Abstract
A chlorobenzene reductive dehalogenase of the anaerobic dehalorespiring bacterium Dehalococcoides sp. strain CBDB1 was identified. Due to poor biomass yields, standard protein isolation procedures were not applicable. Therefore, cell extracts from cultures grown on trichlorobenzenes were separated by native polyacrylamide gel electrophoresis and analyzed directly for chlorobenzene reductive dehalogenase activity within gel fragments. Activity was found in a single band, even though electrophoretic separation was performed under aerobic conditions. Matrix-assisted laser desorption ionization mass spectrometry (MALDI MS) and nano-liquid chromatography-MALDI MS analysis of silver-stained replicas of the active band on native polyacrylamide gels identified a protein product of the cbdbA84 gene, now called cbrA. The cbdbA84 gene is one of 32 reductive dehalogenase homologous genes present in the genome of strain CBDB1. The chlorobenzene reductive dehalogenase identified in our study represents a member of the family of corrinoid/iron-sulfur cluster-containing reductive dehalogenases. No orthologs of cbdbA84 were found in the completely sequenced genomes of Dehalococcoides sp. strains 195 and BAV1 nor among the genes amplified from Dehalococcoides sp. strain FL2 or mixed cultures containing Dehalococcoides. Another dehalogenase homologue (cbdbA80) was expressed in cultures that contained 1,2,4-trichlorobenzene, but its role is unclear. Other highly expressed proteins identified with our approach included the major subunit of a protein annotated as formate dehydrogenase, transporter subunits, and a putative S-layer protein.
Dehalococcoides sp. strain CBDB1 belongs to a phylogenetically isolated cluster of strictly anaerobic bacteria that use chlorinated compounds in their energy metabolism by coupling reductive dehalogenation to electron transport phosphorylation (2, 22). Strain CBDB1 uses polychlorinated benzenes, phenols, and dibenzodioxins as growth-supporting electron acceptors (2, 3, 5, 13). Among the chlorobenzenes, 1,2,3-trichlorobenzene (TCB), 1,2,4-TCB, all tetrachlorobenzene (TeCB) isomers, and penta- and hexachlorobenzene are dechlorinated (2, 10, 13). Reductive dechlorination of chlorinated benzenes was also demonstrated for the Dehalococcoides ethenogenes strain 195 (7), a strain that was originally cultivated with chlorinated ethenes as electron acceptors (22), and for Dehalococcoides-like bacterium DF-1, identified in a mixed culture (38).
Several enzymes catalyzing the respiratory reductive dechlorination of chloroaromatics have been isolated and characterized, e.g., the chlorophenol dehalogenases of Desulfitobacterium spp. (6, 15, 36) and the 3-chlorobenzoate dehalogenase of Desulfomonile tiedjei (30). Attempts to isolate chlorobenzene reductive dehalogenase from Dehalococcoides sp. strain CBDB1 have been hampered by poor biomass yields (10). However, the characterization of chlorobenzene reductive dehalogenase activity in cell extracts of strain CBDB1 showed that this enzyme shared several properties with other purified reductive dehalogenases. Thus, light-reversible inhibition by alkyl iodides and the requirement of low-potential electron donors indicated the involvement of cob(I)alamin as a cofactor. Also, the chlorobenzene reductive dehalogenase of strain CBDB1 was shown to be membrane-associated and could be solubilized by treatment with 0.01% Triton X-100 (10).
Sequencing of the CBDB1 genome (17) revealed the presence of as many as 32 reductive dehalogenase homologous (rdhA) genes that are candidates for the chlorobenzene dehalogenase. In our present study, we describe the purification of a chlorobenzene reductive dehalogenase by native polyacrylamide gel electrophoresis (PAGE) and the identification of the dechlorinating protein and other highly expressed proteins by tandem mass spectrometry, using CBDB1 genome data for gene identification.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strain CBDB1 was cultivated in strictly anaerobic, Ti(III)citrate-reduced, purely synthetic mineral medium with 0.5 mM acetate as the carbon source, hydrogen as the electron donor (a nominal concentration of 7.5 mM), and TCB as the electron acceptor, as described previously (2). Two different procedures were applied to add TCBs to the cultures. In the first experiments, cultures were started with a mixture of 1,2,3- and 1,2,4-TCB (15 μM each), and after all TCB was dechlorinated, cultures were supplied with additional 10 mM 1,2,3-TCB (nominal concentration) added as a 0.2 M solution in hexadecane, providing a steady supply of 1,2,3-TCB to the medium. In later experiments, cultures were fed with successive 30 μM doses of 1,2,3-TCB whenever TCB was depleted. In this way, cultures obtained accumulated nominal concentrations of up to 200 μM 1,2,3-TCB. Cultures were processed when cell numbers reached about 5 × 107 cells ml−1. Cell numbers were quantified by direct cell counting on agarose-covered slides (3).
Preparation of samples for PAGE.
Cells from 200 to 300 ml of culture fluid containing 0.5 to 1 μg protein per ml were harvested under anoxic conditions by centrifugation at 5,500 × g for 30 min at 4°C. Previous centrifugation experiments evaluated by the direct cell-counting method showed a maximum recovery of cells by using these parameters. Higher g values led to lower cell recoveries, probably due to rupturing of the cells. The barely visible pellet was carefully resuspended in 2 ml of residual culture supernatant, using a Pasteur pipette, and centrifugation was repeated once. The final cell pellet was resuspended in 0.1 ml of residual culture supernatant and incubated in taurodeoxycholate (TDC) electrophoresis sample buffer (70% 0.5 M Tris-HCl, pH 6.8, 30% glycerol, 5 mM TDC, 0.01% bromophenol) for 15 min. For the preparation of membrane proteins, cells were broken by three cycles of ultrasonication in buffer 1 (10 mM Tris-HCl, pH 7.5, 1 mM dithiothreitol [DTT]) using a Branson 250 sonicator (at intensity 4; on 20% duty cycle; for 5 min). The lysate was centrifuged at 10,000 × g for 30 min at 4°C, and the supernatant (crude extract) was subjected to ultracentrifugation at 120,000 × g for 1 h at 4°C. The pelleted membranes were solubilized in buffer 1 containing 10 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}. After ultracentrifugation at 120,000 × g for 1 h at 4°C, the supernatant containing solubilized membrane proteins was concentrated by centrifugation through a membrane filter with a molecular cutoff of 50 kDa (Centricon YM-50; Millipore, Bedford, MA). The resulting protein sample (100 μl) was mixed with TDC electrophoresis sample buffer.
