Abstract
Based on phylogenetic analysis of clones retrieved from two nifH gene clone libraries that were created using cDNA from suboxic sediment samples obtained from areas densely vegetated with the high-salt marsh plant Spartina patens, a primer set was designed to target nitrogen-fixing bacteria with sequence similarities to members of the ɛ subclass of Proteobacteria. Nested PCR, denaturing gel electrophoresis, and subsequent sequence analysis of reamplified fragments confirmed the specificity of the primer set by retrieving nifH sequences of only putative members of the ɛ subclass of Proteobacteria, all of which were characterized by a highly divergent 27- or 36-bp insertion in both DNA and cDNA.
Salt marshes are highly productive systems that have received considerable attention from ecologists as a consequence of their importance to the productivity of estuarine waters (9, 40). Marsh productivity is often based on the growth of C4 grass species (10), such as saltmeadow cordgrass [Spartina patens (Ait.) Muhl.], that rely upon the activity of free-living nitrogen-fixing bacteria in sediments to satisfy their nitrogen requirements. Nitrogen fixation by these bacteria may satisfy as much as 50% of a plant's nitrogen requirements (11, 39). The magnitude of nitrogen fixation in salt marshes depends on edaphic conditions, soil physiochemical properties, and patterns of plant growth that affect carbon exudation and release into the sediment (3, 41). Seasonal variation in plant root exudation of carbon resources or ions (13, 41) or changes in soil physiochemical conditions that accompany tidal flooding (7) have been shown to affect the microbial community structure in sediments, as shown previously for the α, β, γ, and δ subclasses of Proteobacteria (7). Although these phylogenetic groups contain many previously identified nitrogen-fixing bacteria (44), changes in the community structure of nitrogen-fixing bacteria have not been observed in marsh sediments so far (7, 30, 31, 33).
Nitrogen-fixing bacteria have been analyzed predominantly using PCR-based profiling tools, such as restriction fragment length polymorphism analysis (6, 7, 33, 42) or denaturing gradient gel electrophoresis (DGGE) (5, 21, 29-31), that typically target nifH, the structural gene for nitrogenase reductase (15). PCR typing protocols, however, can be affected by the limited sensitivity of analyses that generally attempt to retrieve information for all nitrogen-fixing bacteria, by selective amplification (i.e., by primer bias), by the loss of relative abundance information, and by the ambiguity of complex restriction fragment length patterns, since very different organisms can produce similar patterns (8, 18, 33). The failure to detect changes in the community structure of nitrogen-fixing bacteria in salt marshes might therefore be a consequence of methodological issues.
The aim of this study was to develop molecular tools that target members of this functional group on a more specific level. For this purpose, nifH gene clone libraries were created using universal primers and reverse-transcribed mRNA (cDNA) from suboxic sediment samples obtained from areas densely vegetated with S. patens. Rhizosphere samples were retrieved in July 2006 at a depth of 1.5 to 3.5 cm below the surface from two salt marshes with contrasting matrices and histories: Piermont Marsh, a natural salt marsh with high organic matter content, and Harrier Meadow, a restored wetland with low organic matter content (26, 36). Both marshes were brackish, with salinities between 5 and 15 ppt, and were characterized by standing water about 5 cm below the surface which resulted in negative redox potentials at the sampling depth (7).
Nucleic acids were extracted from 0.5-g sediment samples (n = 2 for each site). Cells were lysed by bead beating (14), and nucleic acids were purified by sequential phenol, phenol-chloroform, and chloroform extraction (35) and subsequent precipitation with 2 volumes of 2.5 M NaCl-20% polyethylene glycol 8000 (42), which was followed by additional phenol-chloroform and chloroform extraction and a final isopropanol precipitation. Nucleic acids were washed twice in 70% ethanol, dried, and resuspended in 40 μl distilled water. Duplicate samples were mixed and split into two portions, a DNA sample and an RNA sample. RNA samples were treated with 4 μl DNase I (1 U μl−1; Promega, Madison, WI) at 37°C for 1 h by following the manufacturer's instructions. Ten percent (4 μl) of each RNA solution was subsequently used to transcribe cDNA using 2 μM reverse primer NifHrev (Table 1) (42) and the Reverse-iTMAX reverse transcription blend (ABgene, Rochester, NY) according to the manufacturer's instructions. The controls used to test for DNA contamination of RNA preparations included use of forward primer NifHforA (42) in reverse transcription reactions and attempted 16S rRNA gene amplification from all samples (20).
