Abstract
Microbial communities in a shallow submarine hydrothermal system near Taketomi Island, Japan, were investigated using cultivation-based and molecular techniques. The main hydrothermal activity occurred in a craterlike basin (depth, ∼23 m) on the coral reef seafloor. The vent fluid (maximum temperature, >52°C) contained 175 μM H2S and gas bubbles mainly composed of CH4 (69%) and N2 (29%). A liquid serial dilution cultivation technique targeting a variety of metabolism types quantified each population in the vent fluid and in a white microbial mat located near the vent. The most abundant microorganisms cultivated from both the fluid and the mat were autotrophic sulfur oxidizers, including mesophilic Thiomicrospira spp. and thermophilic Sulfurivirga caldicuralii. Methane oxidizers were the second most abundant organisms in the fluid; one novel type I methanotroph exhibited optimum growth at 37°C, and another novel type I methanotroph exhibited optimum growth at 45°C. The number of hydrogen oxidizers cultivated only from the mat was less than the number of sulfur and methane oxidizers, although a novel mesophilic hydrogen-oxidizing member of the Epsilonproteobacteria was isolated. Various mesophilic to hyperthermophilic heterotrophs, including sulfate-reducing Desulfovibrio spp., iron-reducing Deferribacter sp., and sulfur-reducing Thermococcus spp., were also cultivated. Culture-independent 16S rRNA gene clone analysis of the vent fluid and mat revealed highly diverse archaeal communities. In the bacterial community, S. caldicuralii was identified as the predominant phylotype in the fluid (clonal frequency, 25%). Both bacterial clone libraries indicated that there were bacterial communities involved in sulfur, hydrogen, and methane oxidation and sulfate reduction. Our results indicate that there are unique microbial communities that are sustained by active chemosynthetic primary production rather than by photosynthetic production in a shallow hydrothermal system where sunlight is abundant.
Shallow submarine hydrothermal systems exposed to sunlight are expected to harbor more complex microbial communities than dark deep-sea hydrothermal systems, because there is in situ primary production not only by chemolithotrophs but also by phototrophs. The environmental conditions in shallow submarine hydrothermal systems differ from those in deep-sea hydrothermal systems and terrestrial hot springs with respect to water pressure, temperature, sunlight intensity, salinity, etc. A shallow submarine hydrothermal system in a tropical coral reef location is especially intriguing since photosynthetic biomass production is assumed to complement hydrothermal energy and carbon fluxes. Such a system has been found in Tutum Bay of Ambitle Island in Papua New Guinea (49, 50), although its endemic microbial community remains poorly explored.
Numerous shallow submarine hydrothermal activities have been investigated worldwide (2, 11, 21, 50, 51). However, little is known about entire microbial community structures in shallow submarine hydrothermal systems. A microbial community analysis based on fluorescent in situ hybridization (FISH) targeting thermophilic archaea and bacteria was reported for Vulcano Island, Italy (56). Microbiological investigations of microbial communities have also been reported for Kodakara-shima Island in Japan (21), Milos Island in Greece (6, 28, 58), Vulcano Island in Italy (16), and White Point in California (25). These studies focused on oxidation and reduction of sulfur compounds, while the distribution, diversity, and function of other chemolithotrophic microbial populations have rarely been addressed. In addition, the phylogenetic diversity of archaeal communities has been characterized for Milos in Greece (12), Vulcano in Italy (54), Tachibana Bay in Japan (63), and Tutum Bay in Papua New Guinea (49); however, the physiological diversity and ecological significance of the archaeal phylotypes remain unclear because most of the Archaea detected are still uncultivated and unidentified.
A shallow submarine hydrothermal system associated with a subtropical coral reef has been discovered off Taketomi Island in the Southern Ryukyu Archipelago, Japan. This hydrothermal system is located in and around a craterlike basin structure that is 50 to 60 m in diameter within barrier reefs. Visible fluid emissions with gas bubbles mainly consisting of CH4 and N2 occur, and the highest-temperature fluid flows out through a fissure in the bedrock at the bottom (water depth, 23 m) of the craterlike basin structure. The seafloor adjacent to the shallow submarine hot vents is partially covered with seagrass and live coral colonies (29, 41). Some of the chemical features of the fluids have been reported in previous studies (26, 45). In addition, a novel species of sulfur-oxidizing Gammaproteobacteria, Sulfurivirga caldicuralii, was previously isolated from this shallow hydrothermal system (68).
In addition to hydrothermal fluid emissions, visibly flourishing microbial communities have been observed extensively around the vent sites; these communities include white microbial mats at the deepest vent site and colored microbial mats at shallower vent sites. We have been conducting a series of geochemical and geomicrobiological surveys to elucidate the microbial ecosystem in the shallow submarine hydrothermal system off Taketomi Island. In this report, we identify bacterial and archaeal components inhabiting the hydrothermal fluid and mat formation at the deepest main vent site. Cultivation and cultivation-independent molecular techniques were combined to analyze the population size, phylogenetic diversity, main physiologies of the community, and unique microorganisms in this system.
MATERIALS AND METHODS
Sample collection and measurement of physical properties.
Water samples, microbial mats, and gas components were collected from a shallow submarine hydrothermal system off Taketomi Island, Okinawa, Japan (24°20.9′N, 124°06.10′E), by scuba diving. The samples were transferred to a laboratory within 2 h after sampling, and each sample was appropriately processed as described below. Fluid (205 to 980 ml) and free gas (100 ml) samples were taken in December 2004 and June 2005 from the deepest vent site (the main vent site) at a depth of 23 m in the craterlike basin using gas-tight acrylic syringes. In addition to the hydrothermal vent fluid, seawater samples were collected 1 m above the vent. Reference seawater samples were collected at a site that was 12 m deep and 1.7 km north of the hydrothermal site (the reference site). Free gas was dispensed into a sealed glass vial and preserved at −30°C until analysis. For FISH analysis, a 100 ml-portion of a water sample was fixed with 3.8% formalin and frozen at −30°C before experiments. For DNA extraction, the cells in 450 ml of the main vent fluid and the cells in 175 ml of the reference seawater were collected on 0.22-μm-pore-size cellulose acetate filters and stored at −80°C before experiments. The remaining water was treated for cultivation experiments and chemical analysis as described below.
A microbial mat sample was collected from the main vent site in December 2004. Several man-made objects, such as a sensor and steel pipes, have been planted in the seafloor near the main vent site for a long time. A large amount of a filamentous white mat was growing on the surfaces of these objects and basement rocks. A sensor found 1 m from the main vent was recovered, and the surface mat was collected. A portion of the mat (0.3 g, wet weight) collected for DNA extraction was stored at −80°C before the experiment. For the cultivation experiment, a portion of the mat was anaerobically processed on the boat as described below.
The temperature, pH, oxidation-reduction potential, and dissolved oxygen content at the sampling sites were measured in June 2005 with a portable water quality analyzer (W-23DX; Horiba).
Chemical characterization of water and gas components.
The free gas components were analyzed using gas chromatography with a thermal conductivity detector (model GC-14B; Shimadzu). Ammonium and dissolved sulfide concentrations in the water were determined by the classical indophenol method (60) and the methylene blue method (9), respectively.
Nucleic acid extraction and 16S rRNA gene amplification.
Genomic DNA was extracted from environmental samples (vent fluid, microbial mat, and reference seawater) and isolates with an UltraClean soil DNA kit (Mo Bio Laboratories, Inc.) by following the manufacturer's instructions. Bacterial and archaeal 16S rRNA genes were amplified by PCR as previously described (20), using primers Bac27F and Uni1492R (30) for bacterial rRNA genes and primers Arch21F and Arch958R (13) for archaeal rRNA genes.
Cloning and sequencing of 16S rRNA genes from environmental samples.
The PCR products of 16S rRNA genes from the environmental samples were cloned into the vector pCR2.1 (Invitrogen) to construct libraries, and sequences of single strands that were approximately 0.6 kb long were determined with an Applied Biosystems 3130xl genetic analyzer as described previously (20). Similarity among the clone sequences was analyzed using the FASTA program with DNASIS software (Hitachi Software Engineering), and sequences showing ≥97% identity were assigned to the same phylogenetic clone type (phylotype). Sequences (approximately 1 kb) of both strands were further determined for representative phylotypes. Chimeric sequences were detected with the CHECK-CHIMERA program from RDP II (http://rdp8.cme.msu.edu/html/) and by comparison of the neighbor-joining trees constructed from the first and second halves of each sequence using ARB software.
Sequence analysis of the isolates.
For the microbial isolates, partial sequences (approximately 0.6 kb) of the amplified 16S rRNA gene products were directly determined. Similarity among the sequences was analyzed as described above, and isolates showing ≥98% sequence identity were assigned to the same species. Sequences (0.9 to 1.4 bp) of both strands were further determined for representative isolates that were phylogenetically different from extant species.
Phylogenetic analysis.
The 16S rRNA gene sequences obtained were first compared with the sequences in the nonredundant DDBJ/EMBL/GenBank databases by using the FASTA and gapped-BLAST search programs. The sequences were subsequently imported into the ARB software program and automatically aligned using the fast aligner utility, and the alignment was manually edited. Unambiguously aligned nucleotide positions were used for construction of phylogenetic trees with the neighbor-joining algorithm using CLUSTALX, version 1.83. Bootstrap analysis was performed to assign confidence levels to tree topology.
Cultivation by the liquid serial dilution method.
