Abstract
This report describes the first functional analysis of a bacteriocin immunity gene from Propionibacterium freudenreichii and its use as a selection marker for food-grade cloning. Cloning of the pcfI gene (previously orf5 [located as part of the pcfABC propionicin F operon]) rendered the sensitive host 1,000-fold more tolerant to the propionicin F bacteriocin. The physiochemical properties of the 127-residue large PcfI protein resemble those of membrane-bound immunity proteins from bacteriocin systems found in lactic acid bacteria. The high level of immunity conferred by pcfI allowed its use as a selection marker for plasmid transformation in P. freudenreichii. Electroporation of P. freudenreichii IFO12426 by use of the pcfI expression plasmid pSL102 and propionicin F selection (200 bacteriocin units/ml) yielded 107 transformants/μg DNA. The 2.7-kb P. freudenreichii food-grade cloning vector pSL104 consists of the pLME108 replicon, a multiple cloning site, and pcfI expressed from the constitutive PpampS promoter for selection. The pSL104 vector efficiently facilitated cloning of the propionicin T1 bacteriocin in P. freudenreichii. High-level propionicin T1 production (640 BU/ml) was obtained with the IFO12426 strain, and the food-grade propionicin T1 expression plasmid pSL106 was maintained by ∼91% of the cells over 25 generations in the absence of selection. To the best of our knowledge this is the first report of an efficient cloning system that facilitates the generation of food-grade recombinant P. freudenreichii strains.
Dairy propionibacteria (PAB) are important in the food industry, with a long tradition of use in manufacture of Swiss-type cheese. It has been estimated that the annual production of hard cheeses which undergo propionic acid fermentation in Europe is 560,000 tons (17). Although PAB are applied in production of propionic acid and vitamin B12 and are also used as probiotics, it is only recently that the unexploited potential of PAB as a production host for nutraceuticals, enzymes, and antimicrobials has been considered (13, 14). Recognition of this potential requires further development of efficient molecular tools to implement biotechnologically relevant properties and for generation of PAB strains with improved genetic features (20, 32). Recently, several vector and transformation systems for genetic manipulation of Propionibacterium freudenreichii were published (4, 16, 18). The utility of these systems has been further improved by the characterization of promoter elements and secretion signal sequences (4, 25). Several excellent genetic studies of the production of porphyrins and tetrapyrrole compounds and of vitamin B12 in particular have demonstrated the potential of applying metabolic engineering to P. freudenreichii (19, 26, 27). Such technology is of utmost interest, particularly since P. freudenreichii is a GRAS (generally recognized as safe)-status organism approved by the U.S. Food and Drug Administration (2). However, the safe use of genetically modified strains in food-related applications requires cloning vectors that consist entirely of DNA from food-grade sources and that should be devoid of antibiotic resistance genes (7). Food-grade cloning systems utilizing different selection methods to obtain recombinant strains have been developed for a number of lactic acid bacteria. Preferred methods are complementation of auxotrophic phenotypes (carbohydrate or DNA metabolism) and use of dominant selection markers, such as bacteriocin resistance genes (6, 31).
The propionicin F bacteriocin is a 4.4-kDa negatively charged peptide produced by certain strains of P. freudenreichii (3). The maturation of propionicin F is unique and apparently involves both N- and C-terminal processing of a large proprotein where the mature bacteriocin peptide constitutes amino acids 102 to 145 of the 255-residue PcfA proprotein. Immediately downstream of pcfA reside pcfB, a radical S-adenosyl-methionine transferase, and pcfC, a proline peptidase, both of which are involved in maturation of the peptide bacteriocin. A bacteriocin-type ABC transporter (pcfD) is located further downstream.
Bacteriocin-producing gram-positive bacteria inherit two main mechanisms by which they obtain immunity to their own secreted antimicrobial peptides. Most class II bacteriocin systems contain an immunity gene encoding a small membrane-bound protein that generally confers protection to its cognate bacteriocin alone (12, 21, 24). In addition, some producers of lantibiotic bacteriocins rely on a more complex immunity mechanism consisting of an ABC-transporter system that functions cooperatively with the membrane-bound immunity protein to provide full protection (1, 23, 29). As expected, propionicin F-producing cells have been found to be immune to their own bacteriocin, but growth conditions that induced a bacteriocin-negative phenotype consistently rendered the same strain sensitive to propionicin F. This implied the presence of an immunity factor that is coregulated with the bacteriocin, but the genetic determinant responsible had not been identified (3).
