Abstract
The dissimilatory adenosine-5′-phosposulfate reductase is a key enzyme of the microbial sulfate reduction and sulfur oxidation processes. Because the alpha- and beta-subunit-encoding genes, aprBA, are highly conserved among sulfate-reducing and sulfur-oxidizing prokaryotes, they are most suitable for molecular profiling of the microbial community structure of the sulfur cycle in environment. In this study, a new aprA gene-targeting assay using a combination of PCR and denaturing gradient gel electrophoresis is presented. The screening of sulfate-reducing and sulfur-oxidizing reference strains as well as the analyses of environmental DNA from diverse habitats (e.g., microbial mats, invertebrate tissue, marine and estuarine sediments, and filtered hydrothermal water) by the new primer pair revealed an improved microbial diversity coverage and less-pronounced template-to-PCR product bias in direct comparison to those of the previously published primer set (B. Deplancke, K. R. Hristova, H. A. Oakley, V. J. McCracken, R. Aminov, R. I. Mackie, and H. R. Gaskins, Appl. Environ. Microbiol. 66:2166-2174, 2000). The concomitant molecular detection of sulfate-reducing and sulfur-oxidizing prokaryotes was confirmed. The new assay was applied in comparison with the 16S rRNA gene-based analysis to investigate the microbial diversity of the sulfur cycle in sediment, seawater, and manganese crust samples from four study sites in the area of the Lesser Antilles volcanic arc, Caribbean Sea (Caribflux project). The aprA gene-based approach revealed putative sulfur-oxidizing Alphaproteobacteria of chemolithoheterotrophic lifestyle to have been abundant in the nonhydrothermal sediment and water column. In contrast, the sulfur-based microbial community that inhabited the surface of the volcanic manganese crust was more complex, consisting predominantly of putative chemolithoautotrophic sulfur oxidizers of the Betaproteobacteria and Gammaproteobacteria.
The sulfur cycle is predominated by reductive and oxidative processes of microorganisms: the dissimilatory sulfate reduction is considered as the main anaerobic process in the biomineralization of organic matter in the environment, accounting for up to 50% of its degradation in marine sediments (29), while dissimilatory sulfur oxidation processes occur wherever reduced inorganic sulfur compounds are available from the activity of the sulfate-reducing prokaryotes or from geological sources (5, 16). The sulfate-reducing and sulfur-oxidizing prokaryotes (SRP and SOP, respectively) are phylogenetically and physiologically diverse groups (5, 16, 25, 40, 47). Their polyphyletic nature restricts the concomitant detection of all recognized members by the use of a single 16S rRNA gene-targeting probe or primer pair in environmental analyses and limits the identification of novel lineages. In addition, the 16S rRNA gene-based analysis cannot provide an unambiguous link between the genetic identity of an uncultured microorganism and its physiological or metabolic capacity. Alternative molecular approaches circumvent these limitations by using functional genes that encode key enzymes of the dissimilatory sulfate reduction and sulfur oxidation pathways (ATP sulfurylase, Sat; adenosine-5′-phosphosulfate [APS] reductase, AprBA; sulfite reductase, DsrAB) and thus are much more suited to analyze and determine the phylogenetic complexity of the microbial sulfur cycle in nature.
Indeed, PCR assays have been developed for the amplification of dsrAB genes, but up to now these approaches are restricted to diversity analyses of the SRP community (2, 3, 15, 42, 43, 57). Although multiple events of lateral gene transfer (LGT) have affected the Dsr phylogeny of certain SRP lineages, the usefulness of dsrAB as functional gene markers for environmental analysis was confirmed (32, 62, 67). Recently, new PCR assays have been developed for the amplification of the aprBA gene locus from SRP and sulfur-oxidizing bacteria (SOB). The ubiquitous presence of aprBA genes in SRP was confirmed (38); however, the PCR-based screening among SOB reference strains revealed its restricted distribution to photo- and chemotrophs with strict anaerobic or at least facultative anaerobic lifestyles, e.g., several Chlorobiaceae and most Chromatiaceae species, Thiobacillus, Thiothrix, as well as invertebrate symbionts and their free-living relatives (37). The functional role of a reverse-acting APS reductase in apr-possessing, aerobic members of the alphaproteobacterial SAR11 and SAR116 cluster (19) is unresolved. Phylogenetic studies revealed the occurrence of LGT events with the results showing some anoxygenic photo- and chemolithotrophic representatives (e.g., Thiocapsa roseopersicina, Thiobacillus spp.) harboring a second, SRP-related apr gene locus in addition to a LGT-unaffected gene locus. In some SOB species (e.g., Thiocystis violascens), only the LGT-derived gene copy is present due to loss of the authentic gene (37). The previous analyses allowed the establishment of a comprehensive AprBA database covering most of the genus level diversity of SRP and apr-harboring SOB which provides a high-resolution framework for the assignment of environmentally retrieved gene sequences in molecular ecological studies. Comparative sequence analyses confirmed the reductive- and oxidative-type APS reductases to be highly conserved among SRP and SOB. Although affected by several LGT events involving representatives of SRP and SOB lineages, the aprBA genes have in general been transmitted vertically during evolution, which supports their usage as functional gene markers in microbial diversity surveys (37, 38). Astonishingly, the genes have only been employed by two medical studies to assess the SRP biodiversity in human tissue (13, 65).
In this study, we present a new assay for the functional aprA gene analysis using a combination of PCR and double-gradient denaturing gradient gel electrophoresis (DG-DGGE). The species diversity coverage of the new aprA gene-targeting primer set is evaluated in comparison with a previously published one (13) by phylogenetic investigation of SRP and SOB reference strains as well as several environmental samples derived from distinct habitats (sediment, filtered seawater, microbial mats, and invertebrate tissue). The aprA gene analysis allowed the simultaneous determination of the environmental SRP and SOB diversity, in contrast to the published dsrAB-based assays (32, 62). Finally, the assay was applied to investigate the sulfate-reducing and sulfur-oxidizing microbial communities inhabiting deep-sea sediment, seawater, and manganese crust samples of the volcanic arc of the Lesser Antilles, Caribbean Sea (Caribflux project); the work is complemented with 16S rRNA gene analyses and SRP and SOB cultivation studies.
MATERIALS AND METHODS
Microorganisms.
The investigated SRP and SOB reference strains were either obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) or provided by others as described in Table S1 in the supplemental material.
Study sites and sampling from the Caribbean Sea.
Sediment, seawater, and manganese crusts samples were collected from the volcanic arc of the Lesser Antilles, Caribbean Sea, during the RV Sonne cruise SO-154 (15 January to 8 February 2001) (Table 1). The deep-sea sediment cores and manganese crusts of the Kahouanne Basin (KB) and Montserrat Ridge (MR) were obtained by a multiple corer and a chain bag dredge, respectively. The cores were immediately subsampled by slicing the extruded sediment at 2-cm intervals. Fifteen liters of surface seawater was sampled from the photic water zone near St. Lucia Bay (SLB) by a rosette water sampler and subsequently filtered with Sterivex (pore size, 0.2 μm; Millipore) to collect the pelagic microbial community. For further molecular analyses, the sample material was immediately frozen and stored at −20°C until DNA extraction.
TABLE 1.
Location and characteristics of environmental samples from habitats investigated in this study by aprA and 16S rRNA gene fragment analyses
| Analysis type and sample name(s) from habitat | Sampling site | Position | Depth (m) | Characteristics of the habitat | Sample type(s) |
|---|---|---|---|---|---|
| Comparative aprA gene fragment analysis | |||||
| Spkg | Spiekeroog Island, German Wadden Sea (Germany) | Janssand region, 53°43.45′N, 7°41.80′E | Marine | Sediment surface layer | |
| Ec2, Ec4, Ec9 | Wimereux, Pas-de-Calais (France) | Marine | Echinocardium cordatum tissue, digestive tract | ||
| MatA, MatB, MatD | Cu-Pb-Zn Toyoha underground mine, Hokkaido (Japan) | 500, 550, 550 | Freshwater (53-73°C) | Terrestrial, thermophilic subsurface microbial mats | |
| Vail | Hobo Springs, Vail, CO | Freshwater (50°C) | Terrestrial hot spring, surface water | ||
| Vilm | Vilm Island, Baltic Sea (Germany) | 54°21′N, 13°28′E | Brackish water, estuarine | Sediment surface layer | |
| HR28Begg | Hydrate Ridge, Cascadia margin, OR | 44°34.19′N, 125°08.81′W | 777 | Marine | Beggiatoa field, microbial mats |
| HS2, HS3, HS7, HS8 | Mono Lake, CA | 17.5-28 | Hypersaline (84-95 g liter−1), alkaline | Sulfidogenic and methanogenic enrichment cultures from water column | |
| Comparative aprA and 16S rRNA gene fragment analysis | |||||
| KB-Sed | Kahouanne Basin, Lesser Antilles, Caribbean Sea | Station 15MC, 16°28.83′N, 61°58.81′W | 1,133 | Marine | Sediment surface layer, enrichment cultures |
| MR-Sed | Montserrat Ridge, Lesser Antilles, Caribbean Sea | Station 49MC, 16°39.51′N, 62°20.22′W | 978 | Marine | Sediment surface layer, enrichment cultures |
| MR-MnCr | Montserrat Ridge, Lesser Antilles, Caribbean Sea | Station 52CD, 16°38.85′N, 62°19.97′W begin; 16°37.40′N, 62°19.69′W end | 619-950 | Marine | Manganese crust surface, enrichment cultures |
| SLB-W | St. Lucia Bay, Lesser Antilles, Caribbean Sea | Station 99CTD, 13°51.16′N, 61°04.15′W | 82 | Marine | Filtered seawater (15 liters), enrichment cultures |
MPN counts.
Most probable number (MPN) counts were performed for SRP and SOB by preparing liquid MPN dilution series with media defined for sulfate-reducing and aerobic, thiosulfate-oxidizing bacteria with 1:10 steps as described elsewhere (49). The MPN dilution series were inoculated in parallel with 1 ml of suspended sediment and filtered seawater samples of the Caribbean Sea. Lactate (10 mM), butyrate (5 mM), and propionate (5 mM) were added as carbon sources to the SRP-defined medium.
DNA isolation.