For cultivation experiments, a 30-ml subsample of the main vent fluid was placed into a glass vial with a butyl rubber cap, and the headspace in the vial was filled with 100% N2. A portion of the filamentous white mat (0.1 g) was suspended in 30 ml of sterilized seawater with or without 0.05% (wt/vol) neutralized sodium sulfide in a vial under the 100% N2 gas phase (200 kPa). The mat in the vial was suspended by vigorous shaking before inoculation. The total number of cells in the slurry was determined by 4′,6-diamidino-2-phenylindole (DAPI) staining. To calculate the number of culturable microbial cells, samples were diluted using a liquid decimal dilution series with media targeting a wide range of metabolisms. The cultivation experiment was performed using 3 ml of medium in a 15-ml tube in triplicate.
Aerobic to microaerobic, hydrogen- and/or sulfur-oxidizing chemolithoautotrophs were cultivated at 30, 37, and 55°C in MMJHS medium (65) with four types of headspace gas: (i) H2-CO2-O2 (80:19:1; 200 kPa); (ii) H2-CO2-O2 (75:15:10; 200 kPa); (iii) N2-CO2-O2 (80:19:1; 200 kPa); and (iv) N2-CO2-O2 (75:15:10; 200 kPa). For aerobic methane oxidizers, MJmet medium was used. MJmet medium was prepared by adding 6 mM NaHCO3 and 1 μM CuSO4 to MJ medium consisting of (per liter) 30 g NaCl, 0.14 g K2HPO4, 0.8 g CaCl2, 3.4 g MgSO4·7H2O, 4.18 g MgCl2·6H2O, 0.33 g KCl, 0.25 g NH4Cl, 0.25 g NaNO3, 0.5 mg NiCl2·6H2O, 0.5 mg Na2SeO3·5H2O, 0.1 mg Na2WO4, 20 mg Fe(NH4)2(SO4)2·6H2O, 10 ml of a trace mineral solution (4), and 1 ml of a vitamin solution (4). MJmet medium was prepared using the procedure previously described (19) with headspace gas containing CH4, N2, CO2, and O2 (45:40:10:5; 200 kPa). Methanotrophs were cultivated at 37 and 50°C. For anaerobic, fermentative, sulfur-reducing thermophiles, MJYPS medium (70) and incubation at 55, 70, 85, and 95°C were used. For anaerobic dissimilatory Fe(III) and/or sulfate reducers, MMJHFe medium was used with headspace gas containing H2 and CO2 (80:20; 200 kPa) or N2 and CO2 (80:20; 200 kPa), and it was incubated at 30, 55, 70, and 85°C. MMJHFe medium is MMJ medium (66) supplemented with 0.005% (wt/vol) yeast extract, 5 mM formate, 5 mM acetate, 5 mM pyruvate, 5 mM lactate, 0.2% (wt/vol) sodium sulfate, 10 mM Fe(III) citrate, and 10 mM ferrihydrite (27) and reduced with 0.02% (wt/vol) sodium sulfide. For aerobic and anaerobic heterotrophs, MJYPV medium (57) was used with air or N2-CO2 (80:20; 200kPa) as the headspace gas at cultivation temperatures of 30 and 55°C. For cultivation of strictly anaerobic photoautotrophic microorganisms, MJHS-photo medium was employed; this medium consisted of (per liter) 20 g NaCl, 0.09 g K2HPO4, 0.07g KH2PO4, 1.25 g NH4Cl, 0.8 g CaCl2, 7 g MgCl2·6H2O, 0.33 g KCl, 0.05 g FeCl3, 0.01 g Fe(NH4)2(SO4)2·6H2O, 5 mg NiCl2·6H2O, 5 mg Na2SeO3·5H2O, 1 mg Na2WO4·2H2O, 0.5 mg of resazurin, and 10 ml each of a trace mineral solution (4) and a vitamin solution (4), as well as 0.2% (wt/vol) NaHCO3, 0.1% (wt/vol) Na2S2O3·5H2O, and 0.1% Na2S·9H2O. The final pH was adjusted to 7.0, and the gas phase for this medium was H2-CO2 (80:20; 200 kPa). Cultivation was carried out at 30 and 55°C with a photosynthetic photon flux density of 30 μmol/m2·s.
Some of the microorganisms cultivated in the most diluted media were also isolated subsequently by the dilution-to-extinction method. Partial sequences of the 16S rRNA gene (approximately 700 to 1,400 bp) of the isolates were determined as described below.
FISH and cell counting.
The fixed cells collected on a poly-l-lysine-coated 0.2-μm-pore-size polycarbonate Nuclepore filter were subjected to direct cell counting and phylogenetic quantification by FISH and catalyzed reporter deposition FISH as previously described (48, 61). In addition to the previously reported rRNA-targeted oligonucleotide probes specific for each phylogenetic group, two probes newly designed for bacterial isolates obtained in this hydrothermal system were employed, as shown in Table 1.
TABLE 1.
Oligonucleotide probes used in this study
| Probe | Target microbial group | Sequence (5′-3′) | Target site (rRNA positions)a | Formamide concn (%)b | Salt concn (mM)c | Reference(s) |
|---|---|---|---|---|---|---|
| EUB338 I-III | Bacteria | GCWGCCWCCCGTAGGWGT | 16S (338-355) | 55 | 3 | 1, 10 |
| ARCH915 | Archaea | GTGCTCCCCCGCCAATTCCT | 16S (915-934) | 60 | 0 | 15 |
| CF319 | Cytophaga-Flavobacterium | TGGTCCGTGTCTCAGTAC | 16S (319-336) | 55 | 3 | 32 |
| ALF968 | Alphaproteobacteria | GGTAAGGTTCTGCGCGTT | 16S (968-985) | 55 | 3 | 42 |
| BET42 | Betaproteobacteria | GCCTTCCCACTTCGTTT | 23S (1027-1043) | 55 | 3 | 33 |
| GAM42 | Gammaproteobacteria | GCCTTCCCACATCGTTT | 23S (1027-1043) | 55 | 3 | 33 |
| EPS402 | Epsilonproteobacteria | GAAAKGYGTCATCCTCCACG | 16S (402-423) | 30 | 64 | 69 |
| DELTA495 | Deltaproteobacteria | ARTTAGCCGGYGCTTCCTd | 16S (495-512) | 55 | 3 | 31 |
| Gm705 | Type I methanotrophs | CTGGTGTTCCTTCAGATC | 16S (705-722) | 35 | 42 | 17 |
| Sul994 | S. caldicuralii | CTGGCGCCTTCCGGGGATGT | 16S (994-1013) | 55 | 3 | This study |
| IT9/446 | Isolate IT-9 | GGCCCCAGAGCCCTTCGTCC | 16S (446-464) | 60 | 0 | This study |
Position in 16S or 23S rRNA of E. coli (7).
Formamide concentration in a hybridization solution.
Salt concentration in wash buffer.
The sequence was revised in this study.
Probe IT9/446 was designed for the thermophilic methanotrophic strain IT-9 isolated in this study. Probe Sul994 was designed for S. caldicuralii (68). These probes were designed using ARB software (http://www.arb-home.de), and their specificity was checked by both the gapped BLAST search program on the DDBJ website (http://www.ddbj.nig.ac.jp/) and preliminary FISH experiments. Probe IT9/446 was highly specific to the IT-9 strain and had a 3-base mismatch with two environmental clone sequences obtained from a deep-sea hydrothermal vent at the Mid-Okinawa Trough (accession number AB175557) and from an offshore marine environment in southern California (accession number DQ009352). The 16S rRNA gene sequences in the DDBJ database other than these two clones indicated that there was a mismatch of 5 or more bases with probe IT9/446. For probe Sul994, one environmental clone (accession number AF339744) retrieved from saline water at hydrothermal vents in Vulcano, Italy, had a 1-base mismatch and two Fervidobacterium spp. (accession numbers EF565822 and EF222228) had a 3-base mismatch. Except for this clone and these strains, no 16S rRNA gene sequence having less than a 6-base mismatch with probe Sul994 was found in the DDBJ database. Suitable formamide concentrations in hybridization buffers and suitable salt concentrations in wash buffers for these newly designed probes were determined using cultivated cells of strain IT-9 and S. caldicuralii as the positive controls.