Gel electrophoresis.
Native polyacrylamide gels with a 10% resolving gel and a 4% stacking gel were prepared as described by Läemmli (18), except that the detergent, sodium dodecyl sulfate (SDS), was replaced by 5 mM TDC in all buffers (32). Such native gels were run at 100 to 200 V for 2 h at 4 to 8°C under aerobic conditions. Denaturing gels containing SDS were run under the same conditions but at room temperature.
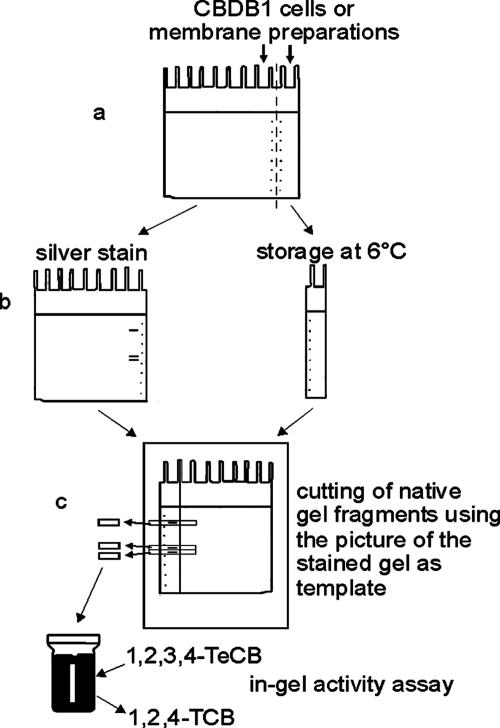
For native PAGE of CBDB1 proteins, aliquots of a sample were applied simultaneously on two lanes of a gel, one of which was later used for activity assays, and the other was used for protein staining and mass spectrometry (Fig. 1). After electrophoresis, two 5-mm scales were stamped into the gel with a sterile Pasteur pipette to compensate for gel shrinkage after the fixing and staining procedures. The gel was then cut vertically into two parts, between the two lanes containing the same sample (Fig. 1). One part of the gel was stained with silver according to the method used by Nesterenko et al. (27). The other part of the gel was left unstained to be able to measure activity. The unstained part of the gel was placed on a printout of a digital mirror-image picture of the silver-stained gel to locate the protein bands in the unstained gel. From the bands that could be visualized on the picture, lanes were cut horizontally into 1- to 5-mm fragments, which were subsequently assayed for dehalogenase activity (see below) or used in a second electrophoresis step. To elute proteins in a second electrophoresis step, gel fragments were cut into 1-mm2 pieces and transferred into Eppendorf cups containing 1 ml of elution buffer (125 mM Tris-HCl, pH 6.8, 0.1% SDS for SDS-PAGE; 125 mM Tris-HCl, pH 6.8, 5 mM TDC for native PAGE). For the elution of proteins from SDS gels, samples were incubated with shaking at 1,300 rpm for 30 min at 95°C. For the elution of proteins from native gels, samples were incubated with shaking at 800 rpm for 30 min at room temperature. The 1-ml supernatants containing eluted proteins were concentrated 20-fold using a membrane filter with a molecular cutoff of 30 kDa (Centricon YM-30). The concentrates were analyzed by SDS-PAGE or a second native PAGE step as described above.
FIG. 1.
Separation and detection scheme of chlorobenzene reductive dehalogenase using native PAGE. First, proteins were separated on a native polyacrylamide gel (a). The gel was then scaled and vertically cut into two parts (b). One part was silver stained and photographed. The mirror image and size-corrected picture was used as a template with which to cut the unstained part of the gel into fragments containing single bands (c). The fragments were analyzed for dechlorination activity in an in vitro activity assay. See text for details.
Assay of dehalogenase activity in gel fragments.
Dehalogenase activity was assayed essentially as described previously (10). Inside an anaerobic chamber, gel fragments from unstained lanes were transferred into 2-ml glass vials. To each vial, 2 ml of assay solution {containing 100 mM potassium acetate [pH 5.8], 1 mM methyl viologen, 2 mM titanium(III)citrate, and 50 μM of a chlorobenzene congener} was added. Vials were sealed with Teflon-lined rubber septa and incubated at 29°C for 24 h. The reaction mixture was then extracted with hexane, and extracts were analyzed for chlorobenzenes by gas chromatography and flame ionization detection (1, 2).
Mass spectrometry. (i) Chemicals.
The peptide calibration standards, angiotensin I, and the human adrenocorticotropic hormone (ACTH) 18-39 were purchased from Bachem (Heidelberg, Germany). Acetonitrile (high-performance liquid chromatography gradient grade) was purchased from Carl Roth GmbH (Karlsruhe, Germany). Trifluoroacetic acid (TFA), tetrahydrofuran, n-octylglucopyranoside (n-OGP), α-cyano-4-hydroxycinnamic acid (CHCA), and water used for high-performance liquid chromatography solvents and matrix-assisted laser desorption ionization (MALDI) matrix solutions were purchased from Fluka Chemie (Buchs, Switzerland). Porcine trypsin was purchased from Promega (Mannheim, Germany); DTT and iodoacetamide (IAA) were from Sigma (Sigma-Aldrich, St. Louis, MO); and citric acid and 2,2′-thiodiethanol were from Aldrich (Sigma-Aldrich, St. Louis, MO).
(ii) In situ trypsinolysis.
