Abstract
Cell culture assays in various formats have been used to study the infectivity of Cryptosporidium spp. as well as to determine the infectivity of naturally occurring oocysts in water. Currently, cell culture assays for infectious Cryptosporidium spp. in water have largely been limited to practice in research laboratories. One obstacle to the routine use of Cryptosporidium cell culture assays for the analysis of water samples is the coordination of water sample collection and processing with readiness of cell culture monolayers. For most Cryptosporidium cell culture assays, monolayers are allowed to develop for 24 to 48 h to reach 80 to 100% confluence prior to inoculation. In this study, we used immunofluorescent assay microscopy to evaluate freshly confluent (2-day-old) and aged (8- to 67-day-old) HCT-8 cell monolayers for their ability to support Cryptosporidium parvum infection. HCT-8 monolayers as old as 67 days were clearly shown to support infection. In two of three experiments, aged monolayers (8- to 11-day-old and 11- to 22-day-old, respectively) developed the same number of C. parvum clusters of infection as freshly confluent monolayers. Results suggest that it may be possible to use cell monolayers from freshly confluent to 3 weeks old on hand for infectivity assays without having to schedule sample processing to coincide with development of freshly confluent monolayers. This would make Cryptosporidium cell culture assays much more feasible for water quality and utility laboratories.
The parasite Cryptosporidium has been responsible for waterborne disease outbreaks worldwide and presents a challenge to the water industry and regulatory agencies. The disease cryptosporidiosis is usually a self-limiting diarrhea in healthy adults but may be life threatening for persons with weakened immune systems. Cryptosporidium oocysts are ubiquitous in environmental waters, are resistant to chlorination, and have been found in treated drinking water (1, 18).
The current U.S. EPA method for the detection of Cryptosporidium in water is based on immunofluorescent assay microscopy (30). Although this method is being used for the current Long-Term 2 Enhanced Surface Water Treatment Rule in the United States, it is not capable of determining the species or infectivity of the detected Cryptosporidium oocysts. Cryptosporidium species capable of infecting both humans and animals are detected frequently in environmental water samples (21, 24, 36). Therefore, these are significant limitations when gauging the public health risk posed by waterborne Cryptosporidium.
Mouse models have frequently been used to determine the infectivity of Cryptosporidium oocysts (4, 9, 16, 20). However, animal infectivity assays are costly and not feasible for the analysis of environmental samples. The use of cell culture to study the intracellular development of Cryptosporidium was described over 2 decades ago (6). In addition to the study of Cryptosporidium biology, cell culture methods have been used to study Cryptosporidium inactivation by disinfectants (4, 13, 23, 26, 28), efficacy of chemotherapeutic agents (3, 5, 34, 37), and detection of infectious oocysts in water samples (1, 7, 10, 17). The developmental stages of the parasite in cell culture are detected using a variety of techniques, including immunofluorescent assay microscopy (27), flow cytometry (31), reverse transcription-PCR (22), and quantitative PCR (5, 8, 11, 15).
One obstacle to the routine use of Cryptosporidium cell culture assays for the analysis of water samples is the coordination of water sample collection and processing with readiness of cell culture monolayers. For most Cryptosporidium cell culture assays, monolayers are allowed to develop for 24 or 48 h to reach 80 to 100% confluence prior to inoculation and then processed for detection of infection 48 to 72 h postinoculation (2, 8, 12, 22, 25, 28, 35). Since it is desirable to process water samples as soon as possible after collection and since most water samples are collected off-site and shipped to a processing laboratory, the required logistics can greatly impede the practical application of Cryptosporidium cell culture assays. Flexibility in readiness of cell culture monolayers would make these assays much more feasible for water quality and utility laboratories.
The human ileocecal adenocarcinoma HCT-8 cell line was previously found to support the greatest amount of Cryptosporidium parvum infection compared to several other cell lines (19, 29) and is commonly used for Cryptosporidium infectivity assays (1, 2, 7, 10, 12, 13, 23, 25, 27, 33). In the present study, we investigated the ability of freshly confluent (2-day-old) and aged (8- to 67-day-old) HCT-8 cell monolayers to support C. parvum infection.
MATERIALS AND METHODS
HCT-8 cell monolayers and media.