Here we identify the propionicin F immunity gene (termed pcfI) that is used in the development of a food-grade cloning system for P. freudenreichii. It is furthermore shown that this system efficiently facilitated cloning and high-level production of the propionicin T1 bacteriocin.
MATERIALS AND METHODS
Bacterial strains, vectors, and media.
The bacterial strains and vectors are shown in Table 1. Escherichia coli was cultivated at 37°C in LB (1% [wt/vol] tryptone, 1% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl) medium supplemented with 100 μg/ml of ampicillin or 50 μg/ml kanamycin for propagation of plasmids. Propionibacteria were grown anaerobically at 30°C in sodium lactate broth (SLB) (10) supplemented with 3.4 μg/ml of chloramphenicol where appropriate.
TABLE 1.
Plasmids and strains used in this studya
| Plasmid or strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Plasmids | ||
| pCR-Blunt II-TOPO | E. coli cloning vector with ccd gene, Kmr + Zeor selection, blue-white screening; 3.5 kb | Invitrogen |
| pAMT1 | E coli-PAB shuttle vector Apr in E. coli and Cmr in PAB, derived from pUC18, pLME108 and cml/cmx PCR product; 6.3 kb | 4 |
| pTD104 | P4E::pctA promoter-gene fusion cloned in pAMT1; 6.9 kb | 4 |
| pTD10 | PAB cloning vector, Cmr in PAB, consisting of pLME108 and cml/cmxA PCR product derived from pAMT1; 3.6 kb | This study |
| pSL101 | PpampE::pcfI promoter-gene fusion cloned in pCR-Blunt II-TOPO; 4.4 kb | This study |
| pSL102 | pTD10 cloned as XbaI fragment into SpeI site of pSL101; 8.0 kb | This study |
| pSL103 | P. freudenreichii food-grade cloning vector, PpampE::pcfI propionicin F resistance marker, derived from pSL102; 3.1 kb | This study |
| pSL104 | P. freudenreichii food-grade cloning vector, propionicin F resistance from PpampS::pcfI, derived from pSL102; 2.7 kb | This study (see Fig. 3) |
| pSL106 | P4E::pctA promoter-gene fusion cloned in BamHI site of pSL104; 3.3 kb | This study |
| Strains | ||
| E. coli Top Ten | F−mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(araleu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| E. coli JM109 | F′ traD36 proA+ proB+ lacIq Δ(lacZ)M15/Δ(lac-proAB) glnV44 e14−gyrA96 recA1 relA1 endA1 thi hsdR17 | New England Biolabs |
| P. freudenreichii LMGT 2946 | Propionicin F producer | 3 |
| P. freudenreichii IFO12426 | High frequency of DNA transformation by electroporation | 4; IFO |
| P. thoenii 419b | Propionicin T1 producer | 10 |
| P. acidipropionici ATCC 4965 | Highly sensitive to propionicin T1 | ATCC |
| P. jensenii LMGT 3032 | pamA+ | 10 |
Abbreviations: ATCC, American Type Culture Collection (Rockville, MD); LMGT, Laboratory of Microbial Gene Technology, Ås, Norway; IFO, Institute of Fermentation Osaka, Japan; Cmr, chloramphenicol selection; Apr, ampicillin selection; Kmr, kanamycin selection; Zeor, zeomycin selection.
P. thoenii 419 was from the Environmental Bacteriology Culture Collection, University of the Orange Free State, Bloemfontein, Republic of South Africa.
General methods.