Bands excised from the silver-stained polyacrylamide gels were washed twice by the addition of 100 μl of 50% ethanol (vol/vol), 50 mM NH4HCO3 (the washing solution), and incubation for 30 min with shaking. The washing solution was removed, and the gel samples were dehydrated by incubation with 100 μl ethanol for 5 min, after which the ethanol was removed. For reduction of cysteine disulfides, 10 μl of 10 mM DTT and 50 mM NH4HCO3 were added, and the samples were incubated for 30 min at 37°C. The supernatants were removed, and the samples were dehydrated by incubation with 100 μl ethanol for 5 min, after which the ethanol was removed. For the alkylation of cysteines, 10 μl of 55 mM IAA and 50 mM NH4HCO3 were added, and the samples were incubated for 30 min at room temperature in the dark. The liquid was removed, and the samples were washed by incubation with 800 μl of washing solution for 5 min. The washing solution was removed, and the washing procedure was repeated once, followed by incubation with 100 μl ethanol for 10 min. After removing the liquid, the samples were dried in a vacuum centrifuge for 60 min. For trypsinolysis, 10 μl trypsin solution (2.5 ng/μl) in 50 mM NH4HCO3, pH 7.8 (digestion buffer), was added to the dry gel pieces. After a 30-min incubation on ice, additional volumes of digestion buffer sufficient to just cover the gel particles were added to the samples, followed by incubation overnight at 37°C. For peptide extraction, 15 μl of 0.1% TFA and 0.2 mM n-OGP were added, and the samples were placed on a shaker for 10 min. The supernatants were collected in fresh vials and stored at −20°C prior to further analysis.
(iii) MALDI MS.
The tryptic digests were prepared for MALDI, using the previously published CHCA affinity sample preparation (9), and analyzed on an Ultraflex II LIFT MALDI-time of flight/time of flight (TOF/TOF) mass spectrometer (Bruker Daltonics, Bremen, Germany). Positively charged ions in the m/z range of 500 to 4,500 Da were analyzed in the reflector mode. After acquisition of mass spectra, tandem mass spectrometry (MS/MS) spectra were acquired for all peptides that could be isolated.
(iv) Nano-LC-MALDI MS.
Two samples for which reliable protein identification could not be achieved by direct MALDI MS analysis of the tryptic digests were analyzed by nano-liquid chromatography (LC)-MALDI MS as described previously (23). In brief, peptide samples were loaded on an 1100 Series model nanoflow LC system (Agilent Technologies, Waldbronn, Germany). The mobile phases used for the reversed-phase separation were buffer A (1% acetonitrile [vol/vol], 0.05% TFA [vol/vol]) and buffer B (90% acetonitrile [vol/vol], 0.04% TFA [vol/vol]). The samples were first loaded onto a trapping column (C18; 0.3 mm by 5 mm; ZORBAX 300 SB; Agilent Technologies), using buffer A, delivered by the loading pump with a flow gradient according to the manufacturer's recommendation. After 5 min, the trapping column was connected to the nanoflow path, and the samples were eluted onto the analytical separation column (C18; 75 μm by 150 mm; ZORBAX 300 SB; Agilent Technologies), using a binary pump operated at 300 nl/min. The binary gradients (given as min/% of buffer B/nl/min flow) were 0/3/300, 5/3/300, 8/15/300, 18/20/300, 38/30/300, 68/45/330, 73/95/350, 78/95/400, and 82/3/300. One hundred ninety-one fractions were collected over a period of 63 min (start time, 14 min; end time, 77 min), using a model 1100 Micro-CF fraction collector (Agilent). The LC effluent was fractionated onto preformed microcrystalline layers of CHCA prepared on prestructured MALDI sample supports (AnchorChip 600/384; Bruker Daltonics, Bremen, Germany). The matrix solution consisted of CHCA (100 g/liter) in 90% tetrahydrofuran, 0.001% TFA (vol/vol), and 50 mM citric acid and contained the two calibration standards angiotensin I (1 pmol/μl) and ACTH 18-39 (2 pmol/μl). Thin layers of CHCA were prepared by spreading 200 μl of matrix solution over the target surface with a Teflon rod (9). Mass analysis of positively charged peptide ions in the m/z range of 500 to 4,500 Da was performed on an Ultraflex II LIFT MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). Sums of 50 single-shot spectra were acquired from 14 different sample spot positions (a total of 700 from each sample). The selection of precursor ions was performed based on the entire MS data set, using a beta version of WarpLC software (in collaboration with Bruker Daltonics, Bremen, Germany), with manual corrections when necessary. Up to 15 MS/MS spectra, each consisting of a total of 4,100 laser single-shot spectra, were acquired from a single MALDI sample. Additionally, manual spectrum acquisition of signals for which parent ion isolation failed in the automatic mode was performed when sample availability permitted.
(v) Data processing.
Automatic detection of the peptide monoisotopic signals was performed using the algorithm SNAP, implemented in FlexAnalysis software (Bruker Daltonics, Bremen, Germany). Internal mass correction was performed using the signals of two peptides (angiotensin I, MH+ 1,296.6853 [monoisotopic mass]), and ACTH (18-39; MH+ 2,465.1989) included in the MALDI matrix solution. Protein identification was performed using Mascot software (Matrixscience, London, United Kingdom) to search a database of the genome of Dehalococcoides strain CBDB1 (GenBank accession number AJ965256). The settings used for the searches were as follows: mass error tolerance for the precursor ions, 30 ppm; mass error tolerance for the fragment ions, 0.8 Da; fixed modification, carbamidomethylation; variable modification, methionine oxidation; number of missed cleavage sites, 1; type of instrument, MALDI-TOF post-source decay.
Sequence analysis.
The predicted amino acid sequence of chlorobenzene reductive dehalogenase was compared to other published sequences using BLASTP (http://www.ncbi.nlm.nih.gov/BLAST), and sequences were aligned with ClustalW (http://www.ebi.ac.uk/clustalw/). Analysis of the putative promoter region was carried out with a Neural Network Promoter Prediction tool (http://www.fruitfly.org/seq_tools/promoter.html), and transmembrane helices in proteins were predicted with a TMHMM tool (http://www.cbs.dtu.dk/services/TMHMM-2.0). Prediction of signal peptide cleavage sites was carried out with SignalP using the hidden Markov model algorithm based on gram-negative bacteria (http://www.cbs.dtu.dk/services/SignalP/) and with TatP (http://www.cbs.dtu.dk/services/TatP/). Theoretical masses of proteins were calculated by using the ExPASy Compute pI/Mw tool (http://expasy.org/tools/pi_tool.html).
Nucleotide sequence accession number.
The whole-genome sequence of Dehalococcoides sp. strain BAV1 has been deposited in the GenBank database under accession number CP000688 (A. Copeland et al., unpublished data).
RESULTS
Protein band pattern and dechlorinating activity in gels.
