 29).
The E. coli chemotaxis machinery that interacts with its MCPs to accomplish signal transduction consists of six proteins and is highly conserved in bacteria. One difference between E. coli and other gram-negative bacteria is that other species tend to have many more MCP genes, on the order of 20 to 60 MCP genes, than E. coli does (2). Most of these genes have unknown functions. We have been interested in defining the functions of some of the 26 MCPs that Pseudomonas aeruginosa encodes (25). So far, chemoeffectors for just six P. aeruginosa MCPs have been reported (17, 22, 26, 28). P. aeruginosa also has a strong energy taxis response that is mediated by its Aer MCP (PA1561) (14). PA1561 mediates aerotaxis, and it is also required for full tactic responses to metabolizable compounds in swarm plates under both aerobic and anaerobic denitrifying conditions (5, 14). In initial work, we screened 18 single MCP mutants for responses to 68 different organic compounds in swarm plates but failed to identify any strains with defective chemotactic responses (A. Ferrández, A. C. Hawkins, and C. S. Harwood, unpublished data). There are several possible reasons for this. One reason is that some of the compounds that we tested do not have a cognate MCP. In these cases, the swarm rings that cells formed likely reflected energy taxis to the oxidizable substrate in the agar, as has been shown for E. coli (8). It is also possible that some of the compounds tested are true chemoattractants but are sensed by more than one P. aeruginosa MCP. If this is true, then a single mutant will not have an observable phenotype. This is, in fact, the case for the P. aeruginosa MCPs PctA, PctB, and PctC, which have overlapping specificities for most of the 20 amino acids that they collectively detect (26). Some MCPs may be specific for inorganic compounds or for repellents. These are classes of compounds for which behavioral responses cannot be easily screened in swarm plate assays. A final possibility, which we consider experimentally here, is that the energy taxis response of P. aeruginosa masks its chemotactic responses in some circumstances. To test this hypothesis, we screened a series of aer mcp double mutants by using swarm plate assays. This allowed us to assign a function to PA2652 as an MCP that senses malate.
Wild-type P. aeruginosa PAO1 and a set of MCP mutants were obtained from the University of Washington Pseudomonas aeruginosa PAO1 transposon mutant collection (16). The position of the transposon insertion was verified for each mutant as suggested by the library creators (http://www.genome.washington.edu/UWGC/pseudomonas/pdf/Mutant_Info.pdf). We constructed a deletion in the MCP PA4310 (pctB) by using an overlap extension PCR (15) because a suitable pctB mutant strain was not represented in the transposon mutant collection. We used pEX19Gm as the P. aeruginosa suicide vector (13), and sucrose counter-selection was used to obtain double recombinant strains, as previously described (7). Gentamicin was used at 50 μg per ml for P. aeruginosa and at 20 μg per ml for E. coli. Swarm plates consisted of a mineral salts medium solidified with 0.3% Noble agar and the appropriate chemoattractant as the sole carbon source (7). The aer pctB double mutant was constructed by the using the aer transposon mutant PTL14586 (Table 1) as the parent strain.
TABLE 1.
Strains constructed and tested using swarm plate assays
| MCP (gene name) | Mutant library strain designation (transposon positions)a | Strain designation for the aer mcp double mutant |
|---|---|---|
| PA0180 | PTL54327 (370/1173) | PAO1334 |
| PA1251 | PTL9939 (670/1626) | PAO1346 |
| PA1561 (aer) | PTL14586 (656/1566) | |
| PA1608 | PTL12842 (334/1626) | PAO1335 |
| PA1646 | PTL18909 (337/1959) | PAO1380 |
| PA2652 | PTL41758 (448/1686) | PAO1384 |
| PA2654 | PTL10830 (147/2145) | PAO1344 |
| PA2788 | PTL14831 (81/1596) | PAO1382 |
| PA4520 | PTL11386 (743/2022) | PAO1342 |
| PA4633 | PTL47884 (338/2139) | PAO1336 |
| PA5072 | PTL16847 (166/1944) | PAO1345 |
Original mutant designation given by the University of Washington P. aeruginosa PAO1 transposon mutant library creators. The nucleotide position of the transposon in the open reading frame followed by the size of the open reading frame in nucleotides is shown in parenthesis.