Phytoplankton were identified and counted by using red autofluorescence with green light excitation, and total cell density was determined by counting DAPI-stained cells. The microbial cells on a polycarbonate filter were immobilized by dipping the filter in a 0.2% low-gelling-temperature agarose gel and were dried at 45°C. The cells were then incubated with lysozyme (10 mg per ml in 0.05 M EDTA and 0.1 M Tris [pH 7.5]) and proteinase K (4 U per ml in 0.05 M EDTA and 0.1 M Tris [pH 7.5]) for 1 h at 37°C to increase cell wall permeability and treated with 0.01 M HCl for 20 min to deactivate these enzymes and endogenous peroxidase. The microbial cells on a filter section were hybridized for 3.5 h at 35°C in a hybridization solution (0.9 M NaCl, 20 mM Tris-HCl [pH 7.5], 10% dextran sulfate, 0.02% sodium dodecyl sulfate, 1% blocking reagent, 0.05% salmon sperm DNA, 0.05% Escherichia coli tRNA) containing 0.1 pmol per μl of each probe labeled with horseradish peroxidase at its 5′ end and the appropriate formamide concentration (Table 1). The filter section was then washed in wash buffer I (5 mM EDTA [pH 8.0], 20 mM Tris-HCl [pH 7.5], 0.01% sodium dodecyl sulfate) containing the appropriate salt concentration for the probe for 10 min at 37°C and subsequently in wash buffer II (1× phosphate-buffered saline with 0.05% Triton X-100) for 15 min at room temperature. The hybridized cells were reacted in 1/50 Cy3-labeled tyramide solution (TSA direct; PerkinElmer) according to the manufacturer's instructions and then washed in wash buffer II and dehydrated with ethanol. The cells were observed with a fluorescence microscope (model BX61; Olympus, Tokyo, Japan) using the WU (excitation at 360 nm and emission at 460 nm) and WIG (excitation at 546 nm and emission at 585 nm) fluorescent filter sets for DAPI- and Cy3-stained microbial cells, respectively. Microbial cell densities were determined by counting more than 5,000 DAPI-stained cells in at least 30 microscopic fields.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of the phylotypes and representative isolates and the sequences of the particulate methane monooxygenase gene pmoA of the methanotrophs have been deposited in the DDBJ/EMBL/GenBank databases. The accession numbers for bacterial phylotypes are as follows: pItb-HW, AB294888 to AB294922; pItb-vmat, AB294923 to AB294979; and pItb-RF, AB294980 to AB295005. The accession numbers for archaeal phylotypes are as follows: pIta-HW, AB301857 to AB301881; pIta-vmat, AB301882 to AB301898; and pIta-RF, AB301899 to AB301906. The accession numbers for 16S rRNA genes of the isolates are shown in Table 3. The accession numbers for pmoA of isolates IT-4 and IT-9 are AB302947 and AB302948, respectively.
TABLE 3.
Representative isolates obtained from the main vent fluid and microbial mat samples
| Representative isolate | Energy acquisition pattern | Phylogenetic affiliation | Related microorganism (% sequence identity) | Isolation temp (°C) | Isolation source | Accession no. |
|---|---|---|---|---|---|---|
| So30-vw | Aerobic sulfur oxidization | γ-Proteobacteria | Thiomicrospira chilensis (96.5) | 30 | Vent fluid and mat | AB301715 |
| VW1 | Microaerobic thiosulfate oxidization | γ-Proteobacteria | Sulfurivirga caldicuralii (100) | 55 | Vent fluid and mat | AB245480 |
| Ho30-mm | Microaerobic hydrogen oxidization | ɛ-Proteobacteria | Sulfurimonas paralvinellae (92.1) | 30 | Mat | AB301716 |
| IT-4 | Microaerobic methane oxidization | γ-Proteobacteria | Methylobacter marinus (92.6) | 30-42 | Mat | AB301717 |
| IT-9 | Microaerobic methane oxidization | γ-Proteobacteria | Methylohalobius crimeensis (94.0) | 50 | Vent fluid | AB301718 |
| An30N-mm | Anaerobic heterotrophic sulfate reduction | δ-Proteobacteria | Desulfovibrio hydrothermalis (94.7) | 30 | Mat | AB301719 |
| An30H-mm | Anaerobic heterotrophic sulfate reduction | δ-Proteobacteria | Desulfovibrio dechloracetivorans (92.5) | 30 | Mat | AB301720 |
| Ag70-vw | Anaerobic mixotrophic Fe(III) reduction | Deferribacterales | Deferribacter desulfuricans (99.3) | 70 | Vent fluid | AB301721 |
| Tc70-vw | Anaerobic heterotrophic sulfur reduction | Thermococcales | Thermococcus kodakaraensis (100) | 70-95 | Vent fluid and mat | AB301722 |
| Ag85-vw | Anaerobic heterotrophic sulfur reduction | Thermococcales | Thermococcus alcaliphilus (99.4) | 85 | Vent fluid | AB301723 |
| Aeh30-vw | Aerobic heterotrophy | γ-Proteobacteria | Pseudoalteromonas ganghwensis (99.6) | 30 | Vent fluid | AB301724 |
| Aeh30-mm | Aerobic heterotrophy | γ-Proteobacteria | Vibrio parahaemolyticus (99.2) | 30 | Mat | AB301725 |
| 5H2 | Photolithotrophy using sulfide | Chlorobiales | Prosthecochloris aestuarii (97.9) | 30 | Mat | AB301726 |
RESULTS
Geographic profiles and physicochemical characterization of the hydrothermal sites.
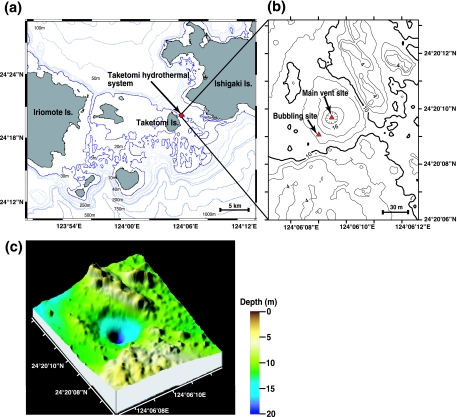
The hydrothermal vent field is located 1 km off the east coast of Taketomi Island in the Southern Ryukyu Archipelago, Japan (Fig. 1a). As shown in a bathymetric map (Fig. 1b and c) created by a multibeam echo sounder system (14), the hydrothermal vent field has a craterlike basin structure that is 50 to 60 m in diameter. Several scattered vent sites were discovered at depths ranging from 10 to 23 m within the basin structure. The main vent site in the deepest part was covered by rock and coarse sand; high-temperature fluid (52°C) containing small amounts of gas bubbles was emitted through a fissure in the bedrock, and white microbial mats were distributed on the surfaces around the main vent (Table 2). The sandy, barren slope within 15 m of the main vent was free of macrobenthos; instead, microbial mats covered the seafloor. On the northwestern edge of the basin, a seagrass meadow extended to a depth of 15 m.
FIG. 1.
Location (a and b) and three-dimensional bathymetric map (c) of the shallow submarine hydrothermal system off Taketomi Island.
TABLE 2.
In situ physical properties, chemical components of the hydrothermal fluids, and microbial mat types observed
| Site | Water depth (m) | Physical and chemical properties of the water
|
Free gas components (%)
|
Microbial mat type | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Temp (°C) | Oxidation-reduction potential (mV) | pH | Dissolved oxygen concn (mg/liter) | Salinity (%) | H2S concn (μM)a | NH4+ concn (μM)a | CH4 | N2 | CO2 | O2 | H2 | |||
| Hydrothermal area | ||||||||||||||
| Main vent site | 1 (surface) | 28 | 91 | 8.2 | 6.4 | 3.8 | NDb | ND | ND | ND | ND | ND | ND | |
| 22 | 34 | −210 | 7.8 | 4.9 | 3.9 | ND | ND | ND | ND | ND | ND | ND | ||
| 23 (bottom) | 52 | −220 | 6.6 | 1.0 | 3.3 | 162-188 | 165-182 | 68.2-69.0 | 29.2-30.1 | 0.45-0.46 | 1.23-1.27 | 0.008-0.011 | White | |
| Bubbling vent site | 12 | 29 | −200 | 8.1 | 5.8 | 3.9 | ND | ND | ND | ND | ND | ND | ND | |
| 13 (bottom) | 41 | −340 | 6.7 | 0.4 | 3.3 | ND | 113 | 68.5-69.5 | 28.1-29.0 | 1.68-1.86 | 0.70-0.73 | 0.011-0.015 | White, green, and brown | |
| Nonhydrothermal area | 1 (surface) | 28 | 55 | 8.2 | 7.3 | ND | ND | ND | ND | ND | ND | ND | ND | |
| (reference site) | 11 | 28 | 53 | 8.2 | 6.4 | ND | ND | ND | ND | ND | ND | ND | ND | |
| 12 (bottom) | 28 | 54 | 8.2 | 6.5 | ND | ND | ND | ND | ND | ND | ND | ND | None | |
Range of values for two samples taken independently from each site.
ND, not determined.
Many prominent gas bubbling points were observed on the southwestern slope 20 to 40 m from the main vent, where debris of dead coral skeletons covered the seafloor. Among these bubbling points, the bubbling vent 35 m from the main vent was the most active, and it was designated “the bubbling vent site.” Thermal fluid at a temperature of 41°C flowed out continuously, and abundant gas bubbles spouted periodically. At the bubbling vent site, various colored (white, brown, and green) microbial mats had developed on the debris-covered seafloor (Table 2). Live corals were found to create small aggregations even in the gas bubbling area. No benthic invertebrates with a visible body size were found in this field.
At the two hydrothermal vent sites, the temperature, pH, dissolved oxygen content, and salinity of the vent fluids were markedly different even 1 m above the vents due to dilution with ambient seawater (Table 2). Only the oxidation-reduction potential values were still low 1 m above the vents. The free gas compositions were similar at the two hydrothermal sites; the gas consisted mainly of CH4 (68.2 to 69.5%) and N2 (28.1 to 30.1%) and there were small amounts of CO2, O2, and H2, which is consistent with the previous results (26). The main vent fluid contained dissolved H2S (162 to 188 μM) in addition to CH4, CO2, H2, and NH4+. The abundant CH4 in this Taketomi system contrasted with the findings for other shallow hydrothermal systems, where CO2 has been reported to be the primary gas component and CH4 has been reported to be a rather minor component (2, 11, 21, 50, 51).
Phylogenetic diversity of 16S rRNA gene clones.
To investigate the phylogenetic diversity of the main vent communities, bacterial and archaeal 16S rRNA gene clone analyses were performed for the vent fluid, white mat, and reference seawater.