Protein preparations of strain CBDB1 were analyzed by native PAGE using TDC as the nondenaturing detergent in the gel. With membrane protein preparations from about 109 cells, a clear and reproducible band pattern with two dominant and several clearly visible but fainter bands was obtained. Essentially the same band pattern was obtained with lysed whole-cell preparations from about 108 cells (Fig. 2a). Dechlorinating activity could be directly recovered from native gels. In former studies, the highest dechlorination rates in crude extracts of strain CBDB1 were achieved with 1,2,3,4-TeCB (10); therefore, this congener was routinely used to assay dehalogenase activity in gel fragments. An experimental setup was designed to correlate dechlorinating activity with distinct bands in the gel (Fig. 1). With this procedure, dechlorinating activity toward 1,2,3,4-TeCB was detected in an area of the gel corresponding to one of the two dominant bands in a parallel silver-stained lane (Fig. 2a). 1,2,3,4-TeCB was dechlorinated to 1,2,4-TCB as the only product. With the native gel fragment containing chlorobenzene dehalogenase, 35 μM 1,2,4-TCB was formed from 50 μM substrate after 24 h of incubation. No activity was detected in other gel fragments. Furthermore, dechlorination of 1,2,3-TCB to 1,3-dichlorobenzene and dechlorination of pentachlorobenzene to 1,2,4,5-TeCB were detected with the respective gel fragment, albeit with very low rates (1 μM 1,3-DCB and below 1 μM 1,2,4,5-TeCB, respectively, after incubation for 24 h).
FIG. 2.
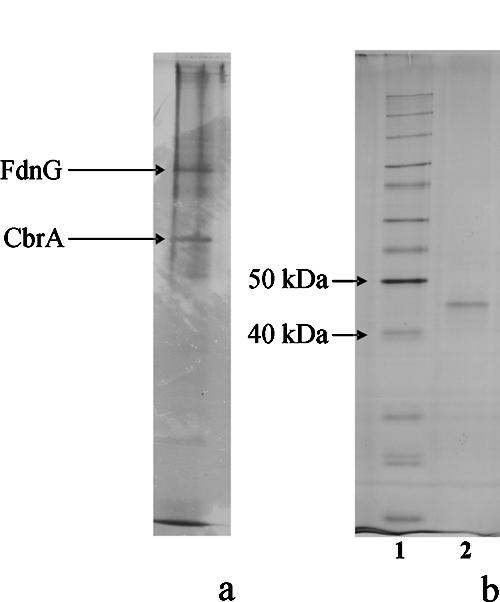
(a) Whole-cell proteins from strain CBDB1 separated by native PAGE. (b) Chlorobenzene dechlorinating protein (CbrA) analyzed by SDS-PAGE. Lanes: 1, molecular mass marker; 2, eluted protein from an active native gel fragment.
In a control experiment, dehalogenase protein was eluted from a native gel fragment and submitted to a second native PAGE procedure or to SDS-PAGE. The silver-stained part of the second native gel displayed a single protein band with the same relative mobility as the active protein from the first native gel (not shown). In the corresponding unstained gel fragment, 1,2,3,4-TeCB dehalogenase activity was again detected. When the active band from the native gels was analyzed by SDS-PAGE, a distinct band in the 45-kDa region was obtained (Fig. 2b). No activity was recovered from SDS-gel fragments.
Mass spectrometric identification of the chlorobenzene reductive dehalogenase.
For mass spectrometric analysis of single bands, the amount of cells applied to native PAGE had to be increased 10- to 100-fold. In our first experiments, we used cultures that were started with a mixture of 1,2,3- and 1,2,4-TCB. After all initial substrate was used up, cultures were overlaid with a hexadecane/1,2,3-TCB layer and incubated for several weeks. For native PAGE, membrane protein preparations from about 1010 cells were used. A band in the silver-stained part of a native gel corresponding to the active fragment in the nonstained part of the gel was carefully excised. The protein contained in the gel was subjected to in situ tryptic digestion, and the eluted cleavage peptides were analyzed by MALDI MS. MS/MS analysis identified two different reductive dehalogenase homologs (RdhAs) from the translated genome of strain CBDB1, cbdbA80 (GenBank accession number CAI82340) and cbdbA84 (GenBank accession number CAI82345). Both genes encode a sequence that contains all features expected for a functional reductive dehalogenase (reviewed in reference 34). However, peptides of both RdhAs were also detected in other bands on the gel, indicating that the gel was overloaded with protein.
In further experiments, cultures that were fed exclusively 1,2,3-TCB as the electron acceptor to avoid possible interferences with other chlorobenzene substrates or their products were analyzed. For mass spectrometric analysis, native gels were loaded with lysed whole-cell preparations from about 109 cells. In a silver-stained band corresponding to the active reductive dehalogenase, the RdhA cbdbA84 was again detected. No other proteins were detected in this gel fragment. A total of 28 signals in the mass spectrum matched that of cbdbA84, resulting in a sequence coverage of 52% for the protein (Fig. 3 and Table 1). No peptide matching the encoded peptide leader in cbdbA84 was found. cbdbA84 was the only RdhA that was found in the gel with this approach, and no smearing of the protein was observed.
FIG. 3.
Protein sequence encoded by the cbdbA84 gene. Peptides identified by mass spectrometry are underlined. The twin-arginine motif and iron-sulfur cluster binding motifs are highlighted in gray; residues of the truncated cobalamin-binding consensus sequence are in bold. The arrow indicates the predicted leader peptide cleavage site.
TABLE 1.