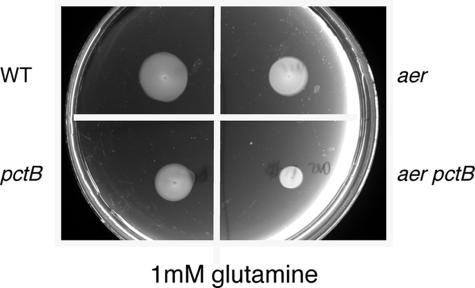
In initial experiments, we found that an aer mutant formed a smaller swarm ring than its wild-type parent in all carbon sources that we tested. This is illustrated in Fig. 1, which shows that an aer mutant formed a noticeably smaller swarm ring than the wild type in plates containing 1 mM glutamine (Fig. 1). PctB is the MCP that has been reported to be responsible for chemotaxis to glutamine (26). In agreement with this, a pctB deletion mutant also formed a smaller swarm ring than the wild type in glutamine swarm plates (Fig. 1). We also observed, however, that an aer pctB double mutant formed a smaller diameter swarm ring than either an aer or a pctB mutant (Fig. 1). Thus, energy taxis and chemotaxis additively contribute to the swarm ring that is formed on glutamine swarm plates.
FIG. 1.
Behavioral responses of P. aeruginosa PAO1 wild-type (WT), aer, aer pctB, and pctB strains as assayed in a mineral medium swarm plate that contained 1 mM glutamine as the sole carbon and energy source. Each quadrant of the plate was stab inoculated with a single colony, and the plate was incubated for 16 h at 37°C.
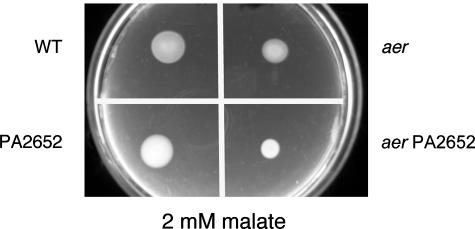
Amino acids are strong chemoattractants for P. aeruginosa (19, 20), and this may explain why energy taxis did not completely mask the chemotactic response to glutamine. We reasoned that energy taxis, however, might completely mask responses to organic compounds, such as succinate, that are relatively weak chemoattractants (19, 20). To investigate this, we constructed a series of different aer mcp double mutants (Table 1) by introducing an in-frame deletion construct of PA1561 (aer) that contained a gentamicin cassette into various mcp mutant strains, using methods similar to those described above. We then tested swarm ring formations on soft agar plates containing malate, succinate, 2-oxoglutarate, citrate, acetate, or glucose. These compounds were present in the soft agar plates at a final concentration of 2 mM. This screen resulted in the identification of a phenotype for 1 of the 10 aer mcp strains examined. The aer PA2652 double mutant strain PAO1384 formed a smaller swarm ring than the aer mutant in 2 mM malate swarm plates (Fig. 2). This suggested that the chemoreceptor encoded by the PA2652 gene senses malate.
FIG. 2.
Behavioral responses of P. aeruginosa PAO1 wild-type (WT), aer, aer PA2652, and PA2652 mutant strains as assayed in a mineral medium swarm plate that contained 2 mM malate as the sole carbon and energy source. Each quadrant of the plate was stab inoculated with a single colony, and the plate was incubated for 16 h at 37°C.