Genomic DNAs from the SRP and SOB reference strains and the enrichment cultures of the MPN series were obtained by using a DNAeasy kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Total environmental DNAs from (i) filtered seawater (SLB), (ii) the 0- to 2- and 4- to 6-cm horizons of the sediments (KB and MR), and (iii) the manganese crust (MR) were extracted in duplicate using a modified version of the original protocol of Zhou et al. (64) (five cycles of freezing [liquid nitrogen] and thawing [at 30°C] of either 2 g of sediment/manganese crust or one filter [in 5.4 ml of the extraction buffer] were included before starting the extraction procedure). Environmental DNAs from (i) the surface sediments of an intertidal sand flat from “Janssand” on Spiekeroog Island, German Wadden Sea, and of a coastal (reed-covered) area from Vilm Island, Baltic Sea, (ii) microbial mats collected from the thermophilic subsurface environment of the Toyoha Mine, Japan, (iii) filtered water of a hydrothermal spring from Vail, CO, (iv) sulfido- and methanogenic enrichment cultures of anoxic bottom waters of Mono Lake, CA, (v) microbial mats of cold-seep Beggiatoa fields at the Hydrate Ridge, Cascadia margin, OR, and (vi) tissue material of Echinocardium cordatum (Table 1) were kindly provided by M. Mussmann, J. Kuever, M. Fukui, J. C. M. Scholten, T. Treude, and S. Gomes da Silva, respectively (see references 20, 28, 34, 42, 48, and 58 for descriptions of sample collection and the DNA extraction procedures). The quality of the isolated DNAs was verified by PCR amplification of the 16S rRNA gene using the primer set GM3F/GM4R and PCR protocol as described by Muyzer et al. (41). The extracted DNAs (dissolved in 10 mM Tris-HCl, 1 mM EDTA, pH 7.5) were stored at −20°C until further molecular analysis.
PCR amplification of partial aprA and 16S rRNA genes for DGGE analysis.
A modified version of the primer set AprA-3-FW/APS-RV, published by Deplancke et al. (13), and a new primer set, AprA-1-FW/AprA-5-RV, were used to amplify an approximately 0.4-kb aprA gene fragment for subsequent DGGE analysis (GC clamp added to the reverse primers) (Table 2). PCR assays were performed with a REDTaq DNA polymerase kit from Sigma-Aldrich (St. Louis, MO). Reaction mixtures (50 μl total volume) contained 5 μl 10× REDTaq PCR buffer (with 11 mM MgCl2), 5 μl 10× bovine serum albumin solution (3 mg/ml), 200 μM deoxynucleoside triphosphates mixture, 1 μM each primer, and 100 ng genomic DNA as template (10 and 100 ng environmental DNA were used in the assays). “Hot start” PCR was performed in a thermocycler (Eppendorf, Hamburg, Germany) followed by a “touchdown” PCR protocol as described in Table 2.
TABLE 2.
aprA gene-targeting primers and thermocycling conditionsa used in the PCR assays
| Primer | Sequence (in 5′→3′ direction) | Primer binding siteb |
|---|---|---|
| New primer set | ||
| AprA-1-FW | TGG CAG ATC ATG ATY MAY GG | 1236-1256 |
| AprA-5-RV | GCG CCA ACY GGR CCR TA | 1615-1631 |
| Modified Deplancke primer set | ||
| AprA-3-FW | TGG CAG ATM ATG ATY MAC GGc | 1236-1256 |
| APS-RV | GGG CCG TAA CCG TCC TTG AA | 1602-1623 |
| GC clamp | CGC CCG CCG CGC CCC GCG CCC GGC CCG CCG CCC CCG CCC G |
PCR thermocycling conditions: initial denaturation step for 5 min at 95°C; followed by 20 cycles of denaturing for 60 s at 95°C, annealing for 60 s at 60 to 50°C (“touchdown” PCR, −0.5°C every new cycle), and elongation for 90 s at 72°C; followed by 15 cycles of 60 s at 94°C, 60 s at 50°C, and 90 s at 72°C; followed by a 10-min final extension step.
Corresponding nucleotide positions of the aprBA operon of Desulfovibrio vulgaris subsp. vulgaris strain Hildenborough (GenBank accession no. Z69372).
AprA-3-FW lacks the false third G at the 3′ end of the primer APS-FW published by Deplancke et al. (13).
Partial 16S rRNA gene amplification from environmental DNAs for subsequent DGGE analysis was performed using the primer sets (i) GM5F (341F) with a GC clamp and 907R for Bacteria (41) and (ii) Arch516F with a GC clamp (K. Knittel, unpublished data) and Arch958R for Archaea (11), following the respective PCR protocols of the former authors.
The PCR products were visually analyzed by electrophoresis of aliquots (10% of the reaction volume) on a 2% (wt/vol) agarose gels run in 1× Tris-borate-EDTA buffer stained with ethidium bromide (0.5 mg liter−1) to verify correct amplicon size. Prior to further analysis, the amplicons of the expected gene fragment size were purified using either a QIAquick gel extraction kit (Qiagen, Hilden, Germany) or a Perfectprep gel cleanup sample kit (Eppendorf, Hamburg, Germany) following the supplier's recommendations.
DG-DGGE analysis of PCR-amplified aprA and 16S rRNA gene fragments.
DG-DGGE analyses (10) of the mixed-template aprA and 16S rRNA gene PCR products were performed using the D-GENE and D-CODE systems (Bio-Rad, München, Germany). For aprA gene fragment analysis, DG-DGGE gels (1.0-mm thick) were poured with an acrylamide gradient from 6 to 8% acrylamide/bis-acrylamide stock solution, 37.5:1 (vol/vol) (Bio-Rad), superimposed over a colinear denaturant gradient from 30 to 60% of denaturant (100% denaturant corresponds to 7 M urea and 40% formamide [vol/vol], deionized with AG501-X8 mixed bed resin [Bio-Rad]). For 16S rRNA gene analysis, DG-DGGE gels (1.0 mm thick) consisted of a 6 to 8% acrylamide gradient superimposed over a colinear 20 to 70% denaturant gradient. Gradients were formed using a Bio-Rad gradient former model 385. Twenty microliters of PCR product was mixed with 6 μl of dye solution (0.1% bromphenol blue [wt/vol], 70% [vol/vol] glycerol) and applied to the gels. The DG-DGGE electrophoresis runs were performed (i) for 2 h at 150 V and subsequently for 2 h at 200 V (aprA gene fragment) and (ii) for 3.5 h at a constant voltage of 200 V (16S rRNA gene fragment) using 1× Tris-acetate-EDTA as buffer and a constant temperature of 60°C. After gel electrophoresis, the gels were stained for 15 min with ethidium bromide (0.5 mg liter−1) and rinsed for 10 min in Milli Q water. The DNA bands were visualized on a UV transillumination table (Biometra, Göttingen, Germany); persisting and dominant bands were excised from the polyacrylamide gel by a scalpel, eluted in 50 μl Tris-HCl, pH 8.0, and reamplified using 1 μl of the eluate as template and PCR conditions as described above. The purity and migration behavior of the reamplification products of the DGGE bands were checked by DG-DGGE. The reamplification products were purified from free PCR primers using either a QIAquick gel extraction kit (Qiagen) or a Perfectprep gel cleanup sample kit (Eppendorf) following the suppliers' recommendations.
Nucleotide sequencing.
The reamplification products of the DG-DGGE bands were sequenced directly in both directions using the respective aprA and 16S rRNA gene amplification primers and an ABI Prism BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Sequencing reactions were run on an ABI PRISM 3100 genetic analyzer (Applied Biosystems).
Comparative analysis of the environmentally derived, partial aprA and 16S rRNA sequences.
The partial 16S rRNA sequences obtained from DGGE analysis were checked using the program CHECK_CHIMERA from the Ribosomal Database Project for detection of chimeric artifacts. The sequence data of each DGGE band were then added into the 16S rRNA sequence database of the Technical University Munich, Germany, using the software program package ARB (http://www.arb-home.de), and the data were assembled and first aligned automatically with the ARB_Align tool; the alignment was then manually corrected. The operational taxonomic units (OTUs) were compared to the GenBank database by the BLAST analysis tools of the National Centre for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST/) in order to determine their phylogenetic affiliation and to identify their closest phylogenetic relatives.
The aprA sequence data obtained from DGGE analysis were assembled and manually corrected using the BioEdit sequence alignment editor (version 7.0.5) (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The deduced, partial AprA sequences of the environmental samples were implemented into the persisting AprBA alignment of SRP and SOB references strains (see the introduction), including all full-length Apr sequences available from the public databases. The AprA data sets were phylogenetically analyzed with the tree inference methods included in the ARB software package and PHYLIP. Regions of ambiguous homology as well as insertions and deletions (indels) were excluded, yielding a final data set of 110 compared amino acid positions. Unrooted phylogenetic trees were calculated based on 411 sequences by performing distance matrix (neighbor-joining), maximum-parsimony, and maximum-likelihood analyses. The global rearrangement and randomized species input order options as well as the JTT matrix as an amino acid replacement model were used in the phylogenetic analysis.
GenBank accession numbers.
The nucleotide sequence data are available under the GenBank accession numbers EF614307 to EF614329 (archaeal 16S rRNA), EF614394 to EF614449 (bacterial 16S rRNA), and EF618618 to EF618673 and EF633037 to EF633105 (aprA).
RESULTS
In this study, the application of a new aprA gene-targeting primer pair (AprA-1-FW/AprA-5-RV) is described for SRP and SOB biodiversity surveys. Prior to environmental analysis, the species coverage and amplification behavior were evaluated and compared to the modified (corrected) version (AprA-3-FW/APS-RV) of a previous primer set published by Deplancke et al. (13); both primer pairs target nearly the same conserved gene region.
Species coverage: PCR amplification results of the new aprA gene-targeting primer pair from reference strains.