Peptides found in native gels with preparations of membrane proteins and whole cells
| Locus tag | GenBank accession no. | No. of peptides found (gel 1/gel 2)a | Mascot score(s) (gel 1/gel 2) | No. of matched m/z values (gel 1/gel 2) | No. of matching MS/MS spectra (gel 1/gel 2) | % of sequence coverage (gel 1/gel 2) | Annotation | Gene | Calculated mass (kDa)b | Presence of signal peptide (name) |
|---|---|---|---|---|---|---|---|---|---|---|
| cbdbA84 | CAI82345 | 1/2 | 263/277 | 24/28 | 2/6 | 50/52 | Chlorobenzene reductive dehalogenase | cbrA | 51.2 | Yes (Tat) |
| cbdbA80 | CAI82340 | 1 | 162 | 11 | 2 | 25 | Putative reductive dehalogenase | rdhA | 52.5 | Yes (Tat) |
| cbdbA195 | CAI82443 | 1/2* | 165/662 | 10/16 | 1/15 | 14/21 | Formate dehydrogenase, major subunit | fdnG | 102.2 | Yes (Tat) |
| cbdbA131 | CAI82389 | 2* | 164 | 4 | 3 | 15 | Putative Ni/Fe hydrogenase, iron-sulfur cluster-binding subunit | 30.4 | No | |
| cbdbA1368 | CAI83414 | 1/2* | 111/255 | 4/7 | 1/6 | 8/11 | Conserved domain protein (putative S-layer protein [24]) | 103.7 | Yes | |
| cbdbA1039 | CAI83145 | 1 | 310 | 20 | 2 | 37 | ABC-type dipeptide/oligopeptide/nickel transport system, periplasmic component | 58.4 | Yes | |
| cbdbA1463 | CAI83489 | 2 | 367 | 10 | 5 | 25 | Peptide ABC transporter, periplasmic peptide-binding protein | 56.8 | Yes | |
| cbdbA1393 | CAI83436 | 1 | 171 | 3 | 3 | 7 | Co-chaperonin GroEL | groEL | 57.1 | No |
| cbdbA960 | CAI83087 | 2* | 164 | 4 | 4 | 14 | Translation elongation factor Tu | tuf | 44.0 | No |
| cbdbA1358 | CAI83407 | 2* | 460 | 11 | 10 | 21 | Chaperone protein DnaK | dnaK | 68.5 | No |
| cbdbA1675 | CAI83683 | 2* | 268 | 8 | 6 | 43 | LemA family protein | 18.9 | Yes | |
| cbdbA1077 | CAI83178 | 2* | 420 | 17 | 15 | 21 | CoA binding ligase | 97.9 | No | |
| cbdbA1729 | CAI83727 | 2* | 73 | 5 | 3 | 15 | TPR domain protein | 30.0 | No |
Peptides were detected with preparations of membrane proteins (gel 1) and whole cells (gel 2). *, peptides identified by nano LC-MALDI MS. Threshold score (P < 0.05) = 44.
Theoretical masses of proteins with leader peptides are given for the processed form. Tat, twin-arginine signal peptide.
Sequence analysis of cbrA and adjacent genes.
The cbdbA84 gene, now identified as chlorobenzene reductive dehalogenase, is one of 32 rdhA genes found in the genome of strain CBDB1 (17). The cbdbA84 gene, now designated cbrA (for chlorobenzene reductive dehalogenase), consists of 1,467 nucleotides encoding 488 amino acids. Potential sigma70-type promoter regions and a ribosome binding site were found upstream of cbrA. A BLASTP search and pairwise alignments using ClustalW revealed that the encoded protein CbrA shows the highest identity (41.2%) with a putative reductive dehalogenase of Dehalococcoides sp. strain BAV1 (accession number AAT48552; see also reference 17 and supplemental material therein). The chlorobenzene reductive dehalogenase gene shares all the sequence characteristics described for other known reductive dehalogenases. Thus, CbrA contains a twin-arginine signal peptide described to mediate transport to or across the cell membrane (Fig. 3). The mature protein has a calculated molecular mass of 51.2 kDa. This is in reasonable agreement with the size of the mature dehalogenase, 45 kDa, determined by SDS-PAGE. Two iron-sulfur cluster binding motifs are encoded near the C-terminal end of the dehalogenase. The cobalamin-binding consensus sequence described for several cobalamin-dependent isomerases and methyltransferases of other prokaryotes (e.g., DXHXXG-X41-SXL-X26-28-GG) (19) and for the three RdhAs in strain CBDB1 [DXXHXXG-X52-SXL-X35-37-G(G)] (11, 17) are absent in CbrA. However, in the corresponding region of CbrA, a truncated version of this consensus sequence, DXH-X26-SXL-X56-G, could be identified.
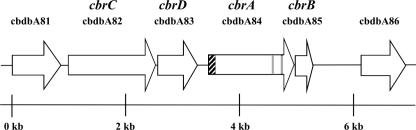
In the genome, cbrA is directly adjacent to and in the same orientation as a gene encoding a putative membrane-anchoring protein (29) with three predicted transmembrane helices, cbrB (cbdbA85). cbrB consists of 273 nucleotides and is located 21 bp downstream of cbrA (Fig. 4). The short stretch between cbrA and cbrB contains a putative ribosome binding site for cbrB but no termination region. As with most putative reductive dehalogenases in strain CBDB1 (17), the cbrAB genes are associated with genes encoding transcription regulators; a two-component regulatory system, composed of a sensor histidine kinase gene (cbrC-cbdbA82) and a response regulator gene (cbrD-cbdbA83), was found directly upstream of cbrA (Fig. 4).
FIG. 4.
Physical map of the cbr gene locus (cbdbA81-cbdbA86). The cbrAB operon is located downstream of a histidine kinase (cbrC) and a response regulator (cbrD). The cbrAB regions encoding the twin-arginine leader peptide and the iron-sulfur cluster binding motif are highlighted. The cbdbA81 and cbdbA86 genes encode amino acid sequences without similarity to characterized proteins.
Identification of further abundant proteins.
Besides the most intense protein band showing dehalogenase activity, several other distinct bands were detected in native electrophoresis gels after silver staining. All clear bands were excised and analyzed by MALDI MS. Bands with low identification scores were analyzed further by nano-LC MALDI MS (Table 1). The second most intense band was present in all gels. The predominant protein in this band was found to be encoded by the cbdbA195 gene, which is presently annotated as the major subunit of formate dehydrogenase (Fig. 2a, FdnG). The gene consists of 2,982 bp potentially encoding a protein of 993 amino acids. Signal peptide prediction tools predicted the presence of a twin-arginine leader peptide of 33 amino acids. After cleavage of this leader peptide, the mature protein is predicted to have a molecular mass of 102.2 kDa. The gene encodes several domains characteristic of the α-subunit of molybdopterin-containing oxidoreductases such as nitrate or formate dehydrogenase. We obtained a mass spectrometric sequence coverage of about 20% for this protein. However, as also found by Morris et al. (25), one of the peptides identified (LSTASSLEALAASFGR) contained a serine residue (underlined) at a crucial position that is occupied by selenocysteine or cysteine in both putative and described formate dehydrogenases (24, 39). In addition, the protein contains an unusual iron-sulfur cluster motif between positions 53 and 64 on the protein, creating doubt about the function of this protein as formate dehydrogenase. In the genome, fdnG is located in the direct vicinity of and in the same orientation as two other genes, annotated as the membrane subunit of formate dehydrogenase and a formate dehydrogenase accessory protein (fdhE), respectively. The latter proteins were not identified in native gels by mass spectrometry.