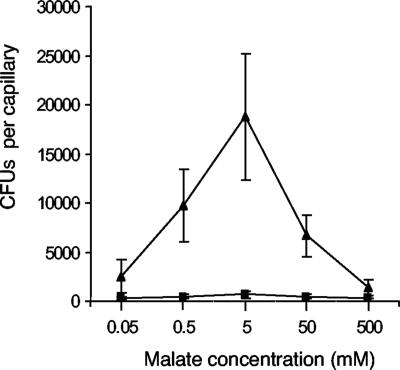
To confirm our initial assignment of PA2652 as an MCP that senses malate, we carried out a quantitative capillary assay (1, 10). This assay, unlike the swarm plate assay, does not depend on metabolism to generate a concentration gradient. Instead, diffusion of the compound from the mouth of a microcapillary tube sets up the concentration gradient. Cells respond to a chemoattractant by swimming up the gradient and into the tube. After a 30-min incubation, the number of cells in the tube was determined by plate counts (10). This assay is quantitative and extremely sensitive. Whereas wild-type cells were chemotactic to malate at concentrations ranging from 0.5 mM to 50 mM, the PA2652 mutant showed no attraction to malate at any of the concentrations tested (Fig. 3). The PA2652 mutant had a strong response to 10 mM arginine (100,000 ± 3,000 cells per capillary), which was on the same order as that observed for wild-type cells (9). Therefore, this strain does not have a generalized chemotactic defect. We generated a plasmid for use in complementing the PA2652 mutation by cloning the PA2652 gene and 500 bp of DNA upstream of the translational start site of PA2652 into pJH1Gm (12). Provision of the PA2652 gene in trans to the PA2652 mutant strain complemented the malate chemotaxis phenotype. The PA2652 mutant carrying the empty vector failed to accumulate to a level above that of the background, using a capillary filled with 10 mM malate, whereas the complemented strain was attracted to 10 mM malate (45,500 ± 7,000 cells per capillary).
FIG. 3.
The chemotactic responses of P. aeruginosa PAO1 (wild type) (▴) and of a PA2652 mutant (strain PTL41758) (▪) to various concentrations of malate in a capillary assay. The data are expressed as CFU and are the averages of six assays ± standard deviations.
Our results demonstrate that the strong energy taxis response of P. aeruginosa can dominate MCP-mediated metabolism-independent chemotactic responses, such as the response to malate. This is in contrast to E. coli, where Aer-mediated taxis to oxidizable carbon sources does not mask metabolism-independent responses (8). By screening P. aeruginosa mcp mutants that are also defective in energy taxis, we found that the sensitivity of the swarm plate screen was increased such that we were able identify PA2652 as a P. aeruginosa chemoreceptor specific for malate. Many flagellated bacteria are highly aerotactic due to energy taxis (3, 27), and energy taxis has been shown to be the dominant behavioral response of some species (3, 4). We therefore anticipate that this screening strategy will be generally useful for identifying the ligand specificities of MCPs from other bacteria. In addition, now that we understand that energy taxis responses can confound responses to specific compounds in swarm plates, it may make sense to turn to other assays, such as the qualitative capillary assay (21), that have not traditionally been used as a screening mode but in which energy taxis does not interfere, to identify MCP functionalities.
Acknowledgments
We thank Michael Jacobs, Colin Manoil, and Maynard Olson for providing the P. aeruginosa mutants from the University of Washington Genome Center P. aeruginosa PAO1 transposon mutant library.
This work was supported by Public Health Service grant GM56665.
Footnotes
Published ahead of print on 12 October 2007.
REFERENCES
- 1.Adler, J. 1973. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74:77-91. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, R. P., and I. B. Zhulin. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. USA 104:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandre, G., S. E. Greer, and I. B. Zhulin. 2000. Energy taxis is the dominant behavior in Azospirillum brasilense. J. Bacteriol. 182:6042-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandre, G., S. Greer-Phillips, and I. B. Zhulin. 2004. Ecological role of energy taxis in microorganisms. FEMS Microbiol. Rev. 28:113-126. [DOI] [PubMed] [Google Scholar]
- 5.Alvarez, C. 2007. Responses to low oxygen and energy taxis by Pseudomonas aeruginosa. Ph.D. thesis. University of Iowa, Iowa City.