By using DNA templates from single SRP and SOB reference strains in the PCR assays, no 0.4-kb aprA amplicon was obtained with AprA-3-FW/APS-RV (with a GC clamp) from the thermophilic sulfate reducer lineages, from several members of the Desulfobulbaceae, Desulfobacteraceae, Syntrophobacteraceae, and the Desulfobacterium anilini-related groups, nor from some Desulfotomaculum and Desulfosporosinus spp., Desulfobacca acetoxidans, Caldivirga maquilingensis, and some aprBA gene-containing, sulfur oxidizer lineages of the Gammaproteobacteria. In contrast, the new primer pair allowed the amplification of correct-sized PCR products from SRP and SOB of a wider phylogenetic range (amplification results are summarized in Table S1 and Fig. S1 in the supplemental material), reflecting its improved species coverage compared to the previous set. Nevertheless, we were still unable to yield amplicons from the Desulfosporosinus spp. tested, which might have been caused by inhibited primer annealing (caused by differential codon usage and/or unusual amino acid substitutions in the target sequence). Indeed, the comparison of the primer sequences to the novel apr database revealed the presence of single mismatches of AprA-1-FW and AprA-5-RV with apr sequences of, e.g., Desulfotomaculum spp. (subcluster Ia), Desulfobacteraceae, and some representatives of sulfur-oxidizing Alphaproteobacteria and Gammaproteobacteria. However, the presence of single mismatches at internal or 5′-end sequence positions in the new PCR primers did not affect their PCR efficiency, as it has been demonstrated for 16S rRNA gene-targeting primers (35, 50). In contrast, the presence of several mismatches at the 3′ ends of AprA-3-FW and APS-RV inhibited an efficient annealing and elongation in the PCR assays and thus caused the limited species coverage of this primer set.
Preferential amplification: PCR amplification results of the new aprA gene-targeting primer from defined DNA template ratio mixtures by DG-DGGE analysis.
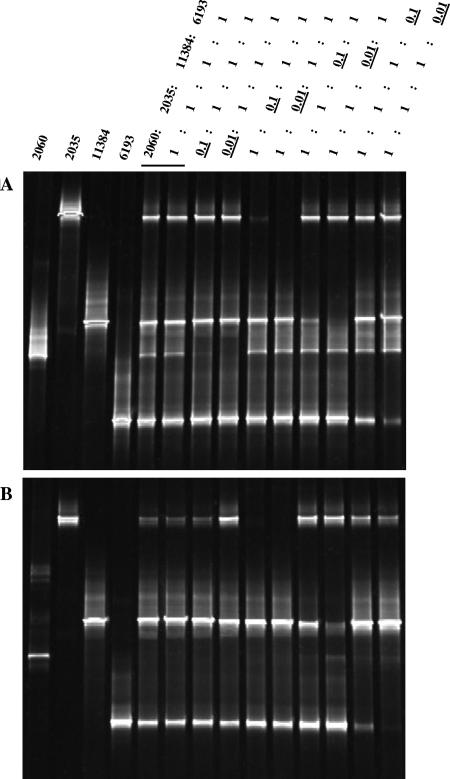
The aprA gene-based approach may suffer from the same methodical constraints as reported for the 16S rRNA gene amplification of mixed-template PCR assays (46, 61), which bias the coverage of phylotypes relative to the original microbial communities they were derived from. To evaluate the template-to-PCR product bias introduced by preferential amplification of certain target sequences, assays were performed using defined template mixtures of genomic DNA from four different SRP reference strains (Desulfosarcina variabilis, Desulfobacter sp. strain DSM 2035, Desulfovibrio profundus, and Desulfotomaculum thermobenzoicum). The DNA concentration of each SRP species was varied to yield relative template mixing ratios of 1:1:1:1, 1:1:1:0.1, or 1:1:1:0.01 in the PCR assays. The denaturing gradient analysis of the obtained mixed-template amplicons indicated that although the AprA-1-FW/AprA-5-RV(GC) primer set appeared to preferentially amplify D. thermobenzoicum (see band intensities on the DGGE gels, Fig. 1), the aprA gene fragments of the remaining SRP species were amplified and detectable in the gels even when the DNA template ratio of one species is only 1:10 relative to the other ones (Fig. 1A). In contrast, the primer pair AprA-3-FW/APS-RV(GC) preferentially amplified D. profundus, leaving only the amplicon of D. thermobenzoicum detectable when the DNA template ratio declined to 1:10 (relative to D. profundus) in the defined mixtures (Fig. 1B). The aprA gene fragment of D. variabilis was only amplified when its template ratio was above 100:1 relative to D. profundus. Thus, the bias introduced by preferential amplification of certain aprA phylotypes appears to be less pronounced with the new primer pair of this study, which is demonstrated by the improved congruence between PCR-detected diversity coverage and actual genetic diversity of SRP in the defined multitemplate mixtures as an example for environmentally derived DNA.
FIG. 1.
Preferential amplification of aprA gene fragments from defined genomic DNA ratio mixtures of the four SRP species Desulfosarcina variabilis (DSM 2060), Desulfobacter sp. (DSM 2035), Desulfovibrio profundus (DSM 11384), and Desulfotomaculum thermobenzoicum (6193) by the primer sets used for DGGE analysis. (A) Primer set AprA-1-FW/AprA-5-RV (GC clamp). (B) Primer set AprA-3-FW/APS-RV (GC clamp). In the PCR assays, the DNA concentration of each SRB species was varied to mixing ratios of 1:1, 1:0.1, or 1:0.01.
Evaluation of the microbial diversity coverage of the new aprA gene-targeting primer by comparative phylogenetic analysis of different habitats.
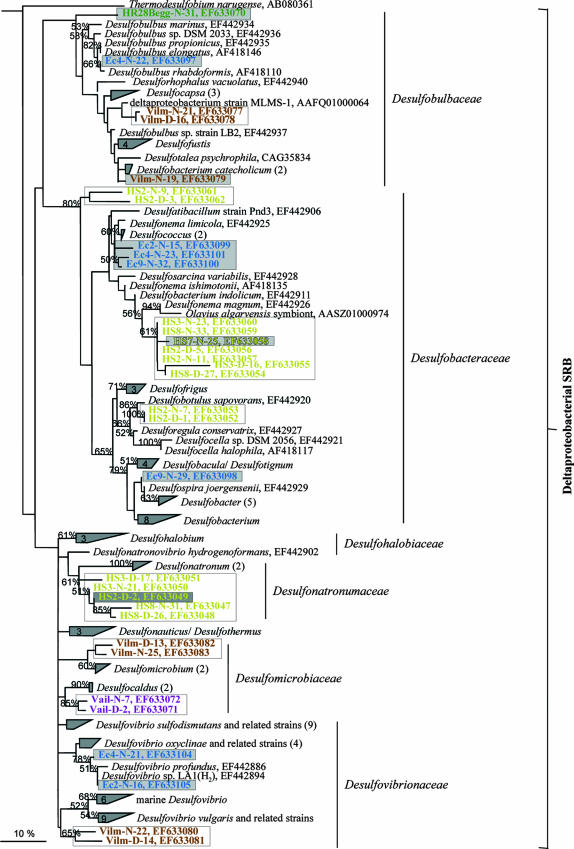
The microbial diversity coverage of the new aprA gene-targeting primer pair was evaluated and compared to that of the modified Deplancke primer set by exploring the environmental community composition of seven divergent habitats (listed in Table 1). The phylogenetic analysis of the received AprA phylotypes revealed congruent results obtained with both primer pairs (marked by open boxes in Fig. 2) for most investigated environmental samples. However, the new primer pair appeared to be generally superior with regard to the environmental SRP and SOB diversity coverage, since several additional phylotypes were only identified by applying AprA-1-FW/AprA-5-RV(GC) in the PCR assays (highlighted by light-gray shaded boxes in Fig. 2). These encompass OTUs related to, e.g., (i) thiotrophic gammaproteobacterial symbionts of Oligochaeta, e.g., Inanidrilus (Spiekeroog Island, oxic sediment surface layer), (ii) Desulfobacterium catecholicum (estuarine area on Vilm Island, anoxic sediment surface layer), (iii) Desulfonema magnum (water column of the hypersaline, alkaliphilic Mono Lake, methanogenic enrichment culture HS7), (iv) thiotrophic gammaproteobacterial symbionts of invertebrates and Desulfobulbaceae (Hydrate Ridge, Cascadia margin, microbial mats of cold-seep Beggiatoa fields), and (v) Desulfonema limicola, Desulfarculus baarsii, “Desulfobacterium anilini,” Desulfomonile tiedjei, Desulfospira joergensenii, Desulfobulbus rhabdoformis, and Desulfovibrio subspecies (Echinocardium cordatum specimen, nodules of the digestive tract). From the latter tissue samples, no aprA amplicon was even obtained using the modified Deplancke primer pair. The received AprA phylotype diversity from the subsurface microbial mats of Toyoha Mine was different for both primer sets, despite generally low microbial diversity present (42). Besides their consistent detection of thermophilic Desulfotomaculum (subclusters Ic/d) in MatB and MatD, the Thermodesulforhabdus- and Thermodesulfobacterium-related OTUs were obtained when applying either the modified or the new primer pair. The Thiobacillus plumbophilus (Apr lineage I) and thiotrophic Oligochaeta symbiont (Apr lineage II) relatives were only identified in mat A with the modified Deplancke primer set (highlighted by dark-gray shaded boxes in Fig. 2).
FIG. 2.
Phylogenetic tree based on 411 SRP/SOB reference strain and environmentally derived partial AprA sequences. The tree was inferred using the maximum-likelihood method. Sequences of the SOB Apr lineage I and Pyrobaculum aerophilum were used as outgroup references. The 16S rRNA gene-based taxonomic classification of SOB and SRP reference strains is indicated. The environmental AprA sequences are highlighted in boldface type and colored according to the habitat they were retrieved from (Spiekeroog Island, orange [Spkg]; Toyoha Mine, red [MatA/B/D]; Hydrate Ridge, green [HR28Begg]; Echinodermata cordatum, blue [Ec2, -4, -9]; Mono Lake, yellow [HS2, -3, -7, -8]; Vilm Island, brown; Vail, violet). The environmental AprA sequences received with primer set AprA-3-FW/APS-RV are termed with a “D” while those obtained with the new primer set AprA-1-FW/AprA-5-RV are termed with a “N” in the sequence name. For comparison of the microbial diversity coverage of both aprA gene-targeting primer pairs, identical phylotypes detected with both primer pairs are marked with open boxes, whereas those phylotypes that were obtained with only one primer pair are highlighted by dark gray (AprA-3-FW/APS-RV) and light gray (AprA-1-FW/AprA-5-RV) shaded boxes. The scale bar corresponds to 10% estimated sequence divergence.