MALDI MS and nano-LC MALDI MS revealed the presence of the gene product of cbdbA1368 (Table 1). The gene encodes a 1,015-amino-acid protein with a 31-amino-acid N-terminal signal peptide, one C-terminal transmembrane helix, five bacterial neuraminidase repeats (BNR/Asp-Box repeats) of unknown function, and a fibronectin type III (FN3) domain. Two homologs of cbdbA1368 are known, DET1407 (accession number AAW39334), carried in Dehalococcoides strain 195, which is speculated to encode the S-layer protein of the strain (24, 25), and DehaBAV1 1214 (accession number ABQ17793), a gene detected in the genome of Dehalococcoides strain BAV1. The overall amino acid identity of cbdbA1368 with DET1407 and DehaBAV1 1214 is 73% and 89%, respectively. However, the degree of conservation of the individual segments of the proteins varies significantly. While the C- and N-terminal parts of the proteins are highly conserved, with amino acid identities of 90 to 100%, several parts in the sequence are much less conserved. The FN3 domain of cbdbA1368 shows sequence identity of only 50% with DET1407 and 25% with DehaBAV1 1214. In addition, insertions of 4 and 13 amino acids are present in the FN3 domains of cbdbA1368 and DehaBAV1 1214, respectively, compared with that of DET1407. Besides the domains identified, five highly conserved repeats of 14 to 20 amino acid residues separated by less conserved stretches of about 15 amino acids can be recognized between amino acid positions 150 and 320 of cbdbA1368 and DET1407.
With GroEL (cbdbA1393), translation elongation factor Tu (cbdbA960), and DnaK (cbdbA1358) dominant cytosolic proteins were also detected. Other proteins detected in our analysis with sequence coverage above 20% were ABC-type transporter periplasmic components (cbdbA1039 and cbdbA1463), a LemA family protein (cbdbA1675), and a gene product originally annotated as “acetyl-coenzyme A (CoA) synthetase” (cbdbA1077) (Table 1). After an up-to-date analysis, we use the annotation “CoA binding ligase” for the cbdbA1077 gene.
DISCUSSION
In this study, for the first time, a chlorobenzene reductive dehalogenase (CbrA) involved in respiratory energy generation was identified. Furthermore, the cbrA gene is the first of 32 rdhA genes in strain CBDB1 to which a function is assigned. Using native PAGE, CbrA was separated from a mixture of concentrated whole-cell proteins. Peptide fragments of the enzyme were identified by MS/MS and correlated with the cbdbA84 gene. CbrA is a novel member of the family of corrinoid/iron-sulfur cluster-containing reductive dehalogenases (34) and the first from Dehalococcoides species that contains, albeit truncated, a cobalamin-binding consensus sequence. A truncated and modified corrinoid-binding motif was also found in a 3,5-dichlorophenol reductive dehalogenase from Desulfitobacterium frappieri PCP-1 (35). The high intensity of the band that represented CbrA demonstrates that CbrA is one of the most abundant proteins in strain CBDB1 when grown on 1,2,3-TCB. In both of our analyses, CbrA had the highest sequence coverage of all proteins detected. The fact that we did not find fragments of the signal peptide with our mass spectrometric analysis indicates that, indeed, the signal peptide is not present in the mature protein. Three other Dehalococcoides reductive dehalogenases were identified following partial purification from enriched mixed cultures, the trichloroethene dehalogenase (TceA) and the tetrachloroethene dehalogenase (PceA) of D. ethenogenes strain 195 (20, 21, 24) and the vinyl chloride reductive dehalogenase of Dehalococcoides sp. strain VS (26). In a recent study, transcription analysis, mass spectrometry of membrane protein fractions, and physiological experiments gave strong evidence that the PceA protein of D. ethenogenes also catalyzes the dechlorination of 2,3-dichlorophenol to monochlorophenol (8).
CbrA was identified in strain CBDB1 grown with 1,2,3-TCB as the sole electron acceptor, suggesting that CbrA is responsible for metabolic dechlorination of 1,2,3-TCB. The massive expression of this protein relative to that of other proteins in strain CBDB1 indicates a crucial role in the metabolism and supports our view that the enzyme is one of the key enzymes of energy conservation in the strain. Dehalogenase activity assays with the CbrA protein band separated by native PAGE supported the assumption that besides the rapidly dechlorinated congener 1,2,3,4-TeCB, 1,2,3-TCB is dechlorinated by CbrA. This indicates that at least some of the previously described dechlorination reactions of strain CBDB1 (10) are catalyzed by one enzyme. No orthologs of the CbrA-encoding gene cbdbA84 were found in the completely sequenced genomes of Dehalococcoides strains 195 (33) and BAV1 nor among the genes amplified from strain FL2 or from mixed cultures containing Dehalococcoides (11, 37). So far, strain CBDB1 is the only pure strain described that is able to grow with 1,2,3-TCB as the sole electron acceptor (2), while cometabolic 1,2,3-TCB dechlorination has been found with strain 195 (7).
A prerequisite for the successful identification of CbrA was the use of methods that allowed detection of dehalogenase activity directly in native PAGE slices. Dechlorination with 1,2,3,4-TeCB was detected even after two successive native PAGE steps under aerobic conditions. In contrast, hydrogenase activity in strain CBDB1 was found to be extremely O2 labile in previous studies (12). Therefore, our findings support the assumption that the very high oxygen sensitivity of the hydrogenotrophic strain CBDB1 is not due to the chlorobenzene dehalogenase but rather to the instability of the hydrogenase(s) toward oxygen. However, other cell components may also be O2 labile.