(i) Bacterial diversity.
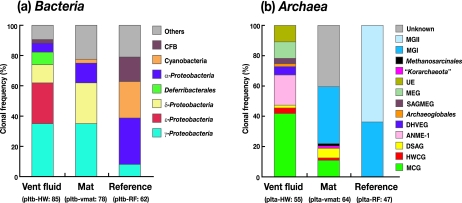
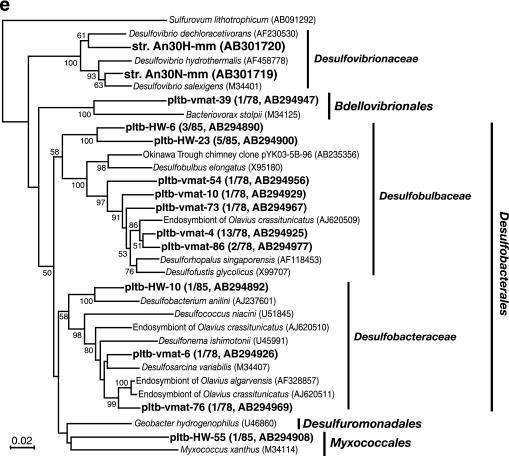
Highly diverse bacterial 16S rRNA gene community structures were found in the vent fluid (35 phylotypes in 85 clones; the library was designated pItb-HW), the mat (57 phylotypes in 78 clones; pItb-vmat), and the reference seawater (29 phylotypes in 62 clones; pItb-RF) (Fig. 2a). The dominant phyla in the vent fluid and adjacent microbial mat were identified as Gamma-, Epsilon-, and Deltaproteobacteria, and then the phylotypes within these groups were phylogenetically characterized (Fig. 3a to e).
FIG. 2.
Microbial community structures based on the bacterial (a) and archaeal (b) 16S rRNA gene clone sequences. The designations of the libraries and the numbers of clones examined are indicated in parentheses. Abbreviations: CFB, Cytophaga-Flavobacterium-Bacteroidetes; HWCG, hot water crenachaeotic group; DSAG, deep-sea archaeal group; SAGMEG, South Africa gold mine euryarchaeotic group; MEG, miscellaneous euryarchaeotic group; UE, unidentified euryarchaeotic phylotypes; MGI, marine crenarchaeotic group I; MGII, marine euryarchaeotic group II.
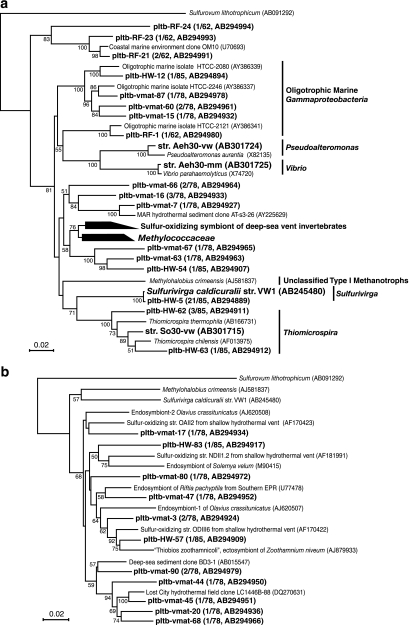
FIG. 3.
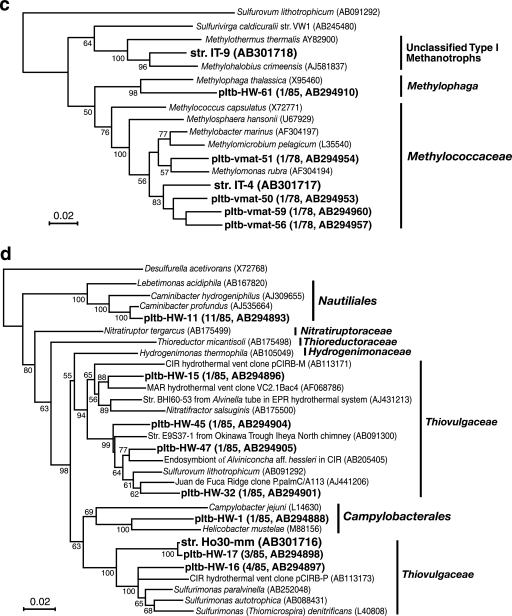
Phylogenetic trees of 16S rRNA gene sequences of bacterial isolates and phylotypes based on neighbor-joining analysis. Trees were constructed using unambiguously aligned homologous nucleotide positions for (a) miscellaneous gammaproteobacterial bacteria (836 positions); (b) gammaproteobacterial bacteria, including symbionts of marine invertebrates (924 positions); (c) gammaproteobacterial methanotrophs and methylotrophs (953 positions); (d) Epsilonproteobacteria (796 positions); and (e) Deltaproteobacteria (753 positions). Bootstrap analysis was performed with 100 repetitions, and only values greater than 50 are indicated at the nodes. Scale bars = 0.02 change per nucleotide position.
(a) Gammaproteobacteria.
Within the Gammaproteobacteria, many phylotypes from the fluid and mat were closely related to free-living and symbiotic sulfur oxidizers from marine habitats, but none of the phylotypes was found in all the libraries (Fig. 3a to c). The pItb-HW-5 phylotype, showing 99.8% sequence similarity with thermophilic thiosulfate-oxidizing organism S. caldicuralii that was previously isolated from this hydrothermal system (68), was the most abundant phylotype in the fluid (clonal frequency, 24.7%) (Fig. 3a). Some phylotypes shown in Fig. 3b were associated with the sulfur-oxidizing isolates NDII1.2, ODIII6, and OAII2 obtained from the Milos shallow hydrothermal system (28). The lineages including these Milos isolates and Taketomi phylotypes also appeared to include many previously uncultivated, sulfur-oxidizing endo- and ectosymbionts of marine invertebrates.
The phylotypes in the methane-oxidizing family Methylococcaceae were recovered only from the mat sample (Fig. 3c). Three phylotypes showed the highest similarity (>94.0%) with methanotrophic isolate IT-4 that was obtained in this study, while one phylotype was similar to the genus Methylomonas. Most of the rest of the gammaproteobacterial phylotypes were previously uncultivated groups of microorganisms, although some of them were phylogenetically associated with environmental clones obtained from deep-sea hydrothermal vent sites.
(b) Epsilonproteobacteria.
The phylotypes in the Epsilonproteobacteria were detected only in the vent fluid (Fig. 2a). The pItb-HW-11 phylotype, the second most dominant phylotype in the fluid (clonal frequency, 12.9%), was affiliated with the thermophilic hydrogen-oxidizing genus Caminibacter (Fig. 3d). One of the phylotypes belonging to the family “Thiovulgaceae” (8), pItb-HW-17, showed 99.9% sequence similarity with the novel mesophilic hydrogen-oxidizing isolate Ho30-mm.
(c) Deltaproteobacteria.
The Deltaproteobacteria was the second most abundant group in the mat (clonal frequency, 26.9%) and the third most abundant group in the fluid (11.7%) (Fig. 2a). This anaerobic group was not detected in the oxidative reference seawater. The deltaproteobacterial phylotypes in the libraries from the fluid and mat were quite different (Fig. 3e). Within the families Desulfobulbaceae and Desulfobacteraceae, some of the phylotypes obtained from the mat library, including the predominant pItb-vmat-4 phylotype (clonal frequency, 16.7%), were closely related to the sulfate-reducing endosymbionts of the marine oligochaetes Olavius spp.
(d) Deferribacterales.
Only one phylotype, phylotype pItb-HW-19, in the thermophilic Deferribacterales was found to be a relatively abundant component of the fluid (clonal frequency, 8.3%; accession no. AB294899). This phylotype was distantly related to the genus Deferribacter, and the data indicated a closest relationship to Flexistipes sp. strain vp180 (92.7% sequence similarity) obtained from a high-temperature offshore oil field (47), whereas we isolated one Deferribacter sp. strain from the same fluid sample.
(ii) Phylogenetic diversity of archaeal 16S rRNA genes.
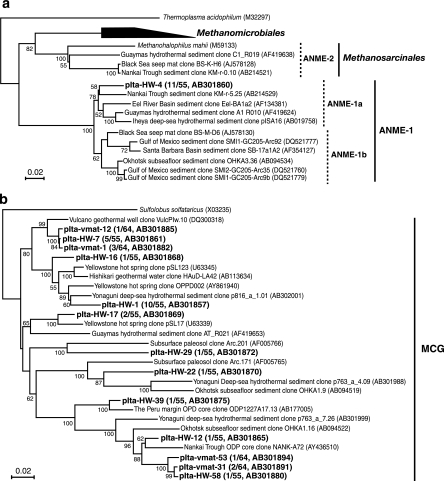
Phylogenetic analysis of archaeal 16S rRNA gene clone sequences revealed that the archaeal community structures in the fluid, mat, and reference seawater might differ greatly (Fig. 2b). In the fluid community (25 phylotypes in 55 clones; pIta-HW), two predominant phylotypes were found: phylotype pIta-HW-4 (clonal frequency, 20.0%) affiliated with anaerobic methane oxidation group 1 (ANME-1) (Fig. 4a) and phylotype pIta-HW-1 (clonal frequency, 18.2%) assigned to the miscellaneous crenarchaeotic group (MCG) (Fig. 4b). Members of ANME-1, which is known as a potential anaerobic methane-oxidizing group, have frequently been detected in various methane-rich marine sediment environments, including methane seeps (44, 46), and even in deep-sea hydrothermal habitats (62, 74). Our results revealed the predominant occurrence of ANME-1 in a shallow submarine hydrothermal system for the first time. The MCG is known to occur in a wide range of marine and terrestrial environments, although the physiological traits remain unknown (73). The phylotypes belonging to the deep-sea hydrothermal vent euryarchaeotic group (DHVEG) and the Archaeoglobales, groups which are specifically found at hydrothermal vent sites (38, 64), were retrieved only from the fluid. The recent successful cultivation of a thermophilic DHVEG archaeon from actively venting deep-sea sulfide structures (52) strongly supports the assumption that the DHVEG consists of thermophiles.