Irrespective of the inference method used, the tree topologies calculated on the basis of the partial AprA sequences and the entire AprBA data set of SRP and SOB reference strains (see the introduction) were highly similar. This confirms that the smaller data set is applicable for the calculation of AprA-based trees and the phylogenetic assignment of environmental AprA sequences. However, as indicated by the low bootstrap values and multifurcations within, e.g., the gamma- and betaproteobacterial SOB group of Apr lineage I or the Desulfovibrionales, the limited capacity of phylogenetic information of the partial AprA sequence is insufficient for higher-level resolutions in certain major phylogenetic groups.
16S rRNA and aprA gene-based analysis: microbial communities of the sulfur cycle from sediment, manganese crust, and seawater samples of the Caribbean Sea.
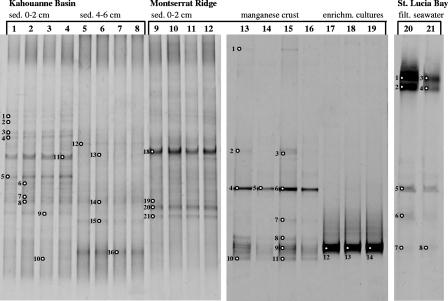
The microbial communities of the sulfur cycle at four study sites in the area of the Lesser Antilles volcanic arc (Table 1) was explored by molecular and cultivation approaches. Because of its improved coverage of environmental species diversity, only the new aprA gene-targeting primer pair was used to investigate the sediment, manganese crust, and filtered seawater samples (Fig. 3) in comparison with the 16S rRNA gene-based DG-DGGE analysis. The 16S rRNA analysis (summarized in Table 3) revealed a homogenous microbial community structure within the oxic sediment surface layers of the KB and MR (temperature, 4.5 to 4.8°C), comprising members of the Crenarchaeota, Gammaproteobacteria and Deltaproteobacteria, Bacteroidetes, low G+C gram-positive organisms, and Acidobacteria. The OTUs were affiliated with environmental sequences derived from various deep-sea sediments (1, 4, 23, 27, 45, 55) but only distantly with validated SOB and SRP. According to the MPN counting technique, the number of SRP increased in the surface sediments of KB and MR with depth from 4.3 × 105 to 2.4 × 106 cells/g, whereas the number of aerobic SOB remained constant between 4.3 × 106 to 7.5 × 106 cells/g. A single thiosulfate-oxidizing isolate revealed distantly related free-living relatives of thiotrophic mussel symbionts (1), whereas most strains belonged to the commonly culturable alphaproteobacterial lineages, e.g., Sphingomonadaceae and Roseobacter (14, 18). Sulfate-reducing relatives of Desulfovibrio acrylicus were isolated from both study sites. Similar to the 16S rRNA analysis, the aprA gene-based results (Fig. 4) demonstrated a homogenous microbial diversity in the sediments of KB and MR; however, all identified environmental AprA phylotypes belonged to novel alphaproteobacterial SOB clusters (AprA lineage I) that lack reference sequences of cultured SOB species. AprA sequences related to further SOB lineages or SRP were not obtained from the sediment samples. Since aprA genes were absent from the thiosulfate-oxidizing isolates, the aprA gene-detected Alphaproteobacteria are not identical to the cultivated ones but represent yet-unrecognized alphaproteobacterial SOB. Consistent with the 16S rRNA analysis, the AprA sequences derived from the sulfidogenic cultures are closely related to the sequence of Desulfovibrio acrylicus.
FIG. 3.
DGGE analysis of aprA gene fragments (amplified with primer pair AprA-1-FW/AprA-5-RV) using DNA samples from sediment (sed.), manganese crust, and seawater of the Caribbean Sea as templates. Lanes 1 to 8, DNA samples from Kahouanne Basin sediment (horizon 0 to 2 cm, lanes 1 to 4; horizon 4 to 6 cm, lanes 5 to 8); lanes 9 to 12, DNA samples from Montserrat Ridge sediment (horizon 0 to 2 cm); lanes 13 to 16, DNA samples from Montserrat Ridge manganese crust; lanes 17 to 19, DNA samples from aerobic sulfur oxidizer enrichment (enrichm.) cultures inoculated with manganese crust; lanes 20 and 21, DNA samples from St. Lucia Bay filtered (filt.) seawater. Bands indicated with numbers were excised from the gels and sequences.
TABLE 3.
List of phylogenetic affiliations of 16S rRNA partial sequences obtained from DGGE bands of environmental samples and enrichment cultures from the Caribbean Sea (collected in January 2001)a
| Investigated study site and environmental sample names | Major phylogenetic group | DGGE band(s) (ID no.) | Closest phylotype/species (acc. no.) | Sim. (%) | Remarks on closest phylotype/relative |
|---|---|---|---|---|---|
| Kahouanne Basin | |||||
| Sediment 0-2 cm | Crenarchaeota | KB-Sed-A3 | Uncultured archaeon clone Napoli-2A-11 (AY592231) | 98 | Environmental sample from sediments of Napoli mud volcano, Eastern Mediterranean, 1,910 m depth |
| KB-Sed-A4 | “Candidatus Nitrosopumilus maritimus” (DQ085097) | 98 | Isolated from a tropical seawater tank, ammonia-oxidizing marine archaeon | ||
| KB-Sed-A33 | Uncultured archaeon clone Urania-1A-36 (AY627460) | 98 | Environmental sample from sediments of a mound near Urania brine lake, Eastern Mediterranean, 3,442 m depth | ||
| Gammaproteobacteria | KB-Sed-B15 | Uncultured bacterium isolate JH12_C30 (AY568871) | 89 | Environmental marine sample | |
| KB-Sed-B14, KB-Sed-B16, KB-Sed-B21 | Uncultured gammaproteobacterium clone BNT-33-01 (AY240721) | 90 | Environmental marine sample from cold-seep sediment, Nankai Trough | ||
| Deltaproteobacteria | KB-Sed-B4 | Uncultured deltaproteobacterium Artic95c95C-5 (AF355039) | 99 | Environmental marine sample from Artic Ocean, SAR324 cluster | |
| Bacteroidetes | KB-Sed-B13 | Uncultured Bacteroidetes bacterium clone PLY-P3-5 (AY354804) | 90 | Environmental sample from coastal seawater (Bacteroidetes “AGG58 cluster”) | |
| Sediment 4-6 cm | Crenarchaeota | KB-Sed-A5, KB-Sed-A6, KB-Sed-A11 | Uncultured archaeon clone Urania-1A-01 (AY627426) | 98 | Environmental sample from sediments of a mound near Urania brine lake, Eastern Mediterranean, 3,442 m depth |
| KB-Sed-A7 | Uncultured archaeon clone Urania-1A-36 (AY627460) | 99 | Environmental sample from sediments of a mound near Urania brine lake, Eastern Mediterranean, 3,442 m depth | ||
| Gammaproteobacteria | KB-Sed-B24 | Uncultured bacterium clone A59 (AY373414) | 94 | Environmental sample from deep-sea sediments of Western Pacific | |
| Low G+C gram positive | KB-Sed-B25 | Uncultured low G+C gram-positive bacterium clone AT-s30 (AY225657) | 90 | Environmental marine sample from hydrothermal sediment of Mid-Atlantic Ridge | |
| Enrichment culture | |||||
| Aerobic SOB | Alphaproteobacteria | KB-Sed-B11 | Pelagibaca bermudensis (DQ178660) | 99 | Isolated from the Bermuda Atlantic Time Series Station, Sargasso Sea |
| KB-Sed-B12 | Uncultured Sphingomonas sp. clone JL-ECS-C44 | 99 | Environmental marine sample, East China Sea | ||
| Gammaproteobacteria | KB-Sed-B9, KB-Sed-B10 | Uncultured gammaproteobacte- rium clone BNT-33-01 (AY240721) | 90 | Environmental marine sample from cold-seep sediment, Nankai Trough | |
| SRB Lactat | Deltaproteobacteria | KB-Sed-B30 | Desulfovibrio sp. strain M2 (AY786352) | 97 | Marine SRB strain |
| Montserrat Ridge | |||||
| Sediment 0-2 cm | Crenarchaeota | MR-Sed-A8, MR-Sed-A9 | “Candidatus Nitrosopumilus maritimus” (DQ085097) | 99 | Isolated from a tropical seawater tank, ammonia- oxidizing marine archaeon |
| MR-Sed-A10 | Uncultured archaeon 19a-5 (AJ294876) | 94 | Environmental sample from sediments of Milos, Aegean Sea | ||
| MR-Sed-A12 | Uncultured archaeon 42-AB6 (AJ867792) | 95 | Environmental sample from sediments of Peru margin, ODP Leg 201, site 1229 | ||