Human ileocecal adenocarcinoma (HCT-8; ATCC CCL244) cell monolayers were maintained in a 5% CO2 atmosphere at 36°C. Cell culture maintenance medium consisted of RPMI 1640 medium with Glutamax (catalog no. 61870-036; Invitrogen, Carlsbad, CA) with 5% defined heat-inactivated fetal bovine serum (catalog no. SH30070.03-HI; HyClone, Logan, UT), 20 mM HEPES buffer, 100,000 U of penicillin G liter−1, 100 mg of streptomycin liter−1, and 250 μg of amphotericin B liter−1. For infectivity assays, eight-well Lab-Tek II chamber slides (catalog no. 154534; Nalge Nunc International, Naperville, IL) were seeded with 500 μl of RPMI maintenance medium containing 4 × 105 cells per ml and incubated at 36°C in a 5% CO2 atmosphere in gas-permeable plastic bags (catalog no. WS14214350; Demco, Madison, WI). Monolayers reached 90% to 100% confluence after 2 days postseeding (baseline), and monolayers aged up to 67 days postseeding were maintained with weekly medium changes.
For inoculation of chamber slides, oocysts were resuspended in immunofluorescence assay (IFA) inoculation medium, which consisted of RPMI 1640 with Glutamax medium with 10% fetal bovine serum, 20 mM HEPES buffer, 100,000 U of penicillin G liter−1, 100 mg of streptomycin liter−1, 250 μg of amphotericin B liter−1, and 100 mg of kanamycin liter−1.
Pretreatment of oocysts.
Three different lots of C. parvum Iowa isolate oocysts (mouse propagated; Waterborne, New Orleans, LA) were used, one for each of three experiments. Oocysts were 30 days, 20 days, and 18 days old postshedding for experiments 1 through 3, respectively.
Oocysts were pretreated for cell culture by suspension in acidified (pH 2.0) Hanks’ balanced salt solution supplemented with trypsin (type II-S porcine pancreas, catalog no. T7409; Sigma Chemical Co., St. Louis, MO) to a final sample concentration of 1% (wt/vol) and then incubated at 37°C for 1 h with 10 s of vortexing every 15 min, as previously described (8). After incubation, an equal volume of IFA inoculation medium was added and samples were centrifuged at maximum speed in a microcentrifuge for 2 min without the brake. Samples were then immediately and carefully aspirated down to 50 μl; 500 μl of IFA inoculation medium was added, and samples were centrifuged as described above. Samples were carefully aspirated to 20 μl and resuspended in 380 μl of prewarmed (37°C) IFA inoculation medium (total volume of 400 μl).
Inoculation of chamber slides.
Just prior to inoculation, maintenance medium was aspirated from cell culture chambers without disturbing the monolayers and 100 μl of prewarmed (37°C) inoculation medium was immediately added to each chamber. Monolayers were inoculated with approximately 1,000, 800, and 1,000 oocysts each for experiments 1 through 3, respectively (approximately 1:200 oocyst:HCT-8 cell ratio). Based on preliminary evaluation of each lot of oocysts, these levels of inocula were expected to produce approximately 100 to 200 clusters of infection per monolayer. Inoculated chamber slides were incubated at 36°C in a 5% CO2 atmosphere in gas-permeable plastic bags for 65 to 72 h to allow for the development of infection.
Staining of monolayers.
