Abstract
Two thermostable lipases were isolated and characterized from Thermosyntropha lipolytica DSM 11003, an anaerobic, thermophilic, alkali-tolerant bacterium which grows syntrophically with methanogens on lipids such as olive oil, utilizing only the liberated fatty acid moieties but not the glycerol. Lipases LipA and LipB were purified from culture supernatants to gel electrophoretic homogeneity by ammonium sulfate precipitation and hydrophobic interaction column chromatography. The apparent molecular masses of LipA and LipB determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis were 50 and 57 kDa, respectively. The temperature for maximal activity of LipA and LipB was around 96°C, which is, so far as is known, the highest temperature for maximal activity among lipases, and the pH optima for growth determined at 25°C (pH25°C optima) were 9.4 and 9.6, respectively. LipA and LipB at 100°C and pH25°C 8.0 retained 50% activity after 6 and 2 h of incubation, respectively. Both enzymes exhibited high activity with long-chain fatty acid glycerides, yielding maximum activity with trioleate (C18:1) and, among the p-nitrophenyl esters, with p-nitrophenyl laurate. Hydrolysis of glycerol ester bonds occurred at positions 1 and 3. The activities of both lipases were totally inhibited by 10 mM phenylmethylsulfonyl fluoride and 10 mM EDTA. Metal analysis indicated that both LipA and LipB contain 1 Ca2+ and one Mn2+ ion per monomeric enzyme unit. The addition of 1 mM MnCl2 to dialyzed enzyme preparations enhanced the activities at 96°C of both LipA and LipB by threefold and increased the durations of their thermal stability at 60°C and 75°C, respectively, by 4 h.
Thermosyntropha lipolytica is an anaerobic, thermophilic, organotrophic, lipolytic, alkali-tolerant, gram-positive-type bacterium. It was isolated from an alkaline hot spring of Lake Bogoria, Kenya, by using a mineral medium supplemented with food-grade olive oil (53), with the specific intent of finding bacterial lipases exhibiting high stability and activity at elevated temperatures and alkaline pH values.
Lipases (carboxyl ester hydrolases; E.C. 3.1.1.3) are enzymes that generally catalyze the synthesis and hydrolysis of long-chain fatty acid esters (14, 45). They are ubiquitous in nature, produced by animals, plants, and fungi, as well as bacteria, but have not yet been reported in archaea. Lipases are activated at the water-lipid interface; they show little activity when the substrate is in the monomeric form, and the activity increases dramatically above the solubility limit, i.e., when the substrate starts to form emulsions. This fact has led to the emergence of a phenomenon known as interfacial activation, which describes substrate emulsions as a necessity for maximum lipolytic activity (8, 14, 45, 56) for the majority of lipases. However, there are a few exceptions that are not interfacially activated despite homology to other known lipases (19, 31, 33).
Lipases have a conserved three-dimensional structure (2), share the α/β hydrolase fold, and have the same catalytic mechanism. The catalytic center contains the catalytic triad serine-histidine-aspartic or glutamic acid. On the other hand, lipases usually have little if any similarity in their primary sequences, molecular masses, pH and temperature optima, substrate and positional specificities, cofactors, and cellular locations (2, 17).
Lipases are among the most versatile of the studied enzyme classes and are used in a number of applications in various industries, including the pharmaceutical, dairy, detergent, cosmetic, oleochemical, fat-processing, leather, textile, cosmetic, and paper industries (4, 7, 21, 22, 46, 57).
Many lipases, including those from mesophilic organisms, have been found to be fairly thermo- and alkali stable. However, it is likely that enzymes produced by thermophilic, alkali-tolerant bacteria will be more thermostable at alkaline pHs while exhibiting high specific activities at elevated temperatures (>70°C) and alkaline pHs (pH of >9). While a few lipases have been reported from aerobic thermoalkaliphilic bacteria (5, 27, 26, 30, 41), here we report on the purification and characterization of the first lipases from an anaerobic thermophilic bacterium, Thermosyntropha lipolytica.
MATERIALS AND METHODS
Culture and growth conditions.
Thermosyntropha lipolytica DSM 11003T was grown in 20- and 100-liter fermentors in a basal medium containing 0.75% yeast extract as the carbon and energy sources under nitrogen gas phase. The basal medium contained (per liter) 0.3 g of K2HPO4, 0.3 g of KCl, 0.5 g of NaCl, 1.0 g of NH4Cl, 0.1 g of MgCl2·6H2O, 0.02 g of CaCl2·2H2O, 3.0 g of NaHCO3, 3.0 g of Na2CO3, 0.5 g of Na2S·9H2O, 0.15 g of cysteine, 2 ml of vitamin solution, and 2.5 ml of trace element solution (15). The pH of the medium for growth determined at 25°C (pH25°C) (57) was adjusted to 8.2, and the growth temperature was 60°C.
Lipase assays.
Lipase was routinely assayed spectrophotometrically using p-nitrophenyl laurate (p-NPL) (Sigma) and p-nitrophenyl palmitate (p-NPP) (Sigma) as substrates (59). The reaction mixture contained 0.3 μM monomeric enzyme, 25 μM MnCl2, 1.088 ml of freshly prepared mixed buffer of 100 mM HEPES (Sigma), 100 mM TAPS (Sigma), and 100 mM CAPS buffer (Sigma), and 12 μl of 300 mM p-NPL or p-NPP in acetonitrile (final concentration in the assay, 3 mM). The assay was typically run for 20 min at 96°C (the assay was linear for up to 45 min). The reaction mixture was cleared by centrifugation before the amount of liberated p-nitrophenol was determined at A405.