| MR-Sed-A36 | Uncultured archaeon clone ODP1227A17.06 (AB177004) | 97 | Environmental sample from sediments of Peru margin, ODP Leg 201, site 1227 | ||
| Gammaproteobacteria | MR-Sed-B40 | Uncultured bacterium isolate JH10_C50 (AY568804) | 96 | Environmental marine sample | |
| Deltaproteobacteria | MR-Sed-B36 | Uncultured deltaproteobacterium clone Therm30-D12 | 92 | Environmental sample from oxic surface sediments of Eastern Mediterranean Sea | |
| Bacteroidetes | MR-Sed-B35 | Uncultured bacterium clone FEMidBac47 (AY769022) | 91 | Environmental sample from sulfide- and methane-rich cold seep, Florida | |
| Acidobacteria | MR-Sed-B39 | Uncultured Acidobacteriaceae bacterium clon VHS-B5-22 (DQ395041) | 99 | Environmental sample from sediment, Victoria Harbor, China | |
| Enrichment culture | |||||
| SRB Lactat | Deltaproteobacteria | MR-Sed-B49 | Desulfovibrio sp. strain M2 (AY786352) | 99 | Marine SRB strain |
| Montserrat Ridge | |||||
| Manganese crust | Crenarchaeota | MR-MnCr-A26, MR-MnCr-A27, MR-MnCr-A28 | Uncultured archaeon clone Urania-2A-31 (AY627509) | 97, 96, 98 | Environmental sample from sediments of a mound near Urania brine lake, Eastern Mediterranean, 3,442 m depth |
| MR-MnCr-A29 | Uncultured archaeon clone 43mENZ.2 (AY661820) | 93 | Environmental sample from metal-rich sediments of Peru margin, ODP Leg 201, site 1231 | ||
| Alphaproteobacteria | MR-MnCr-B7 | Uncultured bacterium (AY753387) | 89 | Environmental sample from biofilms grown on metal surfaces | |
| MR-MnCr-B9 | Uncultured alphaproteobacterium clone 131637 (AY922203) | 92 | Environmental sample from Santa Cruz Basin, Pacific Ocean, 1,674 m depth | ||
| MR-MnCr-B13 | Uncultured alphaproteobacterium clone JTB131 (AB015245) | 94 | Environmental sample from cold-seep area of the Japan Trench | ||
| Gammaproteobacteria | MR-MnCr-B4, MR-MnCr-B6 | Uncultured gammaproteobacterium clone APe4_38 (AB074619) | 91, 92 | Environmental sample | |
| MR-MnCr-B1 | Thiohalomonas nitratireducens (DQ836238) | 93 | Isolated from hypersaline lakes | ||
| MR-MnCr-B5 | Uncultured organism clone ctg_NISA060 (DQ396147) | 99 | Environmental sample from deep-sea octacoral | ||
| Bacteroidetes | MR-MnCr-B2, MR-MnCr-B8 | Uncultured bacterium clone WLB16-200 (DQ015862) | 92, 94 | Environmental sample from water of Lake Bonney, Antarctica, 16 m depth | |
| Enrichment culture | |||||
| Aerobic SOB | Alphaproteobacteria | MR-MnCr-B13, MR-MnCr-B17 | Uncultured alphaproteobacterium JL-ECS_C25 (AY663896) | 98, 97 | Environmental marine sample, East China Sea |
| Gammaproteobacteria | MR-MnCr-B15, MR-MnCr-B16 | Sulfur-oxidizing bacterium ODIII6 (AF170422) | 93 | Environmental sample from shallow-water hydrothermal vent, Milos, Aegean Sea | |
| St. Lucia Bay | |||||
| Filtered seawater | Euryarchaeota | SLB-W-A13 | Uncultured archaeon clone Sd- EA05 (AB194001) | 95 | Environmental sample from hydrothermal vent water, Suiyo Seamount |
| SLB-W-A14, SLB-W-A21 | uncultured marine group II euryarchaeote HF70_97E04 (DQ156478) | 92 | Environmental sample from North Pacific subtropical gyre water (Hawaii), 70 m | ||
| SLB-W-A15 | HF70_25A12 (DQ156469) | 91 | below sea surface | ||
| SLB-W-A20 | HF70_106D07 (DQ156467) | 91 | |||
| SLB-W-A16 | Uncultured marine group II euryarchaeote clone A_08 (DQ299289) | 98 | Environmental marine sample from sponge Reinochalina stalagmites | ||
| SLB-W-A17 | Uncultured marine group II euryarchaeote clone A9D1 (DQ299286) | 96 | Environmental marine sample from sponge Axechina raspailoides | ||
| Alphaproteobacteria | SLB-W-B9 | Uncultured organism ctg_CGOAB89 (DQ395477) | 93 | Environmental marine sample from deep-sea octacoral | |
| Cyanobacteria | SLB-W-B7 | Uncultured Synechococcus sp. strain JL-WNPG-T40 (AY664136) | 98 | Environmental marine sample, West Pacific Gyre | |
| SLB-W-B8 | Uncultured Synechococcus sp. strain JL-ECS-X16 (AY663962) | 97 | Environmental marine sample, East China Sea | ||
| SLB-W-B11 | Uncultured Prochlorococcus sp. strain JL-WNPG-T37 (AY664133) | 97 | Environmental marine sample, West Pacific Gyre | ||
| Bacteroidetes | SLB-W-B1, SLB-W-B4 | Gramella portivictoriae (DQ002871) | 97, 95 | Isolated from marine sediment, Victoria Harbor, East China Sea | |
| Enrichment culture | |||||
| Aerobic SOB LC1 | Alphaproteobacteria | SLB-W-B22 | Erythrobacter sp. strain JL-316 (AY646157) | 99 | Environmental marine sample |
| SLB-W-B23 | Pelagibaca bermudensis (DQ178660) | 99 | Isolated from the Bermuda Atlantic Time Series Station, Sargasso Sea | ||
| SLB-W-B24 | Alphaproteobacterium MBIC3923 (AB016848) | 94 | Environmental marine sample | ||
| Aerobic SOB LA5 | Alphaproteobacteria | SLB-W-B25, SLB-W-B26, SLB-W-B27 | Rhodobacter group bacterium LA5 (AF513437) | 97, 98, 97 | Environmental sample from Hawaiian Archipelago |
| Phototroph 1 | Chlorobia | SLB-W-B9 | Chlorobium bathyomarinum (AY627756) | 99 | Marine isolate from deep-sea hydrothermal vent |
| Bacteroidetes | SLB-W-B48 | Gramella portivictoriae (DQ002871) | 95 | Isolated from marine sediment | |
| Deltaproteobacteria | SLB-W-B12 | Desulfovibrionaceae bacterium MSL79 (AB110541) | 96 | Environmental sample from harbor sediment in Japan | |
| Phototroph 2 | Chlorobia | SLB-W-B13 | Prosthecochloris sp. strain Vk (AJ888467) | 99 | Marine isolate from fishing harbor of Visakhapatnam, India |
| Deltaproteobacteria | SLB-W-B14 | Desulfovibrionaceae bacterium MSL79 (AB110541) | 96 | Environmental sample from harbor sediment in Japan | |
| Phototroph 3 | Chlorobia | SLB-W-B18 | Chlorobium vibrioforme f. sp. thios, NCIB 8346 (AJ290830) | 99 | |
| Deltaproteobacteria | SLB-W-B26 | Desulfovibrionaceae bacterium MSL79 (AB110541) | 97 | Environmental sample from harbor sediment in Japan | |
| SRB H2 LA1 | Deltaproteobacteria | SLB-W-B19 | Desulfovibrionaceae bacterium MSL79 (AB110541) | 98 | Environmental sample from harbor sediment in Japan |
| SRB Lactat-1 LB2 | Deltaproteobacteria | SLB-W-B31 | Desulfovibrio caledonenis (U53465) | 94 | |
| SRB Lactat-2 LA2 | Deltaproteobacteria | SLB-W-B33 | Desulfovibrio brasiliensis (AJ544687) | 98 | |
| SRB Lactat-3 LB1 | Deltaproteobacteria | SLB-W-B30 | Desulfovibrio sp. strain M2 (AY786352) | 97 | Marine SRB strain |
| SRB Propion. LC1 | Deltaproteobacteria | SLB-W-B29 | Desulfobulbus rhabdoformis (U12253) | 100 | |
| SRB Butyrat. LB2 | Deltaproteobacteria | SLB-W-B35 | Uncultured Desulfocapsa sp. clone CBII115 (U12253) | 96 | Environmental sample from marine sediment |
The DGGE band identification number (ID no.) of each received 16S rRNA sequence and the accession number (acc. no.) of its closest phylotype/described species (including their sequence similarity [Sim.]) are given as retrieved by BLAST analysis.
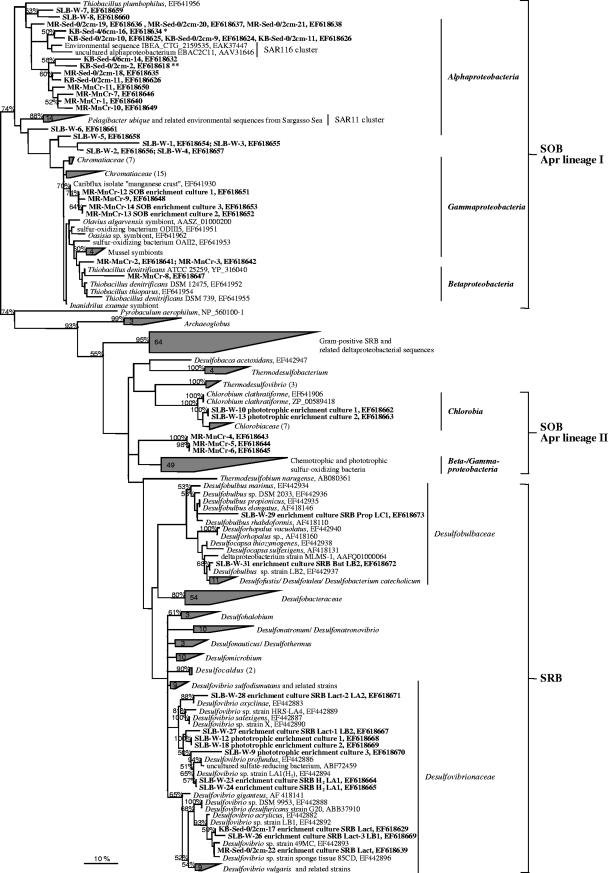
FIG. 4.
Phylogenetic tree based on 411 SRP/SOB reference strain and environmentally derived partial AprA sequences. The tree was inferred using the maximum-likelihood method. Sequences of the Apr lineage I and Pyrobaculum aerophilum were used as outgroup references. The environmental AprA sequences of sediment, seawater, and manganese crust samples of the Caribbean Sea are highlighted in boldface type (for abbreviations used for study sites and enrichment cultures, see Table 1 and Table 3). The 16S rRNA gene-based taxonomic classification of SOB and SRP reference strains is indicated. The scale bar corresponds to 10% estimated sequence divergence.