Monolayers were stained as previously described (27), with minor modifications. Medium was carefully aspirated from chambers, ensuring that monolayers were not disturbed, without a washing step, as this could potentially wash off some Cryptosporidium life stages. Five hundred microliters of room temperature absolute methanol was added to each chamber, and samples were incubated at room temperature for 10 min; then, the methanol was aspirated and monolayers were allowed to air dry. Blocking buffer (500 μl) was added to each chamber. Blocking buffer consisted of 1× phosphate-buffered saline (PBS), 2% goat serum, and 0.002% Tween 20. Slides were then placed on a rocking platform for 30 min at room temperature. After blocking, 150 μl of unlabeled primary rat antisporozoite antibody (catalog no. A600; Waterborne) at 0.008 mg ml−1 in 1× PBS was added to each monolayer and slides placed on a rocking platform for 60 min at room temperature. This polyclonal immunoglobulin G primary antibody is made against sporozoites of C. parvum and is strongly reactive with all in vitro intracellular reproductive stages, including merozoites. After incubation, the primary antibody was aspirated and monolayers were washed twice with 500 μl of 1× PBS. Next, 150 μl of goat anti-rat immunoglobulin G fluorescein isothiocyanate-labeled secondary antibody (30 μl added to 10 ml 1× PBS, catalog no. F6258; Sigma) was added to each monolayer. Slides were then placed on a rocking platform, protected from light, and incubated for 60 min at room temperature. After incubation, the secondary antibody was aspirated and monolayers were washed twice with 500 μl of 1× PBS. After aspiration, 200 μl of U.S. EPA method 1623 DABCO (1,4-diazabicyclo[2.2.2]octane) mounting medium (30) was added to each chamber. Chamber slides were wrapped in parafilm and stored protected from light at 4°C until examined using a Zeiss Axiovert 200 inverted epifluorescent microscope (Zeiss, Inc., Thornwood, NY). A cluster of infection was defined as at least three Cryptosporidium intracellular developmental stages within a 175-μm-diameter area.
Data analysis and statistics.
The significances of differences in the numbers of clusters of infection per fresh or aged monolayer were compared using STATA 9 (College Station, TX) and one-way analysis of variance with post hoc Bonferroni pairwise mean comparisons. An alpha level of 0.05 was used for all statistical tests.
RESULTS
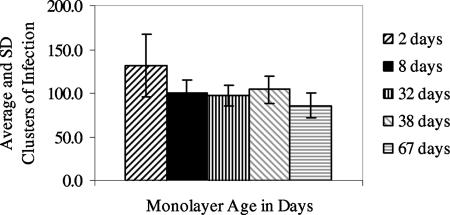
Experiment 1 evaluated baseline 2-day-old freshly confluent monolayers and 8-, 32-, 38-, and 67-day-old monolayers. Monolayers were maintained successfully for extended periods of time by simply changing the cell culture medium once per week, although some replicates of the 32-day-old or older monolayers peeled off partially from the corners of the slides. Based on counts of clusters of infection and numbers of oocysts used to inoculate cell monolayers, infectivity of the lot of oocysts used for this experiment was 10.4%. Aged monolayers were found to be susceptible to infection (Fig. 1). The difference in clusters of infection per monolayer for the baseline 2-day-old monolayers and all other monolayer ages was significant, but some of the differences were small (e.g., P = 0.044 for 2- and 38-day-old monolayers). However, no differences in clusters of infection per monolayer were seen between any of the 8- to 67-day-old monolayers (P > 0.05).
FIG. 1.
Average (±standard deviation [SD]) clusters of infection for experiment 1 with fresh and aged monolayers (n, number of replicate monolayers): 2 day old, n = 10; 8 day old, n = 16; 32 day old, n = 16; 38 day old, n = 8; 67 day old, n = 8.
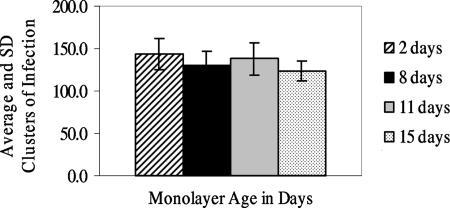
Since 32-day-old and older monolayers showed some peeling, experiments 2 and 3 evaluated monolayers of more practical ages. Experiment 2 evaluated baseline 2-day-old freshly confluent monolayers and 8-, 11-, and 15-day-old monolayers. Based on counts of clusters of infection and numbers of oocysts used to inoculate cell monolayers, infectivity of the lot of oocysts used for this experiment was 16.7%. Means and standard deviations of clusters of infection per monolayer are presented in Fig. 2. A significant difference in the clusters of infection per monolayer was observed only for the baseline 2-day-old and the 15-day-old monolayers (P = 0.008).
FIG. 2.
Average (±standard deviation [SD]) clusters of infection for experiment 2 with fresh and aged monolayers (n, number of replicate monolayers): 2 day old, n = 15; 8 day old, n = 16; 11 day old, n = 13; 15 day old, n = 16.