FIG. 4.
Phylogenetic trees of 16S rRNA gene sequences of the archaeal phylotypes based on neighbor-joining analysis. Trees were constructed in the same manner as the bacterial trees for ANME-1 archaea (768 positions) (a) and MCG archaea (750 positions) (b).
The mat archaeal community showed greater diversity (39 phylotypes in 64 clones; pIta-vmat); only phylotype pIta-vmat-14 affiliated with marine crenarchaeotic group I accounted for more than 10% of the library (clonal frequency, 12.5%). Most of the other diverse archaeal groups found at the vent site, such as the hot water crenachaeotic group, the deep-sea archaeal group, and the South Africa gold mine euryarchaeotic group, have frequently been identified in various marine and/or terrestrial habitats (24, 43, 62, 73), although their physiology and activity are unclear at the present. Meanwhile, there was less archaeal diversity in the reference seawater (pIta-RF). One marine crenarchaeotic group I and seven marine euryarchaeotic group II phylotypes represented the 47 clones sequenced. In the cultivation experiment, a relatively high density of the Thermococcus spp. population was detected in the fluid and mat (see below). However, no phylotype related to Thermococcus was retrieved from the fluid and mat by culture-independent molecular analysis.
Quantitative cultivation and isolation of representative microorganisms.
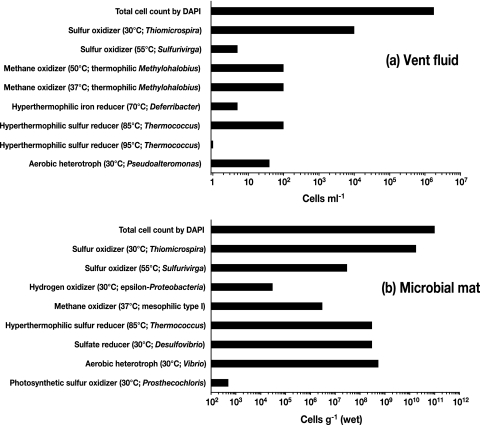
Based on the analyses of chemical characteristics and 16S rRNA gene communities, sulfur- and methane-oxidizing thermophiles and mesophiles were expected to be the predominant organisms in the communities associated with the fluid emission at the main vent site. We estimated the culturability and population density of potentially predominant microbial components in the vent fluid and mat using the liquid serial dilution cultivation technique. Microorganisms showing a variety of types of energy and carbon metabolism were successfully cultivated (Fig. 5). Each representative isolate obtained from the most diluted tube was subjected to a comparative 16S rRNA gene sequence analysis (Table 3 and Fig. 3a to e).
FIG. 5.
Total cell counts determined by direct DAPI staining and minimum population densities determined by the liquid serial (1:10) dilution cultivation technique targeting each type of metabolism. The cultivation temperatures and the microorganisms cultivated in the most dilute series of tubes are indicated in parentheses.
(i) Sulfur-oxidizing microorganisms.
Mesophilic sulfur-oxidizing chemolithoautotrophs had the highest population density in both vent fluid and mat samples (Fig. 5). Six mesophilic sulfur oxidizers were obtained, and all the isolates showed >99% sequence similarity with each other. The representative isolate So30-vw was a member of the genus Thiomicrospira (Fig. 3a). It has often been demonstrated that Thiomicrospira is a typically predominant sulfur-oxidizing genus in shallow and deep-sea hydrothermal environments (6, 28, 37, 40, 55). Thermophilic sulfur-oxidizing chemolithoautotrophs were cultivated at 55°C from both samples, although their population density was considerably less than that of members of the genus Thiomicrospira. Representative isolate VW1, belonging to a novel genus of the Gammaproteobacteria, was previously characterized and described as S. caldicuralii (68) (Fig. 3a). The cultivation experiment demonstrated that S. caldicuralii was relatively less abundant at the main vent site, whereas a culture-independent analysis indicated that a phylotype similar to S. caldicuralii (pItb-HW-5) was predominant in the vent fluid, as mentioned above. Thus, based on the culture-dependent and -independent analyses, members of the sulfur-oxidizing chemolithoautotrophic population might be the most abundant, functionally active microbial components in the vent fluid.
(ii) Methane-oxidizing microorganisms.
Methane oxidizers were estimated to be less abundant than sulfur oxidizers in both samples (Fig. 5); however, novel mesophilic and thermophilic gammaproteobacterial type I methanotrophs were successfully isolated. Methane oxidizers that grew at 50°C were successfully cultivated only from the vent fluid. The thermophilic, methane-oxidizing isolate IT-9 showed the highest sequence similarity (94.0%) with the mesophilic organism Methylohalobius crimeensis (18) (Fig. 3c). Although a methanotrophic strain was cultivated by incubation of the vent fluid at 37°C, the rRNA gene sequence of the strain was identical to the sequence of strain IT-9. IT-9 was the most abundant methanotroph in the vent fluid at both cultivation temperatures (37 and 50°C), whereas thriving mesophilic methanotrophs were found in the mat. Five mesophilic isolates were obtained from the mat at different cultivation temperatures (30 to 42°C), and sequencing analysis revealed that these isolates were phylogenetically identical. The representative isolate IT-4 was moderately related to Methylobacter marinus (92.6% sequence similarity), which was its closest relative (Fig. 3c). The optimum growth temperatures of IT-4 and IT-9 were found to agree with the in situ temperatures of their habitats. The gene encoding particulate methane monooxygenase (pmoA) was successfully amplified from the DNA of IT-4 and IT-9 using the A189/A682 primer set (22) in conditions previously described (20), while soluble methane monooxygenase genes were not detected in either of the isolates when the previously reported primers and conditions were used (34, 35). Electron microscopic analysis of ultrathin sections of IT-4 and IT-9 cells showed the presence of a stack of intracellular membrane disks typical of type I methanotrophs (data not shown).
(iii) Hydrogen-oxidizing microorganisms.
Hydrogen oxidizers were cultivated only from the mat at 30°C, and the population density was relatively low (Fig. 5). The obtained epsilonproteobacterial isolate Ho30-mm was moderately related to Sulfurimonas paralvinellae (92.1% sequence similarity) (71) (Fig. 3d). No thermophilic hydrogen oxidizers were cultivated in this study. Molecular hydrogen was a minor component of the free gas (Table 2), which might be the reason that culturable hydrogen oxidizers were less abundant at the main vent site. In contrast, rRNA gene clone analysis demonstrated the presence of thermophilic, hydrogen-oxidizing, anaerobic to microaerobic Caminibacter in the vent fluid (Fig. 3d).
(iv) (Hyper)thermophilic microorganisms.
In addition to the potential primary producers described above, iron-, sulfur-, and sulfate-reducing, (hyper)thermophilic heterotrophs were also cultivated from the fluid and/or mat, even though their population densities were lower than those of the chemolithotrophic primary producers (Fig. 5). Cultivation of hyperthermophilic sulfur reducers at 70 to 95°C resulted in isolation of seven Thermococcus isolates showing 98 to 100% sequence similarity from both samples. The representative isolate Tc85 showed 100% sequence similarity with Thermococcus kodakaraensis (3). An iron-reducing Deferribacter sp. showing 99.3% sequence similarity with Deferribacter desulfuricans (67) was isolated only from the fluid. Thermococcus spp. have often been cultivated from other shallow hydrothermal systems with high-temperature fluid emissions (21, 23, 56, 63). In contrast, Deferribacter spp. have previously been obtained only from deep-sea hydrothermal habitats (36, 67); therefore, in this study Deferribacter was cultivated for the first time from a shallow hydrothermal habitat.
(v) Other microorganisms.
Anaerobic sulfate-reducing Desulfovibrio spp. were cultivated only from the mat sample, indicating that there were anaerobic microhabitats inside the mat. Aerobic heterotrophs isolated from the fluid and mat were affiliated with Pseudoalteromonas and Vibrio, respectively. The obligately photoautotrophic, strictly anaerobic, sulfur-oxidizing isolate 5H2 was cultivated as a minor component of the mat community. Isolate 5H2 exhibited the closest relationship to a marine photosynthetic bacterium, Prosthecochloris aestuarii in the Chlorobi group (98.1% sequence similarity). The detection of a few phototrophic microorganisms at the main vent site was consistent in both culture-dependent and culture-independent analyses.
Cell densities determined by FISH.