The crenarchaeotic 16S rRNA sequences identified from the manganese crust surface were closely related to the phylotypes of KB and MR sediments. In contrast, the 16S rRNA analysis revealed a higher diversity of OTUs affiliated to Alphaproteobacteria and Gammaproteobacteria that are putatively involved in sulfur oxidation processes within the crust surface. Indeed, several gammaproteobacterial OTUs were related to a newly described nitrate-reducing SOB strain (52) (detected directly in the crust surface) and free-living, thiodenitrifying relatives of marine invertebrate symbionts (34, 44) (thiotrophic cultures). The aprA gene analysis reflected the proposed higher phylogenetic diversity of putative sulfur-oxidizing strains in this habitat in comparison with the sediments (Fig. 4). The identified phylotypes diverged within the SOB-AprA lineage I into sequence clusters of (i) novel alphaproteobacterial SOB (also found in the KB and MR sediments), (ii) Thiobacillus spp. (Betaproteobacteria), and (iii) free-living relatives of thiotrophic invertebrate symbionts (Gammaproteobacteria). The detected phylotypes of AprA lineage II form a basal-branching sequence cluster that is only distantly related to the group of gamma- and betaproteobacterial SOB reference strains. The AprA sequences of the cultivated thiotrophic gammaproteobacterial strains are identical to an OTU directly detected in the manganese crust samples (Fig. 4). These AprA sequences most likely correspond to the 16S rRNA-identified SOB strain ODIII6 (Table 3). As in the sediments, no reductive-type AprA phylotype was detected in the manganese crusts; indeed, no SRP was obtained in enrichment cultures inoculated with sample material from this study site.
The 16S rRNA-based microbial diversity from seawater of SLB (collected from the thermocline zone; temperature, 22°C) was characteristic for bacterioplankton of photic zones in temperate oceanic waters (12, 18, 30) with OTUs related to marine group II euryarchaeota (12, 17) as well as Prochlorococcus and Synechococcus spp. (66), dominating the archaeal and bacterial community; no phylotypes affiliated with validated SOB and SRP were detected. The MPN counting technique, however, pointed to the presence of 2.4 × 109 aerobic SOB cells/ml and 3.9 × 105 SRP cells/ml in the oxic SLB water column. Thiosulfate-oxidizing members of the commonly cultured Alphaproteobacteria, e.g., Erythrobacter, Roseobacter lineage, and Rhodobacter spp. (7, 14, 18), were predominantly cultivated, whereas no gammaproteobacterial SOB were isolated. Besides the Desulfovibrio-, Desulfobulbus- and Desulfocapsa-related SRB strains isolated, three marine green sulfur bacteria (related to Chlorobium vibrioforme f. sp. thios NCBI 8346, Chlorobium bathyomarinum, and Prosthecochloris sp. strain Vk) could be enriched in coculture with a Desulfovibrio species. Consistent with the 16S rRNA, the aprA gene analysis demonstrated the absence of SAR11 or SAR116 cluster-like AprA phylotypes in the SLB water column. The three environmentally derived AprA phylotypes form a separate planktonic sequence cluster within the Alphaproteobacteria (SOB-Apr lineage I) and a novel Apr lineage of yet-uncertain phylogenetic affiliation (distant relationship to the gamma/betaproteobacterial SOB reference strains). Since aprA genes were absent from the cultivated Alphaproteobacteria, the environmental AprA sequences originated from a yet-undiscovered group of planktonic SOB. No aprA gene fragment could be obtained from the C. vibrioforme f. sp. thios relative, whereas the AprA sequences of the C. bathyomarinum and Prosthecochloris sp. cultures represented a novel Apr lineage of marine Chlorobiaceae. No AprA sequences related to SRP were identified from the oxygenated water column, although the results of the MPN cultures pointed to their presence. However, the phylogenetic placements of these cultured sulfate-reducing strains by AprA and 16S rRNA analysis were consistent.
DISCUSSION
In this study, a new PCR- and DGGE-based assay for functional aprA gene analysis is presented which allowed the concomitant molecular determination of the diversity of SRP and SOB in the environment. By screening taxonomically divergent SRP and SOB reference strains in PCR assays, the new primer pair showed an improved species coverage compared to the modified (corrected) version of a previously published primer set (13). In addition, the new primer set presented a higher detection sensitivity in multitemplate PCR assays with defined DNA concentrations; the aprA gene template-to-PCR product bias (generally caused by preferential amplification of certain target sequences [46, 61]) was less pronounced. Finally, comparative phylogenetic analyses of different environmental samples applying both primer pairs demonstrated that the phylotype diversity coverage of the new primer set was superior for most of the investigated habitats (see Results and Fig. 2). Overall, the detected environmental community structures of SRP and SOB based on the identified AprA phylotypes are consistent with previous cultivation and 16S rRNA gene analysis studies of the same or similar study sites/samples (20, 28, 34, 42, 48, 58).
aprA as a functional marker gene for SRP and SOB diversity surveys in comparison with 16S rRNA and dsrAB gene-based studies.
A direct comparison of the diversity analysis results for the aprA and 16S rRNA (and dsrAB) gene approaches is possible in the cases of the subsurface microbial mats of Toyoha Mine (42) and digestive tissue of E. cordatum specimens (20). The aprA gene-based analysis of the microbial mats provided a more complete picture of the dissimilatory microbial sulfur cycle: the Thermodesulforhabdus, Thermodesulfobacterium, and Desulfotomaculum, as well as the (sulfur-oxidizing) Thiobacillus plumbophilus, and free-living invertebrate symbiont relatives remained unidentified by the 16S rRNA gene approaches when applying universal primers (42). According to aprA gene analysis, the symbiotic SRP community of the nodules of E. cordatum is less diverse as it has been suggested by the 16S rRNA data (20). The aprA gene-undetected, putative sulfate-reducing strains were affiliated with sequence clusters of uncultured Deltaproteobacteria, which lack a closely related validated SRP species (only moderate relationships to Desulfobacterium catecholicum, Desulfobacula toluolica, and Desulfotomaculum acetoxidans). This reflects the above-mentioned limitations of the rRNA gene analysis in unambiguously linking the genetic identity of an unknown microorganism to its physiology.
Based on the dsrAB gene analysis of the Toyoha Mine microbial mats, the existence of two novel gram-positive SRP lineages was proposed by Nakagawa and coworkers (42). Indeed, newly discovered Dsr lineages of environmental sequences that lack sequences of cultured reference strains have frequently been found in various ecological studies and used to propose the presence of yet-unrecognized sulfate-respiring representatives (2, 3, 15, 42, 43, 57). However, many sulfite-reducing strains that are incapable of sulfate respiration are closely related to validated SRP, e.g., members of Desulfitobacterium (affiliated with Desulfosporosinus) (53), Sporotomaculum and Pelotomaculum (affiliated with Desulfotomaculum subclusters Ib and Ih) (26), Bilophila and Lawsonia (affiliated with Desulfovibrio), as well as the gram-positive genera Moorella, Carboxydothermus (24, 26), and archaeal Pyrobaculum (25). Since the dsrAB genes of these sulfite reducers have target sites that are fully complementary to the published primer sets (26, 67), the usage of the latter does not allow a differentiation between physiological groups of sulfite and sulfate reducers in PCR amplification. Therefore, the proposal of novel SRP lineages based on Dsr sequences that belong to environmental clusters is ambiguous. In contrast, an identified Apr lineage that lacks a close relationship to SRP reference sequences is a straightforward indication for the presence of an unrecognized phylogenetic lineage of SRP. Thus, our failure to detect additional Desulfotomaculum-related AprA phylotypes in MatB and MatD of Toyoha Mine, besides those affiliated to the thermophilic Desulfotomaculum (the dsrAB-consistent result), might be explained by the presence of gram-positive sulfite respirers.
All published dsr gene-targeting PCR assays are currently restricted to the investigation of SRP diversity because of the low sequence similarity between Dsr in SRP and SOB (32, 62); no universal or SOB-specific amplification primers and no comprehensive DsrAB database of sulfur-oxidizing reference strains exist. The aprA gene-targeting primers of this study, however, allowed the identification of the divergent SOB lineages and even the concomitant investigation of the SRP and SOB species diversity, as demonstrated for some environmental samples e.g., in hydrothermal water (Vail, CO) and microbial mats of Beggiatoa fields (Hydrate Ridge, OR) (Fig. 2). In the latter, the existence of a basal-branching Desulfobulbaceae representative was demonstrated within a microbial community that was otherwise predominated by different chemotrophic SOB species. (Note: a Desulfobulbus relative was identified in these sediment samples by 16S rRNA cloning analysis [T. Losekann, unpublished data]). Nevertheless, the aprA gene-based analysis will underestimate the microbial diversity involved in the oxidative processes of inorganic sulfur compounds in nature as a result of the restricted distribution of the apr genes among the SOB species (see the introduction). As a drawback, this molecular approach will allow their selected detection in environmental samples. However, the low bootstrap values in the SOB lineages show that the partial AprA sequences have only limited phylogenetic information which renders the direct assignment of the oxidative-type environmental AprA sequences to distinct SOB genera/species often difficult. This is also complicated by the LGT-affected evolutionary history of these genes in SOB with the result of some SOB species harboring a second, SRP-related apr gene locus besides an authentic one or only the LGT-derived gene copy (37). Concerning the phylogenetic complexity of SOB in the Beggiatoa field microbial mats, the identified AprA phylotypes pointed to the presence of (at least) one chemolithotrophic sulfur-oxidizing gammaproteobacterium (free-living relative of invertebrate symbionts) and one chemolithotrophic SOB forming a novel lineage of currently uncertain affiliation.
Comparison of 16S rRNA and aprA gene-based analyses: the sulfur-cycling microbial community in the area of the Lesser Antilles volcanic arc (Caribbean Sea).