General molecular biological techniques used in this study were performed as described by Sambrook et al. (28) unless stated otherwise. Transformation of E. coli was performed according to the method of Inoue et al. (15). Plasmid DNA for cloning was purified with QIAprep spin columns (Qiagen, Hilden, Germany), while plasmid DNA for transformation of P. freudenreichii was prepared by use of Midi Prep columns (Qiagen). Restriction enzymes and T4 DNA ligase were purchased from NEB (New England BioLabs, Inc., Beverly, MA) or Fermentas (Vilnius, Lithuania). DNA amplification by PCR for cloning was done using 100-μl reaction mixtures, 2.5 units of Phusion polymerase (New England BioLabs, Inc., Beverly, MA), and 100 pmol of each primer. The PCR conditions included a polymerase activation and template-denaturing step at 98°C (30 s) followed by 35 cycles of denaturing at 98°C (10 s), annealing at 57 to 60°C (30 s), and polymerization at 72°C. DNA fragments from PCR amplification or restriction digests were analyzed by agarose gel electrophoresis and purified on QIAquick purification columns (Qiagen). DNA sequencing was performed with a version 3.1 BigDye Terminator cycle sequencing ready reaction kit and an Applied Biosystems model 3100 genetic analyzer (Applied Biosystems, Foster City, CA). All products were used according to the instructions of the manufacturers.
DNA preparation from P. freudenreichii.
Plasmids from P. freudenreichii were purified using Qiagen MiniPrep or Midi Prep columns as previously described (4) and cells from 5- or 200-ml overnight cultures, respectively. Isolation of total DNA from P. freudenreichii was done using a 5-ml overnight culture (A620 ∼0.5) and Advamax beads according to the recommendation of the manufacturer (Advanced Genetic Technologies Corp., Gaithersburg, MD).
Plasmid constructions.
Propionibacterium replicating cloning vector pTD10 (Table 1; also see Fig. S1 in the supplemental material) was constructed from the pAMT1 shuttle vector by PCR using primers pTD10-f and pTD10-r (see Table S1 in the supplemental material). The 3.6-kb amplified fragment covering the cmlA-cmxA and the pLM108 repAB genes was digested with KpnI and circulated by intramolecular ligation. The ligation reaction mixture was heat inactivated, precipitated, and electroporated into P. freudenreichii IFO12426.
In order to investigate its biological function, the orf5 gene was cloned under the control of the strong PpampE promoter (4) by use of a two-step PCR strategy to construct a promoter-gene fusion as described by Brede et al. (4). The orf5 gene (pcfI) from the propionicin F locus was amplified with primers orf5-f and orf5-r, and the PpampE promoter fragment was generated with primers PAMP4 and PAMP3 (see Table S1 in the supplemental material). In the second PCR step, the PpampE and orf5 fragments were mixed to serve as templates and amplified using primers PAMP4 and orf5-r. In the resulting product, the PpampE promoter element was spliced to the coding sequence of orf5 by an extension overlap at the ATG initiation codon. The PpampE::orf5 fragment was cloned in Topo-zero blunt vector (Invitrogen) to yield the 4.4-kb plasmid pSL101 (Table 1; also see Fig. S1 in the supplemental material). Next, to produce a plasmid for shuttling from E. coli to PAB, XbaI-digested pTD10 vector was inserted into the SpeI site of pSL101. The resulting pSL102 plasmid (Table 1; see also Fig. S1 in the supplemental material) was introduced into the propionicin F-sensitive strain P. freudenreichii IFO12426.
To construct vector pSL103 (Table 1; also see Fig. S1 in the supplemental material) from plasmid pSL102, PCR amplification using primer PSL103-fwd in combination with T7-fwd was carried out. The obtained 3.0-kb fragment was digested with ApaI, circularized by intramolecular ligation, and propagated in P. freudenreichii IFO12426 by use of propionicin F as a selective agent. The 2.7-kb pSL104 vector (see Fig. 3 and Table 1) was generated from pSL102 by use of the phosporylated PCR product obtained with the primer combination PSL103-fwd and PAMP8 (see Table S1 in the supplemental material) to amplify the pLME108 repAB genes and the PpampS promoter (4) fused with the pcfI gene. A fragment containing the P4E promoter and the pctA gene (4) was amplified with primers P4C-BamHI and PT1-419PC-BamHI from the pTD104 plasmid (Table 1; also see Fig. S1 and Table S1 in the supplemental material), digested with BamHI, and cloned in BamHI-treated and dephosporylated pSL104 vector to yield the expression plasmid pSL106 (see Table 1 and Fig. S1 in the supplemental material), which was introduced into P. freudenreichii IFO12426 by use of propionicin F as the selective agent.