TABLE 1.
Primer combinations targeting nifH gene sequences
| Primer | Sequence (5′ → 3′) | Positiona | No. of degeneracies | Size (bp) | Reference |
|---|---|---|---|---|---|
| NifHforA | GCI WTI TAY GGN AAR GGN GG | 19-38 | 128 | 464 | 42 |
| NifHrev | GCR TAI ABN GCC ATC ATY TC | 463-482 | 48 | ||
| NifHforB | GGI TGT GAY CCN AAV GCN GA | 112-132 | 96 | 371 | 42 |
| NifHrev | GCR TAI ABN GCC ATC ATY TC | 463-482 | 48 | ||
| PicenoF | TAC GGI AAR GGB GGI ATY GG | 25-44 | 12 | 428 | 31 |
| PicenoR | SAC GAT GTA GAT YTC CTG | 436-453 | 4 | ||
| PolF | TGC GAY CCS AAR GCB GAC TC | 115-135 | 24 | 362 | 32 |
| PolR | ATS GCC ATC ATY TCR CCG GA | 457-477 | 8 | ||
| ENFBfb | GAT GTA TGT AAA CCT GGT GC | 223-242 | 0 | 252 | This study |
| ENFBr | CTT GTG CTT TTC CTT CAC GG | 419-439 | 0 |
Sequence position in the A. vinelandii nifH coding sequence (GenBank accession number M20568).
The forward primer contained a GC clamp (5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCC CGG GGG GCC) at the 5′ end when it was used for DGGE analyses. Primers without GC clamps were used for reamplification of DGGE fragments for sequence analyses. The fragment sizes are the sizes of fragments generated with primers without the GC clamp.
A nifH gene clone library was created using 10% (2 μl) of each cDNA preparation from the two salt marshes as the template for nested PCR using primer set NifHforA/NifHrev for the initial amplification, followed by amplification with primer set NifHforB/NifHrev (Table 1) (42). The PCR product was ligated into pGEM-T Easy (Promega) and transformed into Escherichia coli TOP-10 (Stratagene, Cedar Creek, TX). Fifteen clones from each library were analyzed for nifH fragments and sequenced using a CEQ 8800 Quickstart kit with 5% dimethyl sulfoxide added to the reaction mixture in a CEQ 8800 sequencer (BeckmanCoulter, Fullerton, CA). This brief census revealed four potential chimeras (28). The remaining 26 nifH sequences were aligned with related sequences resulting from GenBank and EMBL searches (2, 27) using Sequencher 4.2.2 (Gene Codes Corporation, Ann Arbor, MI), CLUSTAL X, and MacClade 4.05 (24, 38). Phylogenetic analyses were performed by maximum parsimony, neighbor joining, and maximum likelihood methods using nucleic acid or amino acid sequences and PAUP*4.0b10 (37). Confidence in tree topologies was gauged using bootstrap resampling methods in PAUP; only values greater than 70% were used (12). Additionally, Bayesian methods in MRBAYES v 3.0 were used (16), and a 95% majority rule consensus tree was generated using PAUP.