The total cell density was greater in the main vent fluid than the ambient seawater 1 m above the vent and the reference seawater (Table 4). Quantitative FISH analyses using several group-specific oligonucleotide DNA probes indicated that the number of Bacteria cells (EUB338 positive) was much greater than the number of Archaea cells (ARCH915 positive) in the vent fluid and reference seawater, whereas the number of Archaea cells slightly exceeded the number of Bacteria cells in the seawater 1 m above the vent (Table 4). At the reference site, probes EPS402 and DELTA495 did not show any signature or showed only a slight signature, indicating that no detectable epsilonproteobacterial cells and few deltaproteobacterial cells were present in the reference seawater. In contrast, at the main vent site, all of the targeted bacterial groups were detected with the group-specific probes, and only the Epsilon- and Deltaproteobacteria populations were considerably larger in the vent fluid. In addition, unclassified cells that were positive with the EUB338 probe but negative with all other probes used were abundant (16.1% of the EUB338-positive cells) in the vent fluid. These results indicate that the Epsilon- and Deltaproteobacteria and the unclassified bacteria prefer the hot vent fluid. The unclassified components may contain additional epsilonproteobacterial populations, because probe EPS402 had 2- or 3-base mismatches specifically with members of the family Nautiliaceae in the Epsilonproteobacteria. Phylotype pItb-HW-11 of the genus Caminibacter was abundant in the main vent fluid (Fig. 3d), and pItb-HW-11 also had a 3-base mismatch with EPS402. Consequently, the presence of Epsilonproteobacteria could have been underestimated in this FISH experiment.
TABLE 4.
Microbial population density determined by DAPI staining of the total cells, autofluorescence of phytoplankton, and FISH analysis using group-specific oligonucleotide DNA probes
| Detection method | Population density (cells/ml) (% of DAPI counts)
|
|||
|---|---|---|---|---|
| Main vent site depth
|
Reference site depth
|
|||
| 23 m (vent) | 22 m (1 m above the vent) | 12 m (bottom) | 11 m (1 m above the bottom) | |
| DAPI (total cell count) | 1.70 × 106 | 8.24 × 105 | 6.68 × 105 | 6.76 × 105 |
| Autofluorescence of phytoplankton | 2.46 × 104 (1.4) | 8.26 × 104 (10.0) | 4.41 × 104 (6.6) | 6.29 × 104 (9.3) |
| Oligonucleotide DNA probes | ||||
| ARCH915 | 7.37 × 105 (43.3) | 3.42 × 105 (41.5) | 1.38 × 105 (20.6) | 2.47 × 105 (36.4) |
| EUB338 | 1.24 × 106 (73.0) | 3.25 × 105 (39.4) | 3.39 × 105 (50.8) | 3.25 × 105 (48.0) |
| ALF968 | 7.03 × 104 (4.1) | 7.48 × 104 (9.1) | 7.25 × 104 (10.9) | 8.33 × 104 (12.3) |
| BET42a | 1.15 × 105 (6.8) | 7.62 × 104 (9.2) | 4.69 × 104 (7.0) | 1.88 × 104 (2.8) |
| GAM42a | 1.33 × 105 (7.8) | 7.79 × 104 (9.5) | 5.66 × 104 (8.5) | 5.09 × 104 (7.5) |
| CF319 | 2.71 × 105 (15.9) | 1.47 × 105 (17.9) | 1.06 × 105 (15.8) | 1.28 × 105 (19.0) |
| EPS402 | 1.66 × 105 (9.8) | 1.26 × 104 (1.5) | 0.00 (0.0) | 0.00 (0.0) |
| DELTA495 | 2.13 × 105 (12.5) | 5.62 × 103 (0.7) | 8.87 × 101 (0.0) | 2.90 × 102 (0.0) |
| Gm705 | 5.47 × 104 (3.2) | 5.02 × 104 (6.1) | 5.42 × 104 (8.1) | 5.02 × 104 (7.4) |
| Sul994 | 6.94 × 104 (4.1) | NDa | ND | ND |
| IT9/446 | 2.18 × 104 (1.3) | ND | ND | ND |
ND, not determined.
To identify the predominantly cultivated chemolithotrophs in the vent fluid, three probes for different targets were employed: (i) probe Sul994 for S. caldicuralii and phylotype pItb-HW-5; (ii) probe Gm705 for diverse type I methanotrophs, including the newly isolated methanotroph IT-4; and (iii) probe IT9/446 for the type I methanotroph IT-9. Gm705-positive methanotrophs were detected in all samples, indicating that type I methanotrophs were ubiquitous in the marine environments. Probes Sul994 and IT9/446 also identified cells in the vent fluid from which S. caldicuralii and isolate IT-9 were cultivated and isolated.
DISCUSSION
Impact of methane-rich fluid on microbial communities.
In many shallow submarine hydrothermal systems described previously, CO2 is the primary component of gas bubbles found at the vent sites and CH4 is generally a minor gas component (2, 11, 21, 50, 51). The abundant CH4 in the fluid of the Taketomi hydrothermal system indicates that this biogeochemical setting likely selects for a largely methane-driven microbial community. However, the FISH cell counts of type I methanotrophs with probes Gm705 and IT9/446 and the results of cultivation experiments indicated that methanotrophs are less abundant than sulfur-oxidizing bacteria. The ANME-1 phylotypes accounted for approximately 20% of the archaeal clone library in the vent fluid sample, but they were not found in other mat or reference water samples. Since ANME-1 archaea require anaerobic, methane- and sulfate-rich conditions, it is likely that the detected ANME-1 archaea are entrained by the vent fluid from an anaerobic, methane- and sulfate-rich subseafloor niche. Anaerobic oxidation of methane could be a major energy-yielding process and could provide abundant energy sources to sustain ecosystems in methane-rich hydrothermal vent environments (62, 74). In the case of the Taketomi hydrothermal system, such an anaerobic methane oxidation process may occur within sediment layers, and the fluid and microbial mat probably include only clues to the subseafloor process.
Implication for subseafloor communities.
In addition to ANME-1 archaea, anaerobic to microaerobic (hyper)thermophilic microorganisms, such as Caminibacter, Deferribacter, Thermococcus, and DHVEG archaea, were found to be abundant components of the microbial community in the main vent fluid (∼52°C). Some inconsistencies among the results of different experiments were found in this study. The phylotype of Caminibacter was abundant in the vent fluid; nevertheless, no thermophilic hydrogen oxidizer was obtained in cultivation experiments. Thermococcus strains were successfully cultivated from the fluid and mat samples, but the sequence of Thermococcus was not retrieved by 16S rRNA gene clone analysis. For Deferribacter and Thermococcus species, the in situ temperature at the main vent site was markedly lower than the optimum growth temperatures of these organisms. Some of the microorganisms detected in the vent fluid were anaerobic to microaerobic despite the presence of a detectable amount of oxygen in the fluid. These inconsistencies suggest that there are more reductive and/or hotter subseafloor habitats and that some microorganisms from these habitats were entrained by hydrothermal fluid flow.
Epsilonproteobacteria habitats expanded to a shallow hydrothermal system.
Although the cultivation analysis did not show prominent culturability of the Epsilonproteobacteria, culture-independent molecular analyses suggested that such organisms were abundant in the hydrothermal fluid and the proximal habitats. Many previous studies have stressed the abundance, phylogenetic and metabolic diversity, and potential ecophysiological significance of the Epsilonproteobacteria in deep-sea hydrothermal environments (8, 38, 39, 53, 61, 64, 65). However, the ecological features of these organisms in shallow submarine hydrothermal systems are very poorly understood. Our results demonstrated their predominance in the shallow Taketomi system. In addition, not only members of the Thiovulgaceae previously cultivated from coastal oxic-anoxic interface zones (5) but also members of the Nautiliaceae that had previously been believed to occur only in deep-sea hydrothermal systems were identified for the first time in a shallow hydrothermal system. This may imply that there is unseen propagation of deep-sea Epsilonproteobacteria and adaptation of these organisms to other habitats outside the deep sea.
Predominance of chemolithoautotrophs and methanotrophs.
Our results indicate that chemolithoautotrophs and methanotrophs, including members of the Epsilonproteobacteria and unique members of the Gammaproteobacteria, predominated at the main vent site in the Taketomi system. There have been few comparable previous investigations of entire microbial communities in shallow submarine hydrothermal systems, although Sievert et al. (59) demonstrated in a study of the Milos shallow hydrothermal system that there were more heterotrophs than autotrophs in the sediment community despite the fact that high numbers of chemolithoautotrophic sulfur-oxidizing bacteria were cultivated from the hydrothermal sediments. Energy sources could be supplied in different ways in sediment and fluid, which should affect microbial community structures. More complex distribution of energy sources is assumed to occur in the sediment environments. Energy and carbon sources, such as H2, H2S, CO2, CH4, and NH4+ derived from geothermal activities, are dissolved in and transferred by the hydrothermal fluid flows, and thus the predominance of chemolithoautotrophs and methanotrophs in the hydrothermal fluid of the Taketomi system is consistent with the expected flow of chemical inputs. In the Taketomi system, analyses of hydrothermal sediment communities around the main vent site are now in progress in our laboratory. Hence, we will soon be able to compare the sediment communities in the Taketomi and Milos systems.
Photosynthetic contribution to shallow hydrothermal systems.