The results of the 16S rRNA analyses revealed that the microbial community compositions from the aerated deep-sea sediment surfaces of KB and MR and the SLB photic water zone are similar to those of other nonhydrothermal, subtropical study sites in the Mediterranean and East China Seas as well as in the Western Pacific (12, 18, 23, 27, 45, 55). Most identified sediment-associated bacteria might have been involved in the decomposition of detritus-derived organic matter, e.g., the Bacteroidetes comprising chemoorganoheterotrophic degraders of various biopolymers (31), while the archaeal community appeared to be dominated by potential chemolithoautotrophic, nitrifying crenarchaeota (33, 63). In contrast, the SLB bacterioplankton consisted predominantly of phototrophic microorganisms, e.g., (putative) proteorhodopsin-containing euryarchaeota (17) and diverse unicellular cyanobacteria (66). However, no environmental 16S rRNA sequence was closely related to a validated SRP or SOB species; thus, the analyses provided no insights into the microbial community of the sulfur cycle at the investigated study sites. Their presence could only be demonstrated by selective cultivation techniques, with the highest taxa diversity apparent in the SLB photic water zone. The collected geochemical data collected during sampling indicated the absence of a direct, external input of reduced inorganic sulfur compounds at these study sites (Caribflux project final report). Interestingly, the aprA gene analysis revealed the bacterioplankton assemblages and the deep-sea sediments of the Caribbean Sea to be dominated by chemotrophic sulfur oxidizers that belong to four distinct and habitat-specific phylotype clusters within the alphaproteobacterial lineage (closest relatives are the SAR116 group members) and at least one separately branching lineage of yet-uncertain taxonomic affiliation. No phylotypes corresponding to SRP and beta/gammaproteobacterial chemolithoautotrophic SOB were detected at the study sites. The wide distribution of apr genes among members of the Alphaproteobacteria was unexpected because previous studies (37) demonstrated their absence in reference strains of this lineage, with the exceptions of Pelagibacter ubique of the SAR11 group (19) and an uncultivated representative of the SAR116 group. The alphaproteobacterial SAR11 cluster and Roseobacter clade members are the numerically dominant bacterioplankton groups in the decomposition of abundant organic sulfur compounds, e.g., dimethylsulfoniopropionate (DMSP) (21, 22, 36), and thus are implicated in sulfur cycling. Several representatives of the Roseobacter clade have also been reported to harbor the ability to oxidize inorganic sulfur intermediates, e.g., sulfite or thiosulfate to facilitate sulfur-based lithoheterotrophy (6, 21, 39), since reduced inorganic sulfur compounds are produced during the degradation of DMSP (21) or organosulfonates (8, 9). Indeed, thiosulfate-oxidizing, sulfate-producing members of this marine heterotrophic SOB group (e.g., Sulfitobacter spp.) have frequently been identified and isolated from various nonhydrothermal (deep-sea) sediments (51, 56, 59). Based on the results of the aprA gene analysis, we postulate that these chemolithoheterotrophic, sulfur-oxidizing members of the Roseobacter clade are also present in the investigated deep-sea sediment and photic water zone samples from the Caribbean Sea. Indeed, thiotrophic Pelagibaca bermudensis relatives were isolated from both habitats. However, no aprA partial sequence was obtained from the latter; this indicates that the culturable members do not represent the abundant species in these habitats. The functional role of the enzyme “dissimilatory APS reductase” in the alphaproteobacterial strains is unclear: a reverse-acting APS reductase might primarily be used for intracellular detoxification of sulfite as an accumulating product of the organic sulfur compound degradation; the oxidation product APS will be incorporated into the assimilatory sulfur metabolism or converted to sulfate (by the activity of a dissimilatory ATP sulfurylase), allowing additional energy conservation as previously proposed for species of SAR11 and SAR116 (37). The dominant role of chemolithoheterotrophic alphaproteobacterial instead of chemolithoautotrophic gammaproteobacterial SOB in the microbial sulfur transformation processes might be realistic for the investigated nonhydrothermal sediment and water column samples that lack a direct input of inorganic sulfur compounds. Nevertheless, the recognized limitations of PCR amplification (46, 61) and the DGGE method (41) might probably have biased the analysis towards the abundant groups in the microbial community, the SOB, with the consequence of the nondetection of minor present sulfate reducers. The 16S rRNA and aprA gene analyses of the sulfidogenic enrichment cultures point consistently to the presence of Desulfovibrio acrylicus-related strains. Interestingly, this SRB species is capable of DMSP cleavage for subsequent reduction of the acrylate moiety to propionate (60).
In contrast to the previous habitats, the geochemical data pointed to a (at least) subrecent, external (hydrothermal) input of inorganic sulfur compounds at the MR manganese crust collection site, reflected in the sulfur-based, resident microbial community structure. Indeed, OTUs affiliated with validated chemolithoautotrophic SOB species (52) were detected directly from the environmental samples by 16S rRNA gene analysis, while close relatives of SOB derived from hydrothermal systems in the Aegean Sea and Mid-Atlantic Ridge (34, 44) could even be isolated. Nevertheless, sulfur-oxidizing members of phylogenetic lineages recognized to be generally dominant at active deep-sea hydrothermal vents, e.g., Epsilonproteobacteria (44, 54), were not identified. The aprA gene analysis revealed the high phylogenetic complexity of SOB in the crust surface which was not discovered by 16S rRNA analysis. The sulfur-cycling microbial community is suggested to consist of (i) putative chemolithoautotrophic SOB strains of the Betaproteobacteria and Gammaproteobacteria, (ii) sulfur-oxidizing representatives of an uncertain-affiliated, novel SOB lineage, and (iii) members of the previously mentioned chemolithoheterotrophic alphaproteobacterial SOB. The betaproteobacterium appeared to be a marine representative of the genus Thiobacillus; however, a clear assignment of the other phylotypes to certain SOB genera/species is not possible with the amount of sequence information of the partial AprAs and the current database because Apr reference sequences of Beggiatoa and chemolithoautotrophic Epsilonproteobacteria (54) are missing. Previous work on Thiomicrospira spp. indicated the general absence of apr genes in this genus (see also Table S1 in the supplemental material), whereas the APS pathway seemed to be restricted to a few chemolithoautotrophic, marine members of Beggiatoa.
Supplementary Material
Acknowledgments
This study was supported by grants of the BMBF (project “Caribflux” under contract number 03G0154C), the DFG (under contract number KU 916/8-1), and the Max Planck Society, Munich, Germany.
Footnotes
Published ahead of print on 5 October 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arakawa, S., T. Sato, Y. Yoshida, R. Usami, and C. Kato. 2006. Comparison of the microbial diversity in cold-seep sediments from different depths in the Nankai Trough. J. Gen. Appl. Microbiol. 52:47-54. [DOI] [PubMed] [Google Scholar]
- 2.Bagwell, C. E., X. Liu, L. Wu, and J. Zhou. 2006. Effects of legacy nuclear waste on the compositional diversity and distributions of sulfate-reducing bacteria in a terrestrial subsurface aquifer. FEMS Microbiol. Ecol. 55:424-431. [DOI] [PubMed] [Google Scholar]
- 3.Bahr, M., B. C. Crump, V. Klepac-Ceraj, A. Teske, M. L. Sogin, and J. E. Hobbie. 2005. Molecular characterization of sulfate-reducing bacteria in a New England salt marsh. Environ. Microbiol. 7:1175-1185. [DOI] [PubMed] [Google Scholar]
- 4.Bano, N., and J. T. Hollibaugh. 2002. Phylogenetic composition of bacterioplankton assemblages from the Artic Ocean. Appl. Environ. Microbiol. 68:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brüser, T., P. N. L. Lens, and H. G. Trüper. 2000. The biological sulfur cycle, p. 47-86. In P. N. L. Lens and L. H. Pol (ed.), Environmental technologies to treat sulfur pollution. IWA Publishing, London, United Kingdom.
- 6.Buchan, A., J. M. Gonzalez, and M. A. Moran. 2005. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, J.-C., and S. J. Giovannoni. 2006. Pelagibaca bermudensis gen. nov., sp. nov., a novel marine bacterium within the Roseobacter clade in the order Rhodobacterales. Int. J. Syst. Evol. Microbiol. 56:855-859. [DOI] [PubMed] [Google Scholar]
- 8.Cook, A. M., and K. Denger. 2002. Dissimilation of the C2 sulfonates. Arch. Microbiol. 179:1-6. [DOI] [PubMed] [Google Scholar]
- 9.Cook, A. M., K. Denger, and T. H. M. Smits. 2006. Dissimilation of C3 sulfonates. Arch. Microbiol. 185:83-90. [DOI] [PubMed] [Google Scholar]
- 10.Cremonesi, L., S. Firpo, M. Ferrari, P. G. Righetti, and C. Gelfi. 1997. Double-gradient DGGE for optimized detection of DNA point mutations. BioTechniques 22:326-330. [DOI] [PubMed] [Google Scholar]
- 11.DeLong, E. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLong, E. F., C. M. Preston, T. Mincer, V. Rich, S. J. Hallam, N. U. Frigaard, A. Martinez, M. B. Sullivan, R. Edwards, B. R. Brito, S. W. Chilsholm, and D. M. Karl. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Nature 311:496-503. [DOI] [PubMed] [Google Scholar]
- 13.Deplancke, B., K. R. Hristova, H. A. Oakley, V. J. McCracken, R. Aminov, R. I. Mackie, and H. R. Gaskins. 2000. Molecular ecological analysis of the succession and diversity of sulfate-reducing bacteria in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 66:2166-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eilers, H., J. Pernthaler, F. O. Gloeckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fishbain, S., J. G. Dillon, H. L. Gough, and D. A. Stahl. 2003. Linkage of high rates of sulfate reduction in Yellowstone hot springs to unique sequence types in the dissimilatory sulfate respiration pathway. Appl. Environ. Microbiol. 69:3663-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedrich, C. G. 1998. Physiology and genetics of sulfur-oxidizing bacteria, p. 235-289. In R. K. Poole (ed.), Advances in microbial physiology, vol. 39. Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 17.Frigaard, N. U., A. Martinez, T. J. Mincer, and E. F. DeLong. 2006. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature 439:847-850. [DOI] [PubMed] [Google Scholar]
- 18.Giovannoni, S. J., and M. S. Rappe. 2000. Evolution, diversity and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley, New York, NY.