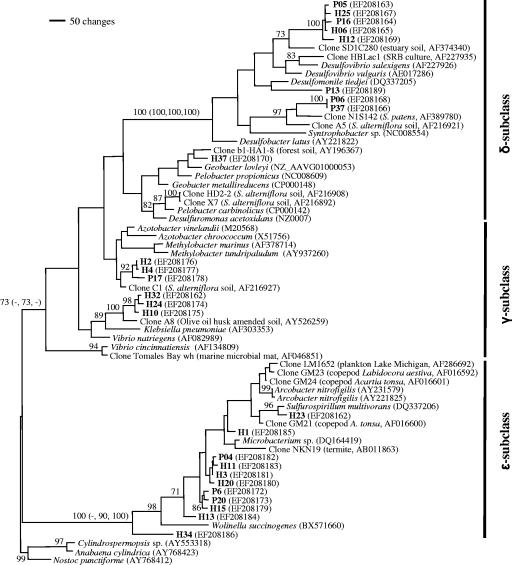
The tree topologies were very similar for the phylogenetic methods employed for both nucleic acid and amino acid sequences and were not affected by the inclusion or exclusion of insertions found in several clones (data not shown). A representative maximum parsimony tree showed that sequences representing bacteria related to the γ, δ, and ɛ subclasses of Proteobacteria were recovered with high bootstrap or probability values supporting the differentiation of these groups (Fig. 1). While nifH gene sequences representing nitrogen-fixing members of the γ and δ subclasses of Proteobacteria have often been retrieved from other environments, such as low-salt marshes (4, 21, 22), retrieval of nifH genes from environmental samples has not been described previously for the ɛ subclass of Proteobacteria.
FIG. 1.
Parsimony-based tree showing the phylogenetic positions of nifH gene clones from two libraries generated from cDNA from rhizosphere samples of S. patens from Piermont Marsh (prefix P) and Harrier Meadow (prefix H) in the γ, δ, and ɛ subclasses of Proteobacteria. The phylogram was created using PAUP*4.0b10 with 10,000 random addition replicates (37). The numbers at the nodes are the bootstrap support values with 10,000 replicates; only values greater than 70% are shown. The numbers in parentheses at nodes are bootstrap support values and posterior probabilities obtained by neighbor joining, maximum likelihood, and Bayesian analyses, respectively. For brevity, these values are shown only for the key phylogenetic nodes in the tree. The outgroups were Cylidrospermopis sp. (accession no. AY553318), Anabaena cylindrica (AY768423), and Nostoc punctiforme (AY768412). SRB, sulfate-reducing bacterium.
In our study, about one-third of all clones analyzed clustered well within the ɛ subclass of Proteobacteria; eight clones were retrieved from sediments of Harrier Meadow, and three clones were retrieved from Piermont Marsh (Fig. 1). Most of the nifH gene sequences of these clones, like the sequences of all cultured and other uncultured relatives, were characterized by a 36-bp insertion; the only exception was clone H34, which had a 27-bp insertion. The sequences of the insertion region were much more variable than the overall sequence of the nifH gene; e.g., the levels of divergence for the insertion between clone H1 and clones H23, H11, and H34 were 33, 42, and 56%, respectively, while the overall sequences showed only 12, 6, and 13% divergence. Since only four nifH gene sequences of cultured bacteria belonging to the ɛ subclass of Proteobacteria could be retrieved from the databases, more specific assignment of clone sequences to clusters within the ɛ subclass of Proteobacteria was limited and not supported by strong bootstrap or probability values (Fig. 1).
nifH genes of members of the ɛ subclass of Proteobacteria have not been detected previously in environmental samples, even though rRNA sequences or isolates demonstrated their presence in these environments (1, 17, 33, 34, 43). For example, a nitrogen-fixing member of the ɛ subclass of Proteobacteria, Arcobacter nitrofigilis ATCC 33309, was originally isolated from roots of S. alterniflora, a common low-salt marsh plant (25). However, nitrogen-fixing members of the ɛ subclass of Proteobacteria were not detected by targeting nifH genes in low-salt marsh environments previously (4, 21, 22). These studies used different primer sets than our study and other studies that detected nifH gene fragments in freshwater and marine samples (23, 45) that grouped with members of the ɛ subclass of Proteobacteria in our analyses (Fig. 1). These observations suggest that primer bias could be a major cause for the failure of detection.