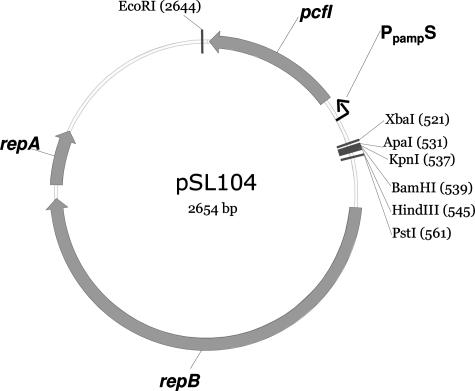
FIG. 3.
Schematic presentation of the pSL104 food-grade cloning vector.
DNA transformation of P. freudenreichii and use of pcfI as a selection marker.
Transformation of P. freudenreichii and selection of plasmid recipient cells were performed by electroporation as described by Brede et al. (4). Plasmids carrying the pcfI gene as a marker were selected using a pour plate method. A 100-μl aliquot of the electroporated cell suspension was mixed with 25 ml of SLB medium (1.2% agar, 55°C) containing 200 bacteriocin units (BU)/ml of propionicin F, where 1 BU is defined as the amount of propionicin F that produced 50% growth inhibition in a 0.2-ml culture of the P. freudenreichii ISU-P59 strain. This mixture was poured into a sterile petri dish and allowed to solidify. The plates were incubated anaerobically at 30°C for 5 to 7 days before transformants appeared in the agar. To confirm the presence of the pSL102 plasmid, 96 randomly picked colonies were replica plated on SLB plates containing chloramphenicol (3.4 μg/ml) in each experiment.
Preparation of propionicin F.
LMGT 2946 was grown anaerobically in MRS broth at 30°C for 72 h before propionicin F was precipitated from the cell-free supernatant by use of 40% wt/vol ammonium sulfate. The bacteriocin (∼5,000 BU/ml) was dissolved in distilled water, filter sterilized (0.2 μM), and stored at −20°C.
Propionicin F immunity assay.
Propionicin F tolerance in P. freudenreichii strains and clones was quantitatively determined by a microtiter plate assay (12). Each well of the microtiter plate contained 50 μl of 2-fold serial dilutions in SLB of the ammonium sulfate-precipitated, filter-sterilized propionicin F and 150 μl of a 100-fold-diluted overnight culture of the test strain or clone. The plates were incubated anaerobically at 30°C for 24 h, and growth was measured spectrophotometrically (A620) using a microtiter plate reader (Multiscan Ascent; Labsystems, Vantaa, Finland). The immunity level was determined as the minimum concentration (BU/ml) of propionicin F that produced 50% growth inhibition of the test bacterium compared to the results obtained with a culture without added bacteriocin.
Nucleotide sequence accession number.
The nucleotide sequence for pcfI was submitted to GenBank under accession no. AY587566.2.
RESULTS AND DISCUSSION
The small-membrane protein PcfI (Orf5) confers immunity to propionicin F.
Bacteriocin producers, including the propionicin F strains, need to protect themselves from the action of the antimicrobial peptide. The P. freudenreichii strain LMGT 2946 exhibited a consistent pattern of an immunity factor coordinately expressed with the bacteriocin. In contrast, a propionicin F-negative culture of this strain was sensitive to the externally added bacteriocin. The propionicin F locus contains several genes with inferred function in bacteriocin maturation and transport, but the cognate immunity system remained unrecognized (3).
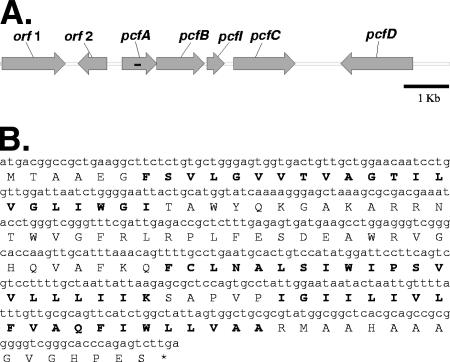
Most bacteriocin loci of gram-positive bacteria contain a dedicated immunity gene that usually resides directly downstream of and is cotranscribed with the bacteriocin structural gene (9). The proteins encoded by these genes are specific and almost exclusively confer immunity to their own bacteriocins alone. These immunity proteins are usually 50 to 150 amino acid residues in length, and they share few or no sequence similarities (9). Investigation of the propionicin F locus for candidate immunity genes brought our attention to orf5, encoding a 127-amino-acid-residue putative membrane protein with three predicted transmembrane helices (Fig. 1B). The pcfABC genes of the propionicin F locus are organized in an operon-like structure in which the orf5 gene resides between pcfB and pcfC (Fig. 1A).