- 6.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrández, A., A. C. Hawkins, D. T. Summerfield, and C. S. Harwood. 2002. Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J. Bacteriol. 184:4374-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greer-Phillips, S. E., G. Alexandre, B. L. Taylor, and I. B. Zhulin. 2003. Aer and Tsr guide Escherichia coli in spatial gradients of oxidizable substrates. Microbiology 149:2661-2667. [DOI] [PubMed] [Google Scholar]
- 9.Guvener, Z. T., D. F. Tifrea, and C. S. Harwood. 2006. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol. Microbiol. 61:106-118. [DOI] [PubMed] [Google Scholar]
- 10.Harwood, C. S., M. Rivelli, and L. N. Ornston. 1984. Aromatic acids are chemoattractants for Pseudomonas putida. J. Bacteriol. 160:622-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazelbauer, G. L., R. E. Mesibov, and J. Adler. 1969. Escherichia coli mutants defective in chemotaxis toward specific chemicals. Proc. Natl. Acad. Sci. USA 64:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hickman, J. W., D. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 102:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 14.Hong, C. S., M. Shitashiro, A. Kuroda, T. Ikeda, N. Takiguchi, H. Ohtake, and J. Kato. 2004. Chemotaxis proteins and transducers for aerotaxis in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 231:247-252. [DOI] [PubMed] [Google Scholar]
- 15.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, H.-E., M. Shitashiro, A. Kuroda, N. Takiguchi, H. Ohtake, and J. Kato. 2006. Identification and characterization of the chemotactic transducer in Pseudomonas aeruginosa PAO1 for positive chemotaxis to trichloroethylene. J. Bacteriol. 188:6700-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meier, V. M., P. Muschler, and B. E. Scharf. 2007. Functional analysis of nine putative chemoreceptor proteins in Sinorhizobium meliloti. J. Bacteriol. 189:1816-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moench, T. T., and W. A. Konetzka. 1978. Chemotaxis in Pseudomonas aeruginosa. J. Bacteriol. 133:427-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moulton, R. C., and T. C. Montie. 1979. Chemotaxis by Pseudomonas aeruginosa. J. Bacteriol. 137:274-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parales, R. E., and C. S. Harwood. 2002. Bacterial chemotaxis to pollutants and plant-derived aromatic molecules. Curr. Opin. Microbiol. 5:266-273. [DOI] [PubMed] [Google Scholar]
- 22.Shitashiro, M., H. Tanaka, C. S. Hong, A. Kuroda, N. Takiguchi, H. Ohtake, and J. Kato. 2005. Identification of chemosensory proteins for trichloroethylene in Pseudomonas aeruginosa. J. Biosci. Bioeng. 99:396-402. [DOI] [PubMed] [Google Scholar]
- 23.Slocum, M. K., and J. S. Parkinson. 1985. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: null phenotypes of the tar and tap genes. J. Bacteriol. 163:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stock, J. B., and M. G. Surette. 1996. Chemotaxis, p. 1103-1129. In F. C. Neidhart, R. Curtiss III, J. I. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 25.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 26.Taguchi, K., H. Fukutomi, A. Kuroda, J. Kato, and H. Ohtake. 1997. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology 143:3223-3229. [DOI] [PubMed] [Google Scholar]
- 27.Taylor, B. L., I. B. Zhulin, and M. S. Johnson. 1999. Aerotaxis and other energy-sensing behavior in bacteria. Annu. Rev. Microbiol. 53:103-128. [DOI] [PubMed] [Google Scholar]
- 28.Wu, H., J. Kato, A. Kuroda, T. Ikeda, N. Takiguchi, and H. Ohtake. 2000. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J. Bacteriol. 182:3400-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhulin, I. B., E. H. Rowsell, M. S. Johnson, and B. L. Taylor. 1997. Glycerol elicits energy taxis of Escherichia coli and Salmonella typhimurium. J. Bacteriol. 179:3196-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]