Due to the limited information available on nitrogen-fixing members of the ɛ subclass of Proteobacteria, we chose to focus on this group and develop more specific primers targeting the nifH gene of the ɛ subclass of Proteobacteria. Forward primer ENFBf and reverse primer ENFBr (Table 1) in the NifHforB/NifHrev primer set range were designed using unique regions in the alignment file of the ɛ subclass of Proteobacteria and were initially checked for specificity using BLAST and Fasta analyses with GenBank and EMBL databases. While the forward primer showed small mismatches (one to three bases) to corresponding sites in the nifH gene sequences of some members of the ɛ subclass of Proteobacteria, such as Arcobacter and Sulfurospirillum sp., the reverse primer was more divergent, with three and six mismatches to the sequences of Wolinella and Arcobacter sp., respectively. The specificity of this reverse primer, however, could not be evaluated with sequences of other organisms, such as Microbacterium and Sulfurospirillum sp., since the sequence information available for these organisms did not cover its binding site. Since these primers were meant to be used in DGGE analyses, the design of degenerate primers was avoided; instead, the specificity of the PCR was reduced by decreasing the annealing temperature arbitrarily to 4°C below the melting temperature.
PCR mixtures contained 2 mM MgCl2, 0.2 mM of each deoxynucleoside triphosphate, 0.4 μM of each primer (with the forward primer carrying a GC clamp), 5 mg ml−1 bovine serum albumin, 1× PCR buffer, 2 μl of template, and 2 U of Taq DNA polymerase (GenScript, Piscataway, NJ). An initial denaturation at 96°C for 10 min was followed by incubation at 80°C for 10 min for Taq polymerase addition, 35 cycles of denaturation at 96°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 45 s, and a final extension at 72°C for 30 min. DNA of A. nitrofigilis ATCC 33309 (25) and Klebsiella oxytoca K10 (Department of Microbiology Culture Collection, Wageningen Agricultural University) were used as positive and negative controls, respectively.
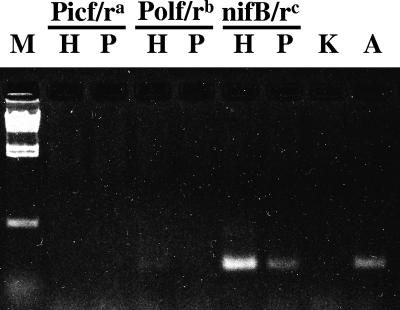
The amplification conditions allowed us to amplify the nifH gene fragment from A. nitrofigilis even though the forward and reverse primers showed one and five mismatches to its target sequence, respectively. However, we failed to amplify nifH gene fragments using DNA from environmental samples. This failure was attributed to low template concentrations in these samples, because a nested approach with amplification products generated either with primer set NifHforA/NifHrev or after nested amplification with primer set NifHforB/NifHrev products as templates for the ENFBf/ENFBr primer set was successful. Since a nested PCR was necessary for detection, amplification products generated with two additional primer sets, PicenoF/PicenoR (31) and PolF/PolR (32) (Table 1), were examined as templates for primer set ENFBf/ENFBr. Both primer sets produced amplification products with DNA from K. oxytoca and the environmental samples but not with DNA from A. nitrofigilis. Nested PCR using the PicenoF/PicenoR primer set that was slightly modified from its published version (31) (Table 1) did not result in generation of any amplification product (Fig. 2), while nested PCR using primer set PolF/PolR yielded very faint double bands not suitable for DGGE (Fig. 2). These results support the speculation that methodological impacts (i.e., primer bias) might have contributed to the failure to detect nifH genes of the ɛ subclass of Proteobacteria in previous studies.
FIG. 2.
Ethidium bromide-stained agarose gel of PCR products generated using the ENFBf/ENFBr primer set in a nested PCR with templates generated using three different primer sets (PicenoF/PicenoR [31], PolF/PolR ([32], and NifH for B/NifHrev [42]) and DNA from Harrier Meadow (lanes H) and Piermont Marsh (lanes P). Lane M contained a λ HindIII size marker, lane K contained a negative control (K. oxytoca), and lane A contained a positive control (A. nitrofigilis).