FIG. 1.
(A) Organization of the pcfA, pcfB, pcfI, pcfC, and pcfD genes involved in propionicin F biosynthesis and immunity (GenBank accession no. AY587566). The black bar inside pcfA indicates the part encoding the propionicin F peptide. For further description, see text. (B) The nucleotide sequence and translation of the pcfI immunity gene (GenBank protein accession no. ABU97165.1). Bold characters indicate predicted transmembrane helical segments of the immunity protein analyzed by use of the Toppred algorithm (33).
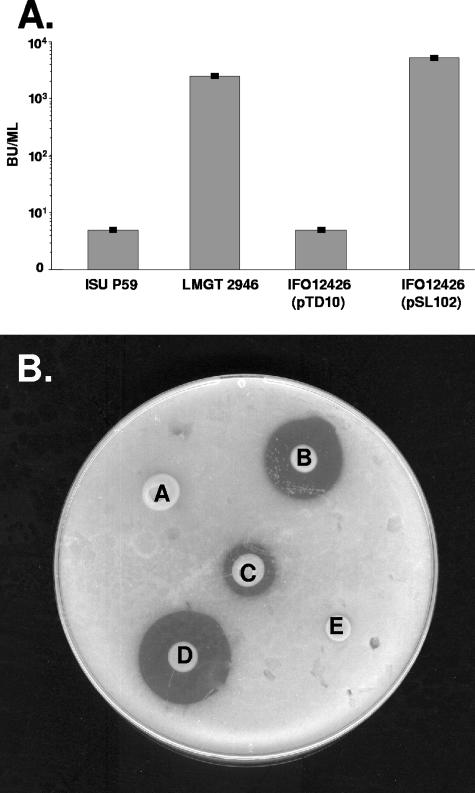
In order to ensure efficient expression, the orf5 gene was cloned behind the constitutive PpampE promoter in the pSL102 plasmid construct. Expression of orf5 in a propionicin F-sensitive strain increased bacteriocin tolerance approximately 1,000-fold (Fig. 2A). This demonstrates that the orf5 gene encodes the cognate immunity protein of the propionicin F bacteriocin; thus, orf5 was annotated pcfI. The immunity assay (Fig. 2A) showed that P. freudenreichii IFO12426 transformants containing pSL102 (PpampE and pcfI) actually tolerated higher concentrations of propionicin F than the wild-type producer strain of P. freudenreichii LMGT 2946. Similar observations have been reported for lacticin 3147, with which it was shown that the protection towards the bacteriocin depended on the level of expression of the itnI immunity gene (22). To further investigate this phenomenon, the MIC for the pSL102 clone was compared to that of clone pSL104, where pcfI is under control of the weaker PpampS promoter (Table 1). This experiment showed that expression of pcfI from the PpampS promoter resulted in fourfold-reduced protection (5,120 BU/ml versus 1,280 BU/ml). This might imply that the concentration of propionicin F tolerated by LMGT 2946 depends on the level at which the pcfI gene is expressed. Whether the propionicin F-sensitive phenotype observed in bacteriocin-negative cultures of LMGT 2946 (3) could have resulted from down-regulated transcription of the pcfABIC operon remains to be addressed.
FIG. 2.
(A) Protection against the antimicrobial action of propionicin F conferred by the pcfI gene. The results represent the averages of the results of three independent experiments, and standard deviations are indicated. (B) Food-grade expression of propionicin T1 from IFO12426 (pSL106) compared to that of wild-type producer strain P. thoenii 419. A, IFO12426; B, P. thoenii419; C, IFO12426 (pTD104); D, IFO12426 (pSL106); E, IFO12426 (pSL104).