- 19.Giovannoni, S. J., H. J. Tripp, S. Givan, M. Podar, K. L. Vergin, D. Baptista, L. Bibbs, J. Eads, T. H. Richardson, M. Noordewier, M. S. Rappe, J. M. Short, J. C. Carrington, and E. J. Mathur. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242-1245. [DOI] [PubMed] [Google Scholar]
- 20.Gomes da Silva, S., D. C. Gillan, N. Dubilier, and C. De Ridder. 2006. Characterization by 16S rRNA gene analysis and in situ hybridization of bacteria living in the hindgut of a deposit-feeding echinoid (Echinodermata). J. Mar. Biol. Assoc. U.K. 86:1209-1213. [Google Scholar]
- 21.Gonzalez, J. M., R. P. Kiene, and M. A. Moran. 1999. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65:3810-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez, J. M., R. Simo, R. Massana, J. S. Covert, E. O. Casamayor, C. Pedros-Alio, and M. A. Moran. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heijs, S. K., G. Aloisi, I. Bouloubassi, R. D. Pancost, C. Pierre, J. S. Sinninghe Damste, J. C. Gottschal, J. D. van Elsas, and L. J. Forney. 2006. Microbial community structure in three deep-sea carbonate crusts. Microb. Ecol. 52:451-462. [DOI] [PubMed] [Google Scholar]
- 24.Henstra, A. M., and A. J. M. Stams. 2004. Novel physiological features of Carboxydothermus hydrogenoformans and Thermoterrabacterium ferrireducens. Appl. Environ. Microbiol. 70:7236-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huber, H., R. Huber, and K. O. Stetter. 2002. Thermoproteales. In M. Dworkin, E. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes. An evolving electronic resource for the microbial community. Springer, New York, NY.
- 26.Imachi, H., Y. Sekiguchi, Y. Kamagata, A. Loy, Y. L. Qiu, P. Hugenholtz, N. Kimura, M. Wagner, A. Ohashi, and H. Harada. 2006. Non-sulfate-reducing, syntrophic bacteria affiliated with Desulfotomaculum cluster I are widely distributed in methanogenic environments. Appl. Environ. Microbiol. 72:2080-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inagaki, F., T. Nunoura, S. Nakagawa, A. Teske, M. Lever, A. Lauer, M. Suzuki, K. Takai, M. Delwiche, F. S. Colwell, K. H. Nealson, K. Horikoshi, S. D'Hondt, and B. B. Joergensen. 2006. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. USA 103:2815-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishii, K., M. Mussmann, M. B. J. MacGregor, and R. Amann. 2004. An improved fluorescence in situ hybridization protocol for the identification of bacteria and archaea in marine sediments. FEMS Microbiol. Ecol. 50:203-212. [DOI] [PubMed] [Google Scholar]
- 29.Joergensen, B. B. 1982. Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature 296:643-645. [Google Scholar]
- 30.Karner, M. B., E. F. DeLong, and D. M. Karl. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507-510. [DOI] [PubMed] [Google Scholar]
- 31.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 32.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koennecke, M., A. E. Bernhard, J. R. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 34.Kuever, J., S. M. Sievert, H. Stevens, T. Brinkhoff, and G. Muyzer. 2002. Microorganisms of the oxidative and reductive part of the sulfur cycle at a shallow-water hydrothermal vent in the Aegean Sea (Milos, Greece). Cah. Biol. Mar. 43:413-416. [Google Scholar]
- 35.Kwok, S., D. E. Kellogg, N. McKinney, D. Spasic, L. Goda, C. Levenson, and J. J. Sninsky. 1990. Effects of primer-template mismatches on the polymerase chain reaction. Nucleic Acids Res. 18:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malmstom, R. R., R. P. Kiene, M. T. Cottrell, and D. L. Kirchman. 2004. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic Ocean. Appl. Environ. Microbiol. 70:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer, B., and J. Kuever. 2007. Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-5′-phosphosulfate reductase-encoding genes (aprBA) among sulfur-oxidizing prokaryotes. Microbiology 153:3478-3498. [DOI] [PubMed] [Google Scholar]
- 38.Meyer, B., and J. Kuever. 2007. Phylogeny of the alpha and beta subunits of the dissimilatory adenosine-5′-phosphosulfate (APS) reductase from sulfate-reducing prokaryotes—origin and evolution of the dissimilatory sulfate-reduction pathway. Microbiology 153:2026-2044. [DOI] [PubMed] [Google Scholar]
- 39.Moran, M. A., J. M. Gonzalez, and R. P. Kiene. 2003. Linking a bacterial taxon to sulfur cycling in the sea: studies of the marine Roseobacter group. Geomicrobiol. J. 20:375-388. [Google Scholar]
- 40.Mori, K., H. Kim, T. Kakegawa, and S. Hanada. 2003. A novel lineage of sulfate-reducing microorganisms: Thermodesulfobiaceae fam. nov., Thermodesulfobium narugense, gen. nov., sp nov., a new thermophilic isolate from a hot spring. Extremophiles 7:283-290. [DOI] [PubMed] [Google Scholar]
- 41.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 42.Nakagawa, T., S. Hanada, A. Maruyama, K. Marumo, T. Urabe, and M. Fukui. 2002. Distribution and diversity of thermophilic sulfate-reducing bacteria within a Cu-Pb-Zn mine. FEMS Microbiol. Ecol. 41:199-209. [DOI] [PubMed] [Google Scholar]
- 43.Nercessian, O., N. Bienvenu, D. Moreira, D. Prieur, and C. Jeanthon. 2005. Diversity of functional genes of methanogens, methanotrophs and sulfate reducers in deep-sea hydrothermal environments. Environ. Microbiol. 7:118-132. [DOI] [PubMed] [Google Scholar]
- 44.Nercessian, O., Y. Fouquet, C. Pierre, D. Prieur, and C. Jeanthon. 2005. Diversity of Bacteria and Archaea associated with a carbonate-rich metalliferous sediment sample from the Rainbow vent field on the Mid-Atlantic Ridge. Environ. Microbiol. 7:698-714. [DOI] [PubMed] [Google Scholar]
- 45.Polymenakou, P. N., S. Bertilsson, A. Tselepides, and E. G. Stephanou. 2005. Bacterial community composition in different sediments from the Eastern Mediterranean Sea: a comparison of four 16S ribosomal DNA clone libraries. Microb Ecol. 50:447-462. [DOI] [PubMed] [Google Scholar]
- 46.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabus, R., T. A. Hansen, and F. Widdel. 1999. Dissimilatory sulfate- and sulfur-reducing prokaryotes, p. 1-87. In M. Dworkin, E. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes. An evolving electronic database for the microbiological community. Springer, New York, NY.
- 48.Scholten, J. C. M., S. B. Joye, J. T. Hollibaugh, and J. C. Murrell. 2005. Molecular analysis of the sulfate reducing and archaeal community in a meromictic soda lake (Mono Lake, California) by targeting 16S rRNA, mcrA, apsA, and dsrAB genes. Microb. Ecol. 50:29-39. [DOI] [PubMed] [Google Scholar]
- 49.Sievert, S. M., T. Brinkhoff, G. Muyzer, V. Ziebis, and J. Kuever. 1999. Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island (Greece). Appl. Environ. Microbiol. 65:3834-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simsek, M., and H. Adnan. 2000. Effect of single mismatches at 3′-end of primers on polymerase chain reaction. Med. Sci. 2:11-14. [PMC free article] [PubMed] [Google Scholar]
- 51.Sorokin, D. Y. 2003. Oxidation of inorganic sulfur compounds by obligately organotrophic bacteria. Microbiology 72:641-653. [PubMed] [Google Scholar]
- 52.Sorokin, D. Y., T. P. Tourova, A. M. Lysenko, and G. Muyzer. 2006. Diversity of culturable halophilic sulfur-oxidizing bacteria in hypersaline habitats. Microbiology 152:3013-3023. [DOI] [PubMed] [Google Scholar]
- 53.Spring, S., and F. Rosenzweig. 2003. The genera Desulfitobacterium and Desulfosporosinus: taxonomy. In M. Dworkin, E. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes. An evolving electronic resource for the microbial community. Springer, New York, NY.
- 54.Takai, K., B. J. Campbell, S. C. Cary, M. Suzuki, H. Oida, T. Nunoura, H. Hirayama, S. Nakagawa, Y. Suzuki, F. Inagaki, and K. Horikoshi. 2005. Enzymatic and genetic characterization of carbon and energy metabolisms by deep-sea hydrothermal chemolithoautotrophic isolates of Epsilonproteobacteria. Appl. Environ. Microbiol. 71:7310-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teske, A. 2006. Microbial communities of deep marine subsurface sediments: molecular and cultivation surveys. Geomicrobiol. J. 23:357-368. [Google Scholar]
- 56.Teske, A., T. Brinkhoff, G. Muyzer, D. P. Moser, J. Rethmeier, and H. W. Jannasch. 2000. Diversity of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents. Appl. Environ. Microbiol. 66:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomsen, T. R., K. Finster, and N. B. Ramsing. 2001. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl. Environ. Microbiol. 67:1646-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Treude, T., A. Boetius, K. Knittel, K. Wallmann, and B. B. Joergensen. 2003. Anaerobic oxidation of methane above gas hydrates at Hydrate Ridge, NE Pacific Ocean. Mar. Ecol. Prog. Ser. 264:1-14. [Google Scholar]
- 59.Tuttle, J. H., and H. W. Jannasch. 1976. Microbial utilization of thiosulfate in the deep sea. Limnol. Oceanogr. 21:697-701. [Google Scholar]
- 60.van der Maarel, M. J. E. C., S. van Bergeijk, A. F. van Werkhoven, A. M. Laverman, W. G. Meijer, W. T. Stam, and T. Hansen. 1996. Cleavage of dimethylsulfoniopropionate and reduction of acrylate by Desulfovibrio acrylicus. Arch. Microbiol. 166:109-115. [Google Scholar]
- 61.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 62.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wuchter, C., B. Abbas, M. J. L. Coolen, L. Herfort, J. van Bleijswijk, P. Timmers, M. Strous, E. Teira, G. J. Herndl, J. J. Middleburg, S. Schouten, and J. S. Sinninghe Damste. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. USA 103:12317-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou, J. Z., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zinkevich, V., and I. B. Beech. 2000. Screening of sulfate-reducing bacteria in colonoscopy samples from healthy and colitic human gut mucosa. FEMS Microbiol. Ecol. 34:147-155. [DOI] [PubMed] [Google Scholar]
- 66.Zinser, E. R., A. Coe, Z. I. Johnson, A. C. Martiny, N. J. Fuller, D. J. Scalan, and S. W. Chilsholm. 2006. Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl. Environ. Microbiol. 72:723-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zverlov, V., M. Klein, S. Lücker, M. W. Friedrich, J. Kellermann, D. A. Stahl, A. Loy, and M. Wagner. 2005. Lateral gene transfer of dissimilatory (bi)sulfite reductase revisited. J. Bacteriol. 187:2203-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.