The specificity of primer set ENFBf/ENFBr for members of the ɛ subclass of Proteobacteria was tested by DGGE analysis. DNA and cDNA obtained from both sites (i.e., Piermont Marsh and Harrier Meadow) were used as templates for primer set NifHforA/NifHrev, and amplification products obtained directly or after nested PCR with primer set NifHforB/NifHrev were used as templates for primer set ENFBf/ENFBr. Products were analyzed with a DCode universal mutation detection system (Bio-Rad Laboratories, Hercules, CA). Samples were electrophoresed at 60οC and 180 V for 10 min and then at 100 V for 16 h (8% polyacrylamide; 37 to 47% denaturant). Gels were stained with ethidium bromide (0.5 μg ml−1) in 0.5× Tris-acetate-EDTA for 30 min, destained in water, and visualized with a UV transilluminator.
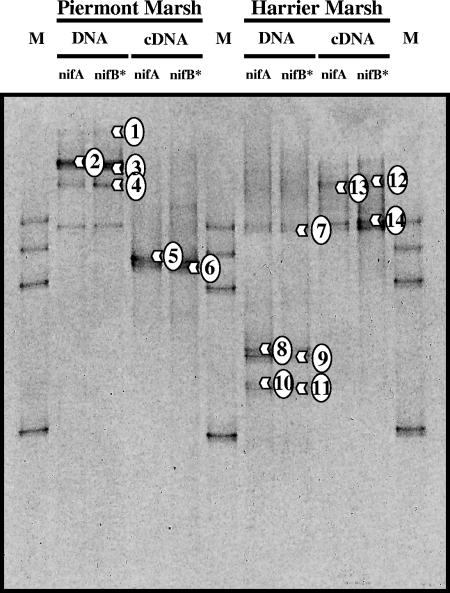
Amplification products obtained with NifHforA/NifHrev or after nested PCR with NifHforB/NifHrev as templates for ENFBf/ENFBr produced very similar profiles in DGGE analyses (Fig. 3), indicating that the additional nested PCR could be used to increase template concentrations without inherently generating any spurious by-products or additional artifactual banding patterns. The DGGE profiles obtained with DNA as the initial PCR template and those obtained with cDNA were distinctly different. The profiles obtained with DNA as the template were generally more complex than those obtained with cDNA as the template, but the profiles did not necessarily overlap (Fig. 3). The validity of these results is supported by studies of nitrogen-fixing bacteria on rice, where the restriction fragment length polymorphism profiles generated for nifH amplicons were very different with DNA and cDNA templates (19), and by the results obtained for nifH gene clone libraries from lake plankton, where most of the cDNA-derived sequences did not group closely with the sequences obtained from DNA (43).
FIG. 3.
DGGE profiles of amplicons generated with primer set ENFBf/ENFBr using amplicons generated either with primer set NifHforA/NifHrev (nifA) or after nested PCR with primer set NifHforB/NifHrev using NifHforA/NifHrev amplicons as the template (nifB*) and DNA or cDNA from rhizosphere samples of S. patens from Piermont Marsh and Harrier Meadow. The marker (lanes M) was a group of clones from the initial nifH gene clone libraries (H15, H11, P04, and H23). Numbers indicate fragments that were extracted and sequenced.
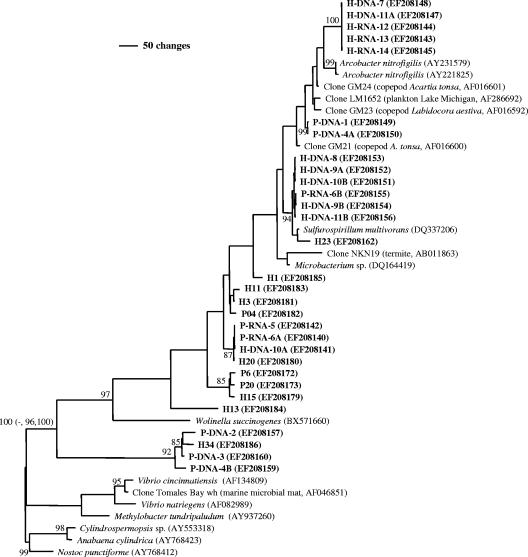
All DGGE bands were excised, reamplified, and cloned, and the clones were reanalyzed by DGGE and subsequently sequenced. Only the bands with numbers in Fig. 3 produced a good sequence. Up to three clones from each DGGE band were sequenced, analyzed for the presence of chimeras, and analyzed by the phylogenetic methods described above. Clones generated from a fragment that had identical sequences were not included in the final phylogenetic analysis. All sequences retrieved harbored a 27- or 36-bp insertion in the nifH gene and showed high bootstrap support (100%) for assignment to the ɛ subclass of Proteobacteria (Fig. 4). More accurate assignment of clones to groups within the ɛ subclass of Proteobacteria, however, was hampered by limited database sequences, demonstrating that the phylogeny of these clones and isolates needs additional work.