It has been shown that bacteriocins bind to specific receptor molecules: for instance, nisin targets lipid II (5), while lactococcin A targets the mannose phosphotransferase system (8). The NisI immunity protein has been shown to interact directly with nisin (30), whereas the LciA immunity protein forms a complex with the receptor and the bacteriocin only when lactococcin A is present (8). The physiochemical properties of PcfI are consistent with either type of immunity mechanism provided by membrane-bound proteins such as LciA or NisI. It is conceivable that PcfI functions in a similar fashion, probably via an interaction between the immunity protein and the propionicin F bacteriocin on the cell surface.
The use of pcfI as a selective marker for plasmid transformation of P. freudenreichii.
Recently, the development of efficient transformation protocols and cloning vectors has demonstrated a potential for generating recombinant P. freudenreichii strains with improved production of vitamins and antimicrobials (4, 27). However, there existed no system for food-grade cloning of P. freudenreichii. Efficient food-grade cloning systems based on bacteriocin immunity genes as dominant resistance markers, and applying the cognate bacteriocin as a selective agent, have been developed for both lactococci and lactobacilli (22, 31). The prospect of food-grade cloning of P. freudenreichii prompted investigation of the utility of pcfI as a selection marker for gene transformation. All transformation experiments were conducted with plasmid prepared from P. freudenreichii due to the high restriction barrier exhibited towards DNA from E. coli (4, 16, 18). Initial experiments were aimed at determining what concentrations of propionicin F provided efficient selection without exhausting the protection obtained from the PcfI protein. By use of a microtiter plate format, twofold serial dilutions of both sterile-filtered propionicin F and chloramphenicol were prepared. Electrocompetent IFO12426 cells were transformed with either 1 μg pAMT1 (control) or 1 μg pSL102 and incubated for 3 h before they were added to the microtiter plate (400-fold final dilution). The plate was incubated anaerobically until control cultures without selection reached stationary phase. Determination of the MIC50 concentration of propionicin F showed that 200 BU/ml of propionicin F provided efficient selection. Therefore, 25-ml SLB-agar plates containing 200 BU/ml of propionicin F were applied in an attempt to select transformants by use of either a spread plate or a pour plate method. On the spread plates, a very high background level of nontransformed cells appeared. Notably, use of the nisin immunity gene nisI as a primary selection marker was possible in liquid but not on solid medium (11, 31). Therefore, the pour plate technique was tested for propionicin F selection, which proved to be very efficient, yielding ∼107 colonies/μg pSL102 plasmid after 5 days of incubation. The presence of the pSL102 plasmid in randomly selected colonies was verified in all transformants tested by replica plating on chloramphenicol. Furthermore, less than a total of 102 background colonies appeared with propionicin F selection from cells transformed with the pAMT1 vector. Thus, compared to chloramphenicol results, the use of propionicin F as a selective agent yielded an identical number of transformants, but a slightly higher background level of nontransformed cells appeared. Encouraged by the efficient combination of propionicin F selection with the pcfI gene as a selective marker, we constructed food-grade cloning vectors devoid of antibiotic resistance genes and other non-PAB DNA. For this objective the 3.0-kb fragment of the pSL102 vector containing only the pLME108 replicon and the pcfI gene with the PpampE promoter was PCR amplified and circularized by intramolecular ligation into the pSL103 vector (see Fig. S1 in the supplemental materials). However, instability in the absence of propionicin F selection impaired utility of the pSL103 vector. In order to overcome the problem of instability, the 2.7-kb pSL104 vector (Fig. 3), in which pcfI expression is directed by the weaker PpampS promoter (4), was designed. The pSL104 recipient cells were easily selected using propionicin F and no signs of vector instability were observed.