The other important energy source for microbial primary production in a shallow-water submarine ecosystem is sunlight. Tarasov et al. (72) revealed that photosynthesis is conspicuously stimulated by the high input of nutrients from hydrothermal fluid in shallow-water hydrothermal environments. In this study, however, phototrophic microorganisms were scarce components of the communities around the main vent site at a depth of 23 m. This may have been the result of less energy input from sunlight into the deepest point of the craterlike basin compared with the greater input of inorganic energy sources by the hydrothermal fluid. In contrast, thick green and brown microbial mats developed at the bubbling vent site at a depth of 13 m. In this report, the community structure of the green and brown microbial mats around the bubbling vent site is not described in detail, but a single phylotype closely related to the phototrophic isolate 5H2 obtained from the main vent site accounted for 95% of the bacterial clone library in the colored mat sample (data not shown). This implies that the microbial mats flourishing around the bubbling vent site are likely dominated by a strictly anaerobic, sulfur-oxidizing photoautotroph belonging to the Chlorobi. The difference in microbial community structures in the mats at different depths might be associated with the photosynthetic photon flux densities in the two habitats.
Numerous reports of microorganisms associated with shallow submarine hydrothermal systems have been published, although none of these reports presented a comprehensive characterization of bacterial and archaeal communities in the systems. In this study, we combined cultivation-based and molecular analytical techniques to obtain an overview of active microbial communities in the Taketomi system. At the moment, it is difficult to provide key microbiological characteristics to differentiate the microbial ecosystems in shallow and deep-sea hydrothermal systems other than the light energy input. This is partially due to the limited information available for shallow submarine hydrothermal systems despite the much greater accessibility of these systems. Nevertheless, this study could undoubtedly contribute to our understanding of unique characteristics of variety of global shallow hydrothermal systems and similarities between shallow-water and deep-sea hydrothermal systems.
Footnotes
Published ahead of print on 5 October 2007.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amend, J. P., K. L. Rogers, E. L. Shock, S. Gurrieri, and S. Inguaggiato. 2003. Energetics of chemolithoautotrophy in the hydrothermal system of Vulcano Island, southern Italy. Geobiology 1:37-58. [Google Scholar]
- 3.Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinkhoff, T., J. Kuever, G. Muyzer, and H. W. Jannasch. 2005. Genus VI. Thiomicrospira Kuenen and Veldkamp 1972, 253AL, p. 193-199. In D. J. Brenner, N. R. Krieg, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2, part B. Springer, New York, NY. [Google Scholar]
- 6.Brinkhoff, T., S. M. Sievert, J. Kuever, and G. Muyzer. 1999. Distribution and diversity of sulfur-oxidizing Thiomicrospira spp. at a shallow-water hydrothermal vent in the Aegean Sea (Milos, Greece). Appl. Environ. Microbiol. 65:3843-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, B. J., A. S. Engel, M. L. Porter, and K. Takai. 2006. The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4:458-468. [DOI] [PubMed] [Google Scholar]
- 9.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 10.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 11.Dando, P., J. Hughes, Y. Leahy, S. Niven, L. Taylor, and C. Smith. 1995. Gas venting rates from submarine hydrothermal areas around the island of Milos, Hellenic Volcanic Arc. Cont. Shelf Res. 15:913-929. [Google Scholar]
- 12.Dando, P., M. Thomm, H. Arab, M. Brehmer, L. Hooper, B. Jochimsen, H. Schlesner, R. Stohr, J. Miquel, and S. Fowler. 1998. Microbiology of shallow hydrothermal vent sites off Palaeochori Bay, Milos (Hellenic Volcanic Arc). Cah. Biol. Mar. 39:369-372. [Google Scholar]
- 13.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furushima, Y., H. Yamamoto, T. Maruyama, T. Ohyagi, Y. Yamamura, S. Imanaga, S. Fujishima, Y. Nakazawa, and A. Shimamura. 2004. Necessity of bottom topography measurements in coral reef regions, p. 930-935. In Proceedings of OCEANS '04 MTS/IEEE TECHNO-OCEAN '04, vol. 2. Marine Technology Society, Columbia, MD. [Google Scholar]
- 15.Giovannoni, S. J., E. F. DeLong, G. J. Olsen, and N. R. Pace. 1988. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J. Bacteriol. 170:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gugliandolo, C., and T. Maugeri. 1993. Chemolithotrophic, sulfur-oxidizing bacteria from a marine, shallow hydrothermal vent of Vulcano (Italy). Geomicrobiol. J. 11:109-120. [Google Scholar]
- 17.Gulledge, J., A. Ahmad, P. A. Steudler, W. J. Pomerantz, and C. M. Cavanaugh. 2001. Family- and genus-level 16S rRNA-targeted oligonucleotide probes for ecological studies of methanotrophic bacteria. Appl. Environ. Microbiol. 67:4726-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heyer, J., U. Berger, M. Hardt, and P. F. Dunfield. 2005. Methylohalobius crimeensis gen. nov., sp. nov., a moderately halophilic, methanotrophic bacterium isolated from hypersaline lakes of Crimea. Int. J. Syst. Evol. Microbiol. 55:1817-1826. [DOI] [PubMed] [Google Scholar]
- 19.Hirayama, H., K. Takai, F. Inagaki, K. H. Nealson, and K. Horikoshi. 2005. Thiobacter subterraneus gen. nov., sp. nov., an obligately chemolithoautotrophic, thermophilic, sulfur-oxidizing bacterium from a subsurface hot aquifer. Int. J. Syst. Evol. Microbiol. 55:467-472. [DOI] [PubMed] [Google Scholar]
- 20.Hirayama, H., K. Takai, F. Inagaki, Y. Yamato, M. Suzuki, K. H. Nealson, and K. Horikoshi. 2005. Bacterial community shift along a subsurface geothermal water stream in a Japanese gold mine. Extremophiles 9:169-184. [DOI] [PubMed] [Google Scholar]
- 21.Hoaki, T., M. Nishijima, H. Miyashita, and T. Maruyama. 1995. Dense community of hyperthermophilic sulfur-dependent heterotrophs in a geothermally heated shallow submarine biotope near Kodakara-jima Island, Kagoshima, Japan. Appl. Environ. Microbiol. 61:1931-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes, A. J., A. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 23.Huber, G., R. Huber, B. E. Jones, G. Lauerer, A. Neuner, A. Segerer, K. O. Stetter, and E. T. Degens. 1991. Hyperthermophilic archaea and bacteria occurring within Indonesian hydrothermal areas. Syst. Appl. Microbiol. 14:397-404. [Google Scholar]
- 24.Inagaki, F., T. Nunoura, S. Nakagawa, A. Teske, M. Lever, A. Lauer, M. Suzuki, K. Takai, M. Delwiche, F. S. Colwell, K. H. Nealson, K. Horikoshi, S. D'Hondt, and B. B. Jørgensen. 2006. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. USA 103:2815-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalanetra, K. M., S. L. Huston, and D. C. Nelson. 2004. Novel, attached, sulfur-oxidizing bacteria at shallow hydrothermal vents possess vacuoles not involved in respiratory nitrate accumulation. Appl. Environ. Microbiol. 70:7487-7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaneshima, K., H. Taira, A. Tokuyama, J. Ossaka, and M. Kimura. 1983. Geochemical studies on gas and hot spring gush out of sea bottom at coastal area of Taketomi-jima. Bull. Fac. Sci. Univ. Ryukyus 36:73-80. [Google Scholar]
- 27.Kostka, J., and K. H. Nealson. 1998. Isolation, cultivation and characterization of iron- and manganese-reducing bacteria, p. 58-78. In R. S. Burlage, R. Atlas, D. Stahl, G. Geesey, and G. Sayler (ed.), Techniques in microbial ecology. Oxford University Press, New York, NY.
- 28.Kuever, J., S. Sievert, H. Stevens, T. Brinkhoff, and G. Muyzer. 2002. Microorganisms of the oxidative and reductive part of the sulphur cycle at a shallow-water hydrothermal vent in the Aegean Sea (Milos, Greece). Cah. Biol. Mar. 43:413-416. [Google Scholar]
- 29.Kuo, J., Shibuno, T., Kanamoto, Z., and T. Noro. 2001. Halophila ovalis (R. Br.) Hook. f. from a submarine hot spring in southern Japan. Aquat. Bot. 70:329-335. [Google Scholar]
- 30.Lane, D. J. 1991. 16S/23S sequencing, p. 115-176. In E. Stackbrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley & Sons, New York, NY.
- 31.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K. H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 33.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 34.McDonald, I. R., E. M. Kenna, and J. C. Murrell. 1995. Detection of methanotrophic bacteria in environmental samples with the PCR. Appl. Environ. Microbiol. 61:116-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miguez, C. B., D. Bourque, J. A. Sealy, C. W. Greer, and D. Groleau. 1997. Detection and isolation of methanotrophic bacteria possessing soluble methane monooxygenase (sMMO) genes using the polymerase chain reaction (PCR). Microb. Ecol. 33:21-31. [DOI] [PubMed] [Google Scholar]
- 36.Miroshnichenko, M. L., A. I. Slobodkin, N. A. Kostrikina, S. L'Haridon, O. Nercessian, S. Spring, E. Stackebrandt, E. A. Bonch-Osmolovskaya, and C. Jeanthon. 2003. Deferribacter abyssi sp. nov., an anaerobic thermophile from deep-sea hydrothermal vents of the Mid-Atlantic Ridge. Int. J. Syst. Evol. Microbiol. 53:1637-1641. [DOI] [PubMed] [Google Scholar]
- 37.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 38.Nakagawa, S., K. Takai, F. Inagaki, H. Chiba, J. Ishibashi, S. Kataoka, H. Hirayama, T. Nunoura, K. Horikoshi, and Y. Sako. 2005. Variability in microbial community and venting chemistry in a sediment-hosted backarc hydrothermal system: impacts of subseafloor phase-separation. FEMS Microbiol. Ecol. 54:141-155. [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa, S., K. Takai, F. Inagaki, H. Hirayama, T. Nunoura, K. Horikoshi, and Y. Sako. 2005. Distribution, phylogenetic diversity and physiological characteristics of epsilon-Proteobacteria in a deep-sea hydrothermal field. Environ. Microbiol. 7:1619-1632. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa, T., K. Takai, Y. Suzuki, H. Hirayama, U. Konno, U. Tsunogai, and K. Horikoshi. 2006. Geomicrobiological exploration and characterization of a novel deep-sea hydrothermal system at the TOTO caldera in the Mariana Volcanic Arc. Environ. Microbiol. 8:37-49. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura, T., S. S. Yamazaki, K. Sakai, H. Yamasaki, Y. Furushima, and H. Yamamoto. 2006. Acroporid corals growing over a methane-bubbling hydrothermal vent, Southern Ryukyu Archipelago. Coral Reefs 25:382. [Google Scholar]
- 42.Neef, A. 1997. Ph.D. thesis. Technische Universität München, Munich, Germany.