FIG. 4.
Parsimony-based tree showing the phylogenetic positions of nifH gene clones from fragments of DGGE profiles generated using the ENFBf/ENFBr primer set (see Fig. 3). In the designations the prefixes P and H indicate Piermont Marsh and Harrier Meadow, respectively; DNA and RNA indicate the template used for initial amplification; and the numbers indicate the fragment numbers in DGGE profiles (Fig. 3). The phylogram was created using PAUP*4.0b10 with 10,000 random addition replicates (37). The numbers at the nodes are bootstrap support values with 10,000 replicates; only values greater than 70% are shown. The numbers in parentheses at nodes are bootstrap support values and posterior probabilities obtained by neighbor joining, maximum likelihood, and Bayesian analyses, respectively. For brevity, these values are shown only for the key phylogenetic nodes in the tree. The outgroups were Cylidrospermopis sp. (accession no. AY553318), A. cylindrica (AY768423), and N. punctiforme (AY768412).
Our study showed that primer set ENFBf/ENFBr and the conditions used in this study were specific enough to retrieve sequence information for defined groups in the ɛ subclass of Proteobacteria. This should enable us to use a more refined and targeted approach focusing on this phylogenetic group that may provide a picture of the dynamics of the nitrogen-fixing microbial community in high-salt marsh sediments that is more accurate than that obtained in previous studies. Such studies could include exploitation of the high level of divergence found in the 36-bp and (for a separate group) 27-bp insertions in the nifH gene sequences of members of the ɛ subclass of Proteobacteria, because the insertion provides an ideal primer or probe target site for future research aimed at examining these bacteria in the environment more specifically.
Nucleotide sequence accession numbers.
The nifH sequences used in this study have been deposited in the GenBank database under accession numbers EF208162 to EF208186 and EF208189. Sequences of DGGE fragments have been deposited under accession numbers EF208140 to EF208160.
Acknowledgments
We are grateful to M. F. Forstner for technical advice and computer software and hardware support.
Footnotes
Published ahead of print on 5 October 2007.
REFERENCES
- 1.Affourtit, J., J. P. Zehr, and H. W. Paerl. 2001. Distribution of nitrogen-fixing microorganisms along the Neuse River Estuary, North Carolina. Microb. Ecol. 41:114-123. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle, C. D., and D. G. Patriquin. 1981. Carbon metabolism of Spartina alterniflora Loisel in relation to that of associated nitrogen-fixing bacteria. New Phytol. 89:275-288. [Google Scholar]
- 4.Brown, M. M., M. J. Friez, and C. R. Lovell. 2003. Expression of nifH genes by diazotrophic bacteria in the rhizosphere of short form Spartina alterniflora. FEMS Microbiol. Ecol. 43:411-417. [DOI] [PubMed] [Google Scholar]