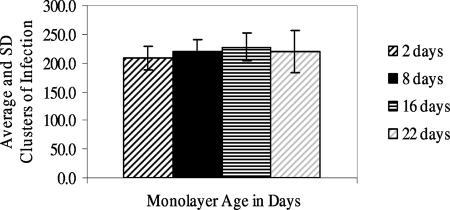
Experiment 3 evaluated baseline 2-day-old freshly confluent monolayers and 11-, 16-, and 22-day-old monolayers (Fig. 3). Infectivity of the lot of oocysts used for this experiment was 21.9%. There were no significant differences in the clusters of infection per monolayer for any of the different aged monolayers.
FIG. 3.
Average (±standard deviation [SD]) clusters of infection for experiment 3 with fresh and aged monolayers (n, number of replicate monolayers): 2 day old, n = 8; 8 day old, n = 8; 16 day old, n = 8; 22 day old, n = 8.
DISCUSSION
In previous Cryptosporidium cell culture studies, monolayers were typically allowed to develop for 24 or 48 h to reach 80 to 100% confluence (freshly confluent) just prior to inoculation (2, 8, 12, 22, 25, 28, 35). This can pose a challenge in coordinating water sample collection and processing with readiness of cell monolayers for inoculation, consequently making it difficult to perform Cryptosporidium cell culture assays routinely. In this study, we evaluated aged HCT-8 cell culture monolayers for their ability to support C. parvum infection in the hopes of progressing to a more convenient, user-friendly method for the routine detection of infectious Cryptosporidium in water. We found that HCT-8 monolayers as old as 67 days clearly supported infection. Further, in two of three experiments, aged monolayers (8 to 11 days old and 11 to 22 days old, respectively) developed the same number of C. parvum clusters of infection as baseline 2-day-old freshly confluent monolayers. Therefore, results suggest that it may be possible to use cell monolayers from freshly confluent to 3 weeks old for infectivity assays without having to schedule sample processing to coincide with development of freshly confluent monolayers. This would make Cryptosporidium cell culture assays much more feasible for water quality and utility laboratories. In addition, many of these laboratories already have the equipment needed to perform Cryptosporidium infectivity assays using chamber slides and immunofluorescent assay microscopy, since the current U.S. EPA method for the detection of Cryptosporidium oocysts in water is based on immunofluorescent assay microscopy (30). While this study clearly demonstrated that aged monolayers will support C. parvum infection, additional studies are needed to determine if other species and genotypes of Cryptosporidium, in particular, C. hominis, exhibit similar behavior.
Interestingly, based on gross microscopic observation, clusters of infection in 8- to 22-day-old monolayers often appeared to contain a greater density of developmental stages than the baseline 2-day-old-monolayer clusters of infection. However, it was not feasible to quantify the number of developmental stages in clusters of infection due to the increased density in the aged monolayers and the different planes of focus in which they appeared. In future studies it may be possible to quantify differences in parasite development in freshly confluent and aged monolayers by using real-time quantitative PCR targeting Cryptosporidium genomic DNA (8) or through the use of alternative microimaging techniques.
To date, no efficient method of producing mature Cryptosporidium oocysts in cell culture has been described, forcing reliance on animal propagation methods. The long-term maintenance of Cryptosporidium in cell culture has been reported previously (12), and recent studies have evaluated the effects of oocyst treatment on excystation (14) and host cell growth phase (32). Future research aimed at optimizing oocyst pretreatment and cell monolayer growth conditions will hopefully allow progress in this important area.
Acknowledgments
This research was supported by the Cooperative State Research, Education, and Extension Service, U.S. Department of Agriculture, under agreement no. 2005-34461-15661 and the Texas Agricultural Experiment Station, Texas A&M University System.
Footnotes
Published ahead of print on 12 October 2007.