To verify the activity obtained with the chromogenic substrate, assays were performed using triglycerides as substrates and measuring the liberated fatty acids by using an NEFA C kit (Waco) as instructed by the vendor. One unit is defined as the amount of the enzyme catalyzing the release of 1 μmol of p-nitrophenol (ɛ = 1.82 × 104 M−1 cm−1) from p-NPL/p-NPP or 1 μmol of free fatty acids from triglycerides per min.
Purification of lipases.
All purification steps were performed at room temperature. Twenty liters of medium was inoculated with 2 liters of an exponentially growing preculture. The pH was adjusted to 8.225°C (7.660°C), and the growth temperature was 60°C. After 18 h, the cells were separated by using an Amicon hollow-fiber filter with a 1-million Da cutoff, and the supernatant was recovered. The extracellular lipase was concentrated from the culture supernatant by filtration with a 10-kDa Amicon hollow-fiber filter (Millipore) and subsequent stepwise saturation to 60% and 75% ammonium sulfate. The precipitate was collected by centrifugation, dissolved in 10 mM sodium phosphate buffer, pH 8.0, and applied to a 30-ml octyl Sepharose fast-flow column (Amersham Biosciences). The column was preequilibrated with 20 mM Tris buffer, pH 8.0, containing 2 M (NH4)2SO4 (buffer A). The bound protein was eluted with a linearly decreasing gradient from 450 ml of buffer A to 450 ml of 20 mM Tris buffer, pH 8.0. Fractions containing high specific lipase activity [around 1.8 M and 0.5 M (NH4)2SO4] were pooled, desalted, and concentrated by using a filter membrane (Millipore). The lipases were tested for purity on native and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels as described by Sambrook et al. and Ausubel et al. (3, 44). The gels were stained with GelCode blue stain reagent (Pierce). The molecular masses of the enzymes were estimated graphically by interpolation from a logarithmic graph of molecular mass versus relative migrations using standard proteins (Sigma).
Protein concentration.
The protein concentrations were determined by using a bicinchoninic acid protein assay kit (Pierce) following the manufacturer's instructions.
The effect of temperature on activity and stability.
A temperature gradient incubator (Scientific Industries, Inc., Bohemia, NY) was used to determine the temperature for maximal activity of both lipases. The enzyme assay was performed at pH25°C 9.4 and 9.6, which were equal (58) to pH80°C 8.5 and 8.8 for LipA and LipB, respectively. The enzyme assays were performed as described above, using a combination buffer containing 100 mM HEPES, 100 mM TAPS, and 100 mM CAPS.
Thermostability was analyzed in triplicate by measuring the residual activity after incubating the enzyme in Tris buffer, pH25°C 8.0, at 60°C, 75°C, and 100°C for various times in series of sealed 2-ml serum bottles from which three bottles were sacrificed for every time point. Each sample contained 15 μg of protein in 0.6 ml of 100 mM Tris buffer, pH 8.0. After the incubation, the samples were concentrated and desalted when necessary by using 10-kDa-cutoff Centricon filter tubes (Millipore); they were then assayed in triplicate for lipase activity and protein concentration.
Determination of substrate specificity.
The substrate specificity and chain length selectivity were determined spectrophotometrically using a variety of p-NP esters (Sigma) and triglycerides. The levels of fatty acids liberated from triglycerides (all from Sigma) were determined by using an NEFA C kit. Long-chain glycerides (≥C12) were dissolved in acetonitrile.
Determination of positional specificity.
The positional specificities of LipA and LipB were analyzed by thin-layer chromatography using a modified version of the procedure reported by Lesuisse et al. (31). The enzyme assays were carried out at the temperature and pH optima of both lipases for 8 h. The assay solution contained 5 mM triolein and 1 mM MnCl2 in a 100 mM TAPS buffer. The assay mixture was sonicated for 30 s before the addition of 50 μg of the purified enzyme and was terminated by rapid cooling in an ice bath. The reaction products were extracted with 2 ml of cold diethylether. The extract was concentrated by evaporation and applied on reversed-phase (multi-K) thin-layer chromatography plates (Whatman). Monolein, 1,3-diolein, 1,2-diolein, and triolein (Sigma) were used as the reference standards. The plates were developed with a mixture of chloroform-acetone-acetic acid (96:4:1). The separated esters were visualized by spraying the plates with 0.1% iodine in chloroform.
Effects of metal ions, EDTA, and PMSF.
To determine the effects of metal ions, EDTA, and phenylmethylsulfonyl fluoride (PMSF) on enzyme activity, different concentrations of up to 10 mM were added directly to the standard p-NPL assay mixture. In addition to the standard assay, the effects of calcium, manganese, and iron, along with EDTA and PMSF, were tested with triolein as the substrate, and the liberated fatty acids quantified using an NEFA C kit.
RESULTS
Lipase level in the culture.
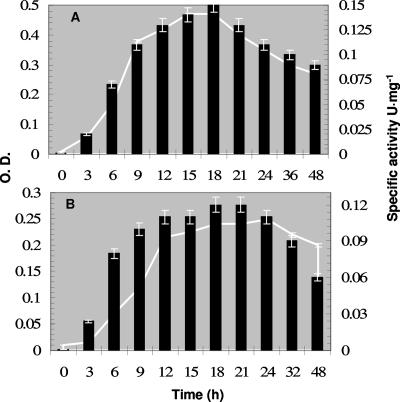
Svetlitshnyi et al. (53) reported that T. lipolytica constitutively produces lipase activity. Here, we report that the maximum formation of lipase activity occurred when the cell biomass reached its maximum (Fig. 1). At pH60°C 7.6, the optimum pH for growth, the maximum specific activity using p-NPL was 0.15 U mg−1 supernatant protein after 15 to 18 h of growth. At pH60°