When CBDB1 cultures set up with a mixture of 1,2,3- and 1,2,4-TCB were used for native PAGE, the dechlorinating band gave signals for two RdhAs, cbdbA80 and cbdbA84. While 1,2,3-TCB dechlorination is linked to the cbdbA84 gene, it could be speculated that cbdbA80 encodes a 1,2,4-TCB reductive dehalogenase. Due to the very slow in vitro dechlorination of 1,2,4-TCB by strain CBDB1 (10), we were not able to directly assay 1,2,4-TCB dehalogenase activity in the gels. Therefore, we attempted to analyze electrophoretically separated proteins from cultures grown exclusively on 1,2,4-TCB; however, mass spectrometric analysis did not yield signal intensities significantly above our detection threshold.
In a recent study, Morris et al. performed mass spectrometric analysis of membrane-enriched fractions from pure and mixed cultures of strain 195 and strain CBDB1 (25). In fractions from strain CBDB1 grown with 2,3-dichlorophenol as the sole electron acceptor, they detected the PceA ortholog cbdbA1588 (31% protein sequence coverage) as the most abundant RdhA, and two additional RdhAs, cbdbA80 and cbdbA88 (13% and 10% protein sequence coverage, respectively). CbrA (cbdbA84) was not detected, corroborating evidence that this protein is specifically associated with chlorobenzene dechlorination. The detection of cbdbA80 as the second most abundant RdhA in 2,3-dichlorophenol-grown CBDB1 cells might indicate, however, that the expression of cbdbA80 is not induced specifically by one chlorinated compound but that expression is either broadly induced or even constitutive. A highly similar ortholog of cbdbA80, DET1559, was expressed in a tetrachloroethene-grown mixed culture containing strain 195, albeit to a much lower degree than that of TceA and PceA (25), supporting this hypothesis.
Besides the reductive dehalogenase, the oxidation of external hydrogen and channeling of the electrons into the membrane-bound electron chain play pivotal roles in the respiratory metabolism of strain CBDB1, as described previously (12). The most abundant protein besides CbrA, however, was a protein (cbdbA195) annotated as the major subunit of formate dehydrogenase (fdnG). The high abundance, a twin-arginine leader peptide targeting the protein to the twin-arginine translocation pathway (4), and the presence of several redox cluster-binding domains suggest that the gene encodes an important protein in the respiratory metabolism of strain CBDB1. The annotation of cbdbA195 as formate dehydrogenase, however, is contradictory to physiological experiments with strain CBDB1, which clearly demonstrated that formate is not used by strain CBDB1 as an electron donor (2). The presence of a serine residue in cbdbA195 at a crucial position that contains selenocysteine or cysteine as described in the formate dehydrogenases of other species (24, 39) points toward a function other than formate utilization. Similar results have been described for Dehalococcoides strain 195 (22, 24, 25). In fact, no single selenocysteine protein and no protein of the selenium incorporation pathways (SelA, SelB, SelD, or YbbB) were detected in Dehalococcoides species in a recent comparative genome analysis (39).
Subunits of the predicted hydrogen uptake complex hupL/hupS previously speculated to be responsible for hydrogen uptake in strain CBDB1 (17) and detected in the proteomes of Dehalococcoides strains 195 and CBDB1 in another study (25) were not identified in the protein extract, with our approach. Only the increased sensitivity of a reverse transcription-PCR approach resulted in the detection of the mRNA of hupS (G. Jayachandran and L. Adrian, unpublished data).
With both whole-cell and membrane protein preparations, membrane-associated and periplasmic proteins constituted the major portion of proteins identified by mass spectrometry. Dehalococcoides cells are disc shaped and therefore have a high surface to volume ratio. Thus, membrane-associated proteins most likely constitute a major portion of total protein in Dehalococcoides cells. This might explain why the band pattern obtained with membrane protein preparations or whole cells was essentially the same. Cytosolic proteins such as GroEL, elongation factor Tu, DnaK, and the gene product of cbdbA1077 (CoA binding ligase) were detected in faint bands after native PAGE of membrane protein preparations, indicating significant concentrations of these proteins in the cytosol.
Acknowledgments
We thank Hans Lehrach for continuing support of our work. We thank Bernd Krostitz-Schroeer for cultivation of strain CBDB1 and Beata Lukaszewska-McGreal and Dorothea Theiss for technical assistance with mass spectrometry. JGI is acknowledged for making available genome data of strain BAV1 prior to publication.
The mass spectrometric analyses were performed as part of an external collaboration with the project SMP Protein of the National Genome Research Network (NGFN), funded by the Germany Ministry of Education and Research (BMBF). This work was funded by the National Genome Research Network (NGFN) of the German Ministry of Education and Research (BMBF) and the Zukunftsfonds of the Technologiestiftung Berlin (TSB) and the Structural Funds of the European Union within the project 2D/3D-ProteinChips.
Footnotes
Published ahead of print on 12 October 2007.
REFERENCES
- 1.Adrian, L., W. Manz, U. Szewzyk, and H. Görisch. 1998. Physiological characterization of a bacterial consortium reductively dechlorinating 1,2,3- and 1,2,4-trichlorobenzene. Appl. Environ. Microbiol. 64:496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 3.Adrian, L., S. K. Hansen, J. M. Fung, H. Görisch, and S. H. Zinder. 2007. Growth of Dehalococcoides strains with chlorophenols as electron acceptor. Environ. Sci. Technol. 41:2318-2323. [DOI] [PubMed] [Google Scholar]
- 4.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 5.Bunge, M., L. Adrian, A. Kraus, M. Opel, W. G. Lorenz, J. R. Andreesen, H. Görisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. [DOI] [PubMed] [Google Scholar]
- 6.Christiansen, N., B. K. Ahring, G. Wohlfarth, and G. Diekert. 1998. Purification and characterization of the 3-chloro-4-hydroxy-phenylacetate reductive dehalogenase of Desulfitobacterium hafniense. FEBS Lett. 436:159-162. [DOI] [PubMed] [Google Scholar]
- 7.Fennell, D. E., I. Nijenhuis, S. F. Wilson, S. H. Zinder, and M. M. Häggblom. 2004. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ. Sci. Technol. 38:2075-2081. [DOI] [PubMed] [Google Scholar]
- 8.Fung, J. M., R. M. Morris, L. Adrian, and S. H. Zinder. 2007. Expression of reductive dehalogenase genes in Dehalococcoides ethenogenes strain 195 growing on tetrachloroethene, trichloroethene, or 2,3-dichlorophenol. Appl. Environ. Microbiol. 73:4439-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gobom, J., M. Schuerenberg, M. Mueller, D. Theiss, H. Lehrach, and E. Nordhoff. 2001. Alpha-cyano-4-hydroxycinnamic acid affinity sample preparation. A protocol for MALDI-MS peptide analysis in proteomics. Anal. Chem. 73:434-438. [DOI] [PubMed] [Google Scholar]
- 10.Hölscher, T., H. Görisch, and L. Adrian. 2003. Reductive dehalogenation of chlorobenzene congeners in cell free extracts of Dehalococcoides sp. strain CBDB1. Appl. Environ. Microbiol. 69:2999-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hölscher, T., R. Krajmalnik-Brown, K. M. Ritalahti, F. v. Wintzingerode, H. Görisch, F. E. Löffler, and L. Adrian. 2004. Multiple nonidentical reductive-dehalogenase-homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70:5290-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jayachandran, G., H. Görisch, and L. Adrian. 2003. Studies on hydrogenase activity and chlorobenzene respiration in Dehalococcoides sp. strain CBDB1. Arch. Microbiol. 180:411-416. [DOI] [PubMed] [Google Scholar]
- 13.Jayachandran, G., H. Görisch, and L. Adrian. 2004. Dehalorespiration with penta- and hexachlorobenzene by Dehalococcoides sp. strain CBDB1. Arch. Microbiol. 182:498-504. [DOI] [PubMed] [Google Scholar]
- 14.Reference deleted.