REFERENCES
- 1.Aboytes, R., G. D. Di Giovanni, F. A. Abrams, C. Rheinecker, W. McElroy, N. Shaw, and M. W. LeChevallier. 2004. Detection of infectious Cryptosporidium in filtered drinking water. J. Am. Water Works Assoc. 96:88-98. [Google Scholar]
- 2.Arrowood, M. J. 2002. In vitro cultivation of Cryptosporidium species. Clin. Microbiol. Rev. 15:390-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrowood, M. J., J. R. Mead, L.-T. Xie, and X. U. You. 1996. In vitro anticryptosporidial activity of dinitroaniline herbicides. FEMS Microbiol. Lett. 136:245-249. [DOI] [PubMed] [Google Scholar]
- 4.Arrowood, M. J., L. T. Xie, K. Rieger, and J. Dunn. 1996. Disinfection of Cryptosporidium parvum oocysts by pulsed light treatment evaluated in an in vitro cultivation model. J. Eukaryot. Microbiol. 43:88S. [DOI] [PubMed] [Google Scholar]
- 5.Cai, X., K. M. Woods, S. J. Upton, and G. Zhu. 2005. Application of quantitative real-time reverse transcription-PCR in assessing drug efficacy against the intracellular pathogen Cryptosporidium parvum in vitro. Antimicrob. Agents Chemother. 49:4437-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Current, W. L., and T. B. Haynes. 1984. Complete development of Cryptosporidium in cell culture. Science 224:603-605. [DOI] [PubMed] [Google Scholar]
- 7.Di Giovanni, G. D., F. H. Hashemi, N. J. Shaw, F. A. Abrams, M. W. LeChevallier, and M. Abbaszadegan. 1999. Detection of infectious Cryptosporidium parvum oocysts in surface and filter backwash water samples by immunomagnetic separation and integrated cell culture-PCR. Appl. Environ. Microbiol. 65:3427-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Giovanni, G. D., and M. W. LeChevallier. 2005. Quantitative PCR assessment of Cryptosporidium parvum cell culture infection. Appl. Environ. Microbiol. 71:1495-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finch, G., E. Black, L. Gyürék, and M. Belosevic. 1993. Ozone inactivation of Cryptosporidium parvum in demand-free phosphate buffer determined by in vitro excystation and animal infectivity. Appl. Environ. Microbiol. 58:4203-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gennaccaro, A. L., M. R. McLaughlin, W. Quintero-Betancourt, D. E. Huffman, and J. B. Rose. 2003. Infectious Cryptosporidium parvum oocysts in final reclaimed effluent. Appl. Environ. Microbiol. 69:4983-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godiwala, N. T., A. Vandewalle, H. D. Ward, and B. A. Leav. 2006. Quantification of in vitro and in vivo Cryptosporidium parvum infection by using real-time PCR. Appl. Environ. Microbiol. 72:4484-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hijjawi, N. S., B. P. Meloni, U. M. Morgan, and R. C. Thompson. 2001. Complete development and long-term maintenance of Cryptosporidium parvum human and cattle genotypes in cell culture. Int. J. Parasitol. 31:1048-1055. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, A. M., K. Linden, K. M. Ciociola, R. De Leon, G. Widmer, and P. A. Rochelle. 2005. UV inactivation of Cryptosporidium hominis as measured in cell culture. Appl. Environ. Microbiol. 71:2800-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato, S., M. B. Jenkins, W. C. Ghiorse, and D. D. Bowman. 2001. Chemical and physical factors affecting the excystation of Cryptosporidium parvum oocysts. J. Parasitol. 87:575-581. [DOI] [PubMed] [Google Scholar]
- 15.Keegan, A. R., S. Fanok, P. T. Monis, and C. P. Saint. 2003. Cell culture-Taqman PCR assay for evaluation of Cryptosporidium parvum disinfection. Appl. Environ. Microbiol. 69:2505-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korich, D. G., J. R. Mead, M. S. Madore, N. A. Sinclair, and C. R. Sterling. 1990. Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. Appl. Environ. Microbiol. 56:1423-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeChevallier, M. W., G. D. Di Giovanni, J. L. Clancy, Z. Bukhari, S. Bukhari, J. S. Rosen, J. Sobrinho, and M. M. Frey. 2003. Comparison of method 1623 and cell culture-PCR for detection of Cryptosporidium spp. in source waters. Appl. Environ. Microbiol. 69:971-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeChevallier, M. W., W. D. Norton, and R. G. Lee. 1991. Giardia and Cryptosporidium spp. in filtered drinking water supplies. Appl. Environ. Microbiol. 57:2617-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meloni, B. P., and R. C. Thompson. 1996. Simplified methods for obtaining purified oocysts from mice and for growing Cryptosporidium parvum in vitro. J. Parasitol. 82:757-762. [PubMed] [Google Scholar]