- 43.Nunoura, T., H. Hirayama, H. Takami, H. Oida, S. Nishi, S. Shimamura, Y. Suzuki, F. Inagaki, K. Takai, K. H. Nealson, and K. Horikoshi. 2005. Genetic and functional properties of uncultivated thermophilic crenarchaeotes from a subsurface gold mine as revealed by analysis of genome fragments. Environ. Microbiol. 7:1967-1984. [DOI] [PubMed] [Google Scholar]
- 44.Nunoura, T., H. Oida, T. Toki, J. Ashi, K. Takai, and K. Horikoshi. 2006. Quantification of mcrA by quantitative fluorescent PCR in sediments from methane seep of the Nankai Trough. FEMS Microbiol. Ecol. 57:149-157. [DOI] [PubMed] [Google Scholar]
- 45.Oomori, T. 1987. Chemical compositions of submarine hot spring water and associated bottom sediments near Taketomi-jima at southern part of the Ryukyu Island Arc, North-west Pacific. J. Earth Sci. Nagoya Univ. 35:325-340. [Google Scholar]
- 46.Orphan, V. J., K.-U. Hinrichs, W. Ussler III, C. K. Paull, L. T. Taylor, S. P. Sylva, J. M. Hayes, and E. F. DeLong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orphan, V. J., L. T. Taylor, D. Hafenbradl, and E. F. DeLong. 2000. Culture-dependent and culture-independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl. Environ. Microbiol. 66:700-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pichler, T., J. P. Amend, J. Garey, P. Hallock, N. P. Hsia, D. J. Karlen, D. R. Meyer-Dombard, B. J. McCloskey, and R. E. Price. 2006. A natural laboratory to study arsenic geobiocomplexity. EOS Trans. Am. Geophys. Union 87:221-225. [Google Scholar]
- 50.Pichler, T., J. Veizer, and G. E. M. Hall. 1999. The chemical composition of shallow-water hydrothermal fluids in Tutum Bay, Ambitle Island, Papua New Guinea and their effect on ambient seawater. Mar. Chem. 64:229-252. [Google Scholar]
- 51.Prol-Ledesma, R., C. Canet, M. Torres-Vera, M. Forrest, and M. Armienta. 2004. Vent fluid chemistry in Bahía Concepción coastal submarine hydrothermal system, Baja California Sur, Mexico. J. Volcanol. Geotherm. Res. 137:311-328. [Google Scholar]
- 52.Reysenbach, A.-L., Y. Liu, A. B. Banta, T. J. Beveridge, J. D. Kirshtein, S. Schouten, M. K. Tivey, K. L. Von Damm, and M. A. Voytek. 2006. A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature 442:444-447. [DOI] [PubMed] [Google Scholar]
- 53.Reysenbach, A.-L., K. Longnecker, and J. Kirshtein. 2000. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent. Appl. Environ. Microbiol. 66:3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers, K. L., and J. P. Amend. 2005. Archaeal diversity and geochemical energy yields in a geothermal well on Vulcano Island, Italy. Geobiology 3:319-332. [Google Scholar]
- 55.Ruby, E. G., C. O. Wirsen, and H. W. Jannasch. 1981. Chemolithotrophic sulfur-oxidizing bacteria from the Galapagos Rift hydrothermal vents. Appl. Environ. Microbiol. 42:317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rusch, A., and J. P. Amend. 2004. Order-specific 16S rRNA-targeted oligonucleotide probes for (hyper)thermophilic archaea and bacteria. Extremophiles 8:357-366. [DOI] [PubMed] [Google Scholar]
- 57.Sako, Y., S. Nakagawa, K. Takai, and K. Horikoshi. 2003. Marinithermus hydrothermalis gen. nov., sp. nov., a strictly aerobic, thermophilic bacterium from a deep-sea hydrothermal vent chimney. Int. J. Syst. Evol. Microbiol. 53:59-65. [DOI] [PubMed] [Google Scholar]
- 58.Sievert, S. M., T. Brinkhoff, G. Muyzer, W. Ziebis, and J. Kuever. 1999. Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island (Greece). Appl. Environ. Microbiol. 65:3834-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sievert, S. M., J. Kuever, and G. Muyzer. 2000. Identification of 16S ribosomal DNA-defined bacterial populations at a shallow submarine hydrothermal vent near Milos Island (Greece). Appl. Environ. Microbiol. 66:3102-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Solorzano, L. 1969. Determination of ammonia in natural waters by the phenolhypochlorite method. Limnol. Oceanogr. 14:799-801. [Google Scholar]
- 61.Sunamura, M., Y. Higashi, C. Miyako, J. Ishibashi, and A. Maruyama. 2004. Two bacterial phylotypes are predominant in the Suiyo seamount hydrothermal plume. Appl. Environ. Microbiol. 70:1190-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takai, K., and K. Horikoshi. 1999. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics 152:1285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takai, K., and Y. Sako. 1999. A molecular view of archaeal diversity in marine and terrestrial hot water environments. FEMS Microbiol. Ecol. 28:177-188. [Google Scholar]
- 64.Takai, K., T. Gamo, U. Tsunogai, N. Nakayama, H. Hirayama, K. H. Nealson, and K. Horikoshi. 2004. Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles 8:269-282. [DOI] [PubMed] [Google Scholar]
- 65.Takai, K., F. Inagaki, S. Nakagawa, H. Hirayama, T. Nunoura, Y. Sako, K. H. Nealson, and K. Horikoshi. 2003. Isolation and phylogenetic diversity of members of previously uncultivated epsilon-Proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol. Lett. 218:167-174. [DOI] [PubMed] [Google Scholar]
- 66.Takai, K., A. Inoue, and K. Horikoshi. 2002. Methanothermococcus okinawensis sp. nov., a thermophilic, methane-producing archaeon isolated from a Western Pacific deep-sea hydrothermal vent system. Int. J. Syst. Evol. Microbiol. 52:1089-1095. [DOI] [PubMed] [Google Scholar]
- 67.Takai, K., H. Kobayashi, K. H. Nealson, and K. Horikoshi. 2003. Deferribacter desulfuricans sp. nov., a novel sulfur-, nitrate- and arsenate-reducing thermophile isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 53:839-846. [DOI] [PubMed] [Google Scholar]
- 68.Takai, K., M. Miyazaki, T. Nunoura, H. Hirayama, H. Oida, Y. Furushima, H. Yamamoto, and K. Horikoshi. 2006. Sulfurivirga caldicuralii gen. nov., sp. nov., a novel microaerobic, thermophilic, thiosulfate-oxidizing chemolithoautotroph, isolated from a shallow marine hydrothermal system occurring in a coral reef, Japan. Int. J. Syst. Evol. Microbiol. 56:1921-1929. [DOI] [PubMed] [Google Scholar]
- 69.Takai, K., H. Oida, Y. Suzuki, H. Hirayama, S. Nakagawa, T. Nunoura, F. Inagaki, K. H. Nealson, and K. Horikoshi. 2004. Spatial distribution of marine crenarchaeota group I in the vicinity of deep-sea hydrothermal systems. Appl. Environ. Microbiol. 70:2404-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takai, K., A. Sugai, T. Itoh, and K. Horikoshi. 2000. Palaeococcus ferrophilus gen. nov., sp. nov., a barophilic, hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney. Int. J. Syst. Evol. Microbiol. 50:489-500. [DOI] [PubMed] [Google Scholar]
- 71.Takai, K., M. Suzuki, S. Nakagawa, M. Miyazaki, Y. Suzuki, F. Inagaki, and K. Horikoshi. 2006. Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 56:1725-1733. [DOI] [PubMed] [Google Scholar]
- 72.Tarasov, V. G., A. V. Gebruk, V. M. Shulkin, G. M. Kamenev, V. I. Fadeev, V. N. Kosmynin, V. V. Malakhov, D. A. Starynin, and A. I. Obzhirov. 1999. Effect of shallow-water hydrothermal venting on the biota of Matupi Harbour (Rabaul Caldera, New Britain Island, Papua New Guinea). Cont. Shelf Res. 19:79-116. [Google Scholar]
- 73.Teske, A. P. 2006. Microbial communities of deep marine subsurface sediments: molecular and cultivation surveys. Geomicrobiol. J. 23:357-368. [Google Scholar]
- 74.Teske, A., K.-U. Hinrichs, V. Edgcomb, A. de Vera Gomez, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]