In order to test the utility and robustness of the cloning system, it was decided to clone and express the propionicin T1 bacteriocin. A previous study showed that high amounts of propionicin T1 were produced by P. freudenreichii when the pctA gene was expressed from the P4E promoter (4). A PCR-generated P4E::pctA fragment was ligated into the BamHI site of the pSL104 vector, and the ligation mixture was electroporated into IFO12426. The transformation was diluted into proper aliquots, and propionicin F-resistant transformants were selected using the pour plate technique. The colonies were screened for production of the propionicin T1 bacteriocin by agar overlay of the P. acidipropionici ATCC 4965 indicator strain. Approximately 0.2% of the colonies produced propionicin T1. Two transformants were isolated, and the integrity of the pSL106 plasmid carrying the P4E::pctA insert was confirmed by restriction analysis and DNA sequencing. Bacteriocin production by the pSL106 and the pTD104 clones was compared to that of the wild-type propionicin T1 producer strain, P. thoenii 419 (Fig. 2B). In SLB broth the pSL106 clone produced 640 BU/ml, which is equal to the amount presented in previous reports on P4E-controlled propionicin T1 expression in P. freudenreichii (4). The utility of a food-grade cloning system greatly depends on stable maintenance of the recombinant gene. In order to assess segregational stability, the pSL106 clone was cultivated without selection for 25 generations (four serial culture transfers using 1% inoculums every 48 h). Throughout this period, plate samples were analyzed, with results showing that the proportion of propionicin T1-positive colonies decreased linearly from 100 to 91%, with a calculated plasmid loss of 0.33% per generation. This demonstrates that the stability of the pSL104 vector is compatible with industrial applications.
Concluding remarks.
This report presents the first molecular characterization of a bacteriocin immunity system from Propionibacterium species. Furthermore, these results were utilized to develop an efficient system that offers high-level food-grade recombinant gene expression in P. freudenreichii with a potential for generation of strains with improved industrial features.
Supplementary Material
Acknowledgments
D. A. Brede and T. Faye were funded by the Norwegian Research Council.
Footnotes
Published ahead of print on 12 October 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aso, Y., K. Okuda, J. Nagao, Y. Kanemasa, N. Thi Bich Phuong, H. Koga, K. Shioya, T. Sashihara, J. Nakayama, and K. Sonomoto. 2005. A novel type of immunity protein, NukH, for the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. Biosci. Biotechnol. Biochem. 69:1403-1410. [DOI] [PubMed] [Google Scholar]
- 2.Barefoot, S. F., and C. G. Nettles. 1993. Antibiosis revisited: bacteriocins produced by dairy starter cultures. J. Dairy Sci. 76:2366-2379. [DOI] [PubMed] [Google Scholar]
- 3.Brede, D. A., T. Faye, O. Johnsborg, I. Odegard, I. F. Nes, and H. Holo. 2004. Molecular and genetic characterization of propionicin F, a bacteriocin from Propionibacterium freudenreichii. Appl. Environ. Microbiol. 70:7303-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brede, D. A., T. Faye, M. P. Stierli, G. Dasen, A. Theiler, I. F. Nes, L. Meile, and H. Holo. 2005. Heterologous production of antimicrobial peptides in Propionibacterium freudenreichii. Appl. Environ. Microbiol. 71:8077-8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breukink, E., I. Wiedemann, C. van Kraaij, O. P. Kuipers, H. Sahl, and B. de Kruijff. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361-2364. [DOI] [PubMed] [Google Scholar]
- 6.Bron, P. A., M. G. Benchimol, J. Lambert, E. Palumbo, M. Deghorain, J. Delcour, W. M. de Vos, M. Kleerebezem, and P. Hols. 2002. Use of the alr gene as a food-grade selection marker in lactic acid bacteria. Appl. Environ. Microbiol. 68:5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vos, W. M. 1999. Safe and sustainable systems for food-grade fermentations by genetically modified lactic acid bacteria. Int. Dairy J. 9:3-10. [Google Scholar]
- 8.Diep, D. B., M. Skaugen, Z. Salehian, H. Holo, and I. F. Nes. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. USA 104:2384-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ennahar, S., T. Sashihara, K. Sonomoto, and A. Ishizaki. 2000. Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol. Rev. 24:85-106. [DOI] [PubMed] [Google Scholar]