- 20.Neumann, N. F., L. L. Gyurek, G. R. Finch, and M. Belosevic. 2000. Intact Cryptosporidium parvum oocysts isolated after in vitro excystation are infectious to neonatal mice. FEMS Microbiol. Lett. 183:331-336. [DOI] [PubMed] [Google Scholar]
- 21.Nichols, R. A. B., B. M. Campbell, and H. V. Smith. 2006. Molecular fingerprinting of Cryptosporidium oocysts isolated during water monitoring. Appl. Environ. Microbiol. 72:5428-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rochelle, P. A., D. M. Ferguson, T. J. Handojo, R. De Leon, M. H. Stewart, and R. L. Wolfe. 1997. An assay combining cell culture with reverse transcriptase PCR to detect and determine the infectivity of waterborne Cryptosporidium parvum. Appl. Environ. Microbiol. 63:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rochelle, P. A., M. M. Marshall, J. R. Mead, A. M. Johnson, D. G. Korich, J. S. Rosen, and R. De Leon. 2002. Comparison of in vitro cell culture and a mouse assay for measuring infectivity of Cryptosporidium parvum. Appl. Environ. Microbiol. 68:3809-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruecker, N. J., N. Bounsombath, P. Wallis, C. S. Ong, J. L. Isaac-Renton, and N. F. Neumann. 2005. Molecular forensic profiling of Cryptosporidium species and genotypes in raw water. Appl. Environ. Microbiol. 71:8991-8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schets, F. M., G. B. Engels, M. During, and A. M. de Roda Husman. 2005. Detection of infectious Cryptosporidium oocysts by cell culture immunofluorescence assay: applicability to environmental samples. Appl. Environ. Microbiol. 71:6793-6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin, G.-A., K. G. Linden, M. J. Arrowood, and M. D. Sobsey. 2001. Low-pressure UV inactivation and DNA repair potential of Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 67:3029-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slifko, T. R., D. Friedman, J. B. Rose, and W. Jakubowski. 1997. An in vitro method for detecting infectious Cryptosporidium oocysts with cell culture. Appl. Environ. Microbiol. 63:3669-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slifko, T. R., D. E. Huffman, and J. B. Rose. 1999. A most-probable-number assay for enumeration of infectious Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 65:3936-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Upton, S. J., M. Tilley, and D. B. Brillhart. 1994. Comparative development of Cryptosporidium parvum (Apicomplexa) in 11 continuous host cell lines. FEMS Microbiol. Lett. 118:233-236. [DOI] [PubMed] [Google Scholar]
- 30.U.S. EPA. 2005. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. Office of Research and Development, Government Printing Office, Washington, DC.
- 31.Widmer, G., E. A. Corey, B. Stein, J. K. Griffiths, and S. Tzipori. 2000. Host cell apoptosis impairs Cryptosporidium parvum development in vitro. J. Parasitol. 86:922-928. [DOI] [PubMed] [Google Scholar]
- 32.Widmer, G., Y. L. Yang, R. Bonilla, S. Tanriverdi, and K. M. Ciociola. 2006. Preferential infection of dividing cells by Cryptosporidium parvum. Parasitology 133:131-138. [DOI] [PubMed] [Google Scholar]
- 33.Woods, K. M., M. V. Nesterenko, and S. J. Upton. 1995. Development of a microtitre ELISA to quantify development of Cryptosporidium parvum in vitro. FEMS Microbiol. Lett. 128:89-94. [DOI] [PubMed] [Google Scholar]
- 34.Woods, K. M., M. V. Nesterenko, and S. J. Upton. 1996. Efficacy of 101 antimicrobials and other agents on the development of Cryptosporidium parvum in vitro. Ann. Trop. Med. Parasitol. 90:603-615. [DOI] [PubMed] [Google Scholar]
- 35.Woods, K. M., and S. J. Upton. 2007. In vitro development of Cryptosporidium parvum in serum-free media. Lett. Appl. Microbiol. 44:520-523. [DOI] [PubMed] [Google Scholar]
- 36.Xiao, L., A. Singh, J. Limor, T. K. Graczyk, S. Gradus, and A. Lal. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.You, X., M. J. Arrowood, M. Lejkowski, L. Xie, R. F. Schinazi, and J. R. Mead. 1996. In vitro evaluation of anticryptosporidial agents using MDCK cell culture and chemiluminescence immunoassay. J. Eukaryot. Microbiol. 43:87S. [DOI] [PubMed] [Google Scholar]