- 5.Burgmann, H., S. Meier, M. Bunge, F. Widmer, and J. Zeyer. 2005. Effects of model root exudates on structure and activity of a soil diazotroph community. Environ. Microbiol. 7:1711-1724. [DOI] [PubMed] [Google Scholar]
- 6.Burgmann, H., F. Widmer, W. V. Sigler, and J. Zeyer. 2004. New molecular screening tools for analysis of free-living diazotrophs in soil. Appl. Environ. Microbiol. 70:240-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke, D., E. P. Hamerlynck, and D. Hahn. 2002. Interactions among plant species and microorganisms in salt marsh sediments. Appl. Environ. Microbiol. 68:1157-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casamayor, E. O., R. Massana, S. Benlloch, L. Ovreas, B. Diez, V. J. Goddard, J. M. Gasol, I. Joint, F. Rodriguez-Valera, and C. Pedros-Alio. 2002. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar system. Environ. Microbiol. 4:338-348. [DOI] [PubMed] [Google Scholar]
- 9.Currin, C. A., S. Y. Newell, and H. W. Paerl. 1995. The role of standing dead Spartina alterniflora and benthic micoralgae in salt marsh food webs; considerations based on multiple stable isotope analysis. Mar. Ecol. Prog. Ser. 121:99-116. [Google Scholar]
- 10.Dame, R. F. 1989. The importance of Spartina alterniflora to Atlantic Coast estuaries. Rev. Aquat. Biol. 1:639-660. [Google Scholar]
- 11.DeLaune, R. D., T. C. Feijtel, and W. H. Patrick. 1989. Nitrogen flows in a Louisiana gulf coast salt marsh: spatial considerations. Biogeochemistry 8:25-37. [Google Scholar]
- 12.Hillis, D. M., and J. J. Bull. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42:182-192. [Google Scholar]
- 13.Hines, M. E., R. S. Evans, B. R. Sharak Genthner, S. G. Willis, S. Friedman, J. N. Rooney-Varga, and R. Devereux. 1999. Molecular phylogenetic and biogeochemical studies of sulfate-reducing bacteria in the rhizosphere of Spartina alterniflora. Appl. Environ. Microbiol. 65:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hönerlage, W., D. Hahn, and J. Zeyer. 1995. Detection of mRNA of nprM in Bacillus megaterium ATCC 14581 grown in soil by whole cell hybridization. Arch. Microbiol. 163:235-241. [Google Scholar]
- 15.Howard, J. B., and D. C. Rees. 1996. Structural basis of biological nitrogen fixation. Chem. Rev. 96:2965-2982. [DOI] [PubMed] [Google Scholar]
- 16.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinform. Appl. Notes 17:754-755. [DOI] [PubMed] [Google Scholar]
- 17.Izquierdo, J. A., and K. Nusslein. 2006. Distribution of extensive nifH gene diversity across physical soil microenvironments. Microb. Ecol. 51:441-452. [DOI] [PubMed] [Google Scholar]
- 18.Kisand, V., and J. Wikner. 2003. Limited resolution of 16S rDNA DGGE caused by melting properties and closely related DNA sequences. J. Microbiol. Methods 54:183-191. [DOI] [PubMed] [Google Scholar]
- 19.Knauth, S., T. Hurek, D. Brar, and B. Hurek. 2005. Influence of different Oryza cultivars on expression of nifH gene pools in different roots of rice. Environ. Microbiol. 7:1725-1733. [DOI] [PubMed] [Google Scholar]
- 20.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-176. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY.