- 10.Faye, T., D. A. Brede, T. Langsrud, I. F. Nes, and H. Holo. 2002. An antimicrobial peptide is produced by extracellular processing of a protein from Propionibacterium jensenii. J. Bacteriol. 184:3649-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrington, A., and C. Hill. 1991. Construction of a bacteriophage-resistant derivative of Lactococcus lactis subsp. lactis 425A by using the conjugal plasmid pNP40. Appl. Environ. Microbiol. 57:3405-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holo, H., O. Nilssen, and I. F. Nes. 1991. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J. Bacteriol. 173:3879-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugenholtz, J., J. Hunik, H. Santos, and E. Smid. 2002. Nutraceutical production by propionibacteria. Lait 82:103-112. [Google Scholar]
- 14.Hugenholtz, J., and E. J. Smid. 2002. Nutraceutical production with food-grade microorganisms. Curr. Opin. Biotechnol. 13:497-507. [DOI] [PubMed] [Google Scholar]
- 15.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 16.Jore, J. P., N. van Luijk, R. G. Luiten, M. J. van der Werf, and P. H. Pouwels. 2001. Efficient transformation system for Propionibacterium freudenreichii based on a novel vector. Appl. Environ. Microbiol. 67:499-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerjean, J. R., S. Condon, R. Lodi, G. Kalantzopoulos, J. F. Chamba, T. Suomalainen, T. Cogan, and D. Moreau. 2000. Improving the quality of European hard-cheeses by controlling of interactions between lactic acid bacteria and propionibacteria. Food Res. Int. 33:281-287. [Google Scholar]
- 18.Kiatpapan, P., Y. Hashimoto, H. Nakamura, Y.-Z. Piao, H. Ono, M. Yamashita, and Y. Murooka. 2000. Characterization of pRGO1, a plasmid from Propionibacterium acidipropionici, and its use for development of a host-vector system in propionibacteria. Appl. Environ. Microbiol. 66:4688-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiatpapan, P., N. Kanamnuay, B. Jongserijit, Y. Z. Piao, M. Yamashita, and Y. Murooka. 2003. Production of 5-aminolevulinic acid and vitamin B12 using metabolic engineering of Propionibacterium freudenreichii, p. 221-232. In B. C. Saha (ed.), ACS Symposium no. 862: fermentation biotechnology. Oxford University Press, New York, NY.
- 20.Kiatpapan, P., and Y. Murooka. 2002. Genetic manipulation system in propionibacteria. J. Biosci. Bioeng. 93:1-8. [PubMed] [Google Scholar]
- 21.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. De Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 22.McAuliffe, O., C. Hill, and R. P. Ross. 2000. Identification and overexpression of itnl, a novel gene which confers immunity to the two-component lantibiotic lacticin 3147. Microbiology 146:129-138. [DOI] [PubMed] [Google Scholar]
- 23.McAuliffe, O., R. P. Ross, and C. Hill. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25:285-308. [DOI] [PubMed] [Google Scholar]
- 24.Nes, I. F., and H. Holo. 2000. Class II antimicrobial peptides from lactic acid bacteria. Biopolymers 55:50-61. [DOI] [PubMed] [Google Scholar]
- 25.Piao, Y., N. Kawaraichi, R. Asegawa, P. Kiatpapan, H. Ono, M. Yamashita, and Y. Murooka. 2004. Molecular analysis of promoter elements from Propionibacterium freudenreichii. J. Biosci. Bioeng. 97:310-316. [DOI] [PubMed] [Google Scholar]
- 26.Piao, Y., P. Kiatpapan, M. Yamashita, and Y. Murooka. 2004. Effects of expression of hemA and hemB genes on production of porphyrin in Propionibacterium freudenreichii. Appl. Environ. Microbiol. 70:7561-7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piao, Y., M. Yamashita, N. Kawaraichi, R. Asegawa, H. Ono, and Y. Murooka. 2004. Production of vitamin B12 in genetically engineered Propionibacterium freudenreichii. J. Biosci. Bioeng. 98:167-173. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Siegers, K., and K. D. Entian. 1995. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 61:1082-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takala, T. M., and P. E. Saris. 2006. C terminus of NisI provides specificity to nisin. Microbiology 152:3543-3549. [DOI] [PubMed] [Google Scholar]
- 31.Takala, T. M., and P. E. Saris. 2002. A food-grade cloning vector for lactic acid bacteria based on the nisin immunity gene nisI. Appl. Microbiol. Biotechnol. 59:467-471. [DOI] [PubMed] [Google Scholar]
- 32.Van Luijk, N., M. P. Stierli, S. M. Schwenninger, C. Herve, G. Dasen, J. P. M. Jore, P. H. Pouwels, M. J. van der Werf, M. Teuber, and L. Meile. 2002. Genetics and molecular biology of propionibacteria. Lait 82:45-57. [Google Scholar]
- 33.von Heijne, G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.