- 15.Krasotkina, J., T. Walters, K. A. Maruya, and S. W. Ragsdale. 2001. Characterization of the B12- and iron-sulfur-containing reductive dehalogenase from Desulfitobacterium chlororespirans. J. Biol. Chem. 276:40991-40997. [DOI] [PubMed] [Google Scholar]
- 16.Reference deleted.
- 17.Kube, M., A. Beck, S. H. Zinder, H. Kuhl, R. Reinhardt, and L. Adrian. 2005. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23:1269-1273. [DOI] [PubMed] [Google Scholar]
- 18.Läemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Ludwig, M. L., and R. G. Matthews. 1997. Structure-based perspectives on B12-dependent enzymes. Annu. Rev. Biochem. 66:269-313. [DOI] [PubMed] [Google Scholar]
- 20.Magnuson, J. K., R. V. Stern, J. M. Gossett, S. H. Zinder, and D. R. Burris. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maymó-Gatell, X., Y.-T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 23.Mirgorodskaya, E., C. Braeuer, P. Fucini, H. Lehrach, and J. Gobom. 2005. Nanoflow liquid chromatography coupled to matrix-assisted laser desorption/ionization mass spectrometry: sample preparation, data analysis, and application to the analysis of complex peptide mixtures. Proteomics 5:399-408. [DOI] [PubMed] [Google Scholar]
- 24.Morris, R. M., S. Sowell, D. Barofsky, S. H. Zinder, and R. E. Richardson. 2006. Transcription and mass-spectroscopic proteomic studies of electron transport oxidoreductases in Dehalococcoides ethenogenes. Environ. Microbiol. 8:1499-1509. [DOI] [PubMed] [Google Scholar]
- 25.Morris, R. M., J. M. Fung, B. G. Rahm, S. Zhang, D. L. Freedman, S. H. Zinder, and R. E. Richardson. 2007. Comparative proteomics of Dehalococcoides spp. reveals strain-specific peptides associated with activity. Appl. Environ. Microbiol. 73:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller, J. A., B. M. Rosner, G. v. Abendroth, G. Meshulam-Simon, P. L. McCarty, und A. Spormann. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70:4880-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nesterenko, M. V., M. Tilley, and S. J. Upton. 1994. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods 28:239-242. [DOI] [PubMed] [Google Scholar]
- 28.Reference deleted.
- 29.Neumann, A., G. Wohlfarth, and G. Diekert. 1998. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J. Bacteriol. 180:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni, S., J. K. Fredrickson, and L. Xun. 1995. Purification and characterization of a novel 3-chlorobenzoate-reductive dehalogenase from the cytoplasmic membrane of Desulfomonile tiedjei DCB-1. J. Bacteriol. 177:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reference deleted.
- 32.Schrattenholz, A. (ed.). 2001. Methoden der Proteomforschung: Molekulare Analyse der Proteinexpression. Spektrum Akademischer Verlag GmbH Heidelberg, Berlin, Germany.
- 33.Seshadri, R., L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methe, N. L. Ward, W. C. Nelson, R. T. DeBoy, H. M. Khouri, J. F. Kolonay, R. J. Dodson, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. T. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Zinder, and J. F. Heidelberg. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105-108. [DOI] [PubMed] [Google Scholar]
- 34.Smidt, H., A. D. L. Akkermans, J. van der Oost, and W. M. de Vos. 2000. Halorespiring bacteria: molecular characterization and detection. Enzyme Microb. Technol. 27:812-820. [DOI] [PubMed] [Google Scholar]
- 35.Thibodeau, J., A. Gauthier, M. Duguay, R. Villemur, F. Lépine, P. Juteau, and R. Beaudet. 2004. Purification, cloning, and sequencing of a 3,5-dichlorophenol reductive dehalogenase from Desulfitobacterium frappieri PCP-1. Appl. Environ. Microbiol. 70:4532-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van de Pas, B. A., H. Smidt, W. R. Hagen, J. van der Oost, G. Schraa, A. J. M. Stams, and W. M. de Vos. 1999. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J. Biol. Chem. 274:20287-20292. [DOI] [PubMed] [Google Scholar]
- 37.Waller, A. S., R. Krajmalnik-Brown, F. E. Loffler, and E. A. Edwards. 2005. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing cultures. Appl. Environ. Microbiol. 71:8257-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, Q., C. E. Milliken, G. P. Meier, J. E. Watts, K. R. Sowers, and H. D. May. 2002. Dechlorination of chlorobenzenes by a culture containing bacterium DF-1, a PCB dechlorinating microorganism. Environ. Sci. Technol. 1:3290-3294. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, Y., H. Romero, G. Salinas, and V. N. Gladyshev. 2006. Dynamic evolution of selenocysteine utilization in bacteria: a balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 7:R94. [DOI] [PMC free article] [PubMed] [Google Scholar]