- 21.Lovell, C. R., M. J. Friez, J. W. Longshore, and C. E. Bagwell. 2001. Recovery and phylogenetic analysis of nifH sequences from diazotrophic bacteria associated with dead aboveground biomass of Spartina alterniflora. Appl. Environ. Microbiol. 67:5308-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lovell, C. R., Y. M. Piceno, J. M. Quattro, and C. E. Bagwell. 2000. Molecular analysis of diazotroph diversity in the rhizosphere of the smooth cordgrass, Spartina alterniflora. Appl. Environ. Microbiol. 66:3814-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacGregor, B. J., B. Van Mooy, B. J. Baker, M. Mellon, P. H. Moisander, H. W. Paerl, J. Zehr, D. Hollander, and D. A. Stahl. 2001. Microbiological, molecular biological and stable isotopic evidence for nitrogen fixation in the open waters of Lake Michigan. Environ. Microbiol. 3:205-219. [DOI] [PubMed] [Google Scholar]
- 24.Maddison, W. P., and D. R. Maddison. 1999. MacClade: analysis of phylogeny and character evolution. Sinauer Associates, Sunderland, MA. [DOI] [PubMed]
- 25.McClung, C. R., and D. G. Patriquin. 1980. Isolation of a nitrogen-fixing Campylobacter species from the roots of Spartina alterniflora Loisel. Can. J. Microbiol. 26:881-886. [DOI] [PubMed] [Google Scholar]
- 26.Montalto, F. A., T. S. Steenhuis, and J.-Y. Parlange. 2006. The hydrology of Piermont Marsh, a reference for tidal marsh restoration in the Hudson River estuary, New York. J. Hydrol. 316:108-128. [Google Scholar]
- 27.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pester, M., M. W. Friedrich, B. Schink, and A. Brune. 2004. pmoA-based analysis of methanotrophs in a littoral lake sediment reveals a diverse and stable community in a dynamic environment. Appl. Environ. Microbiol. 70:3138-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piceno, Y. M., and C. R. Lovell. 2000. Stability in natural bacterial communities. I. Nutrient addition effects on rhizosphere diazotroph assemblage composition. Microb. Ecol. 39:32-40. [DOI] [PubMed] [Google Scholar]
- 30.Piceno, Y. M., and C. R. Lovell. 2000. Stability in natural bacterial communities. II. Plant resource allocation effects on rhizosphere diazotroph assemblage composition. Microb. Ecol. 39:41-48. [DOI] [PubMed] [Google Scholar]
- 31.Piceno, Y. M., P. A. Noble, and C. R. Lovell. 1999. Spatial and temporal assessment of diazotroph assemblage composition in vegetated salt marsh sediments using denaturing gradient gel electrophoresis analysis. Microb. Ecol. 38:157-167. [DOI] [PubMed] [Google Scholar]
- 32.Poly, F., L. J. Monrozier, and R. Bally. 2001. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152:95-103. [DOI] [PubMed] [Google Scholar]
- 33.Poly, F., L. Ranjard, S. Nazaret, F. Gourbiere, and L. J. Monrozier. 2001. Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl. Environ. Microbiol. 67:2255-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosch, C., A. Mergel, and H. Bothe. 2002. Biodiversity of denitrifying bacteria and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68:3818-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Seigel, A., C. Hatfield, and J. M. Hartman. 2005. Avian response to restoration of urban tidal marshes in the Hackensack Meadowlands, New Jersey. Urban Habitats 3:87-116. [Google Scholar]
- 37.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 38.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White, D. S., and B. L. Howes. 1994. Translocation, remineralization, and turnover of nitrogen in the roots and rhizomes of Spartina alterniflora. Am. J. Bot. 81:1225-1234. [Google Scholar]
- 40.Whiting, G. J. 1989. Nitrogen exchange between a portion of vegetated salt marsh and the adjoining creek. Limnol. Oceanogr. 34:463-473. [Google Scholar]
- 41.Whiting, G. J., E. L. Gandy, and D. C. Yoch. 1986. Tight coupling of root-associated nitrogen fixation and plant photosynthesis in the salt marsh grass Spartina alterniflora and carbon dioxide enhancement of nitrogenase activity. Appl. Environ. Microbiol. 52:108-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Widmer, F., B. T. Shaffer, L. A. Porteous, and R. J. Seidler. 1999. Analysis of nifH gene pool complexity in soil and litter at a Douglas fir forest site in the Oregon Cascade mountain range. Appl. Environ. Microbiol. 65:374-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zani, S., M. T. Mellon, J. L. Collier, and J. P. Zehr. 2000. Expression of nifH genes in natural microbial assemblages in Lake George, New York, detected by reverse transcriptase PCR. Appl. Environ. Microbiol. 66:3119-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zehr, J. P., B. D. Jenkins, S. M. Short, and G. F. Steward. 2003. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5:539-554. [DOI] [PubMed] [Google Scholar]
- 45.Zehr, J. P., M. T. Mellon, and S. Zani. 1998. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl. Environ. Microbiol. 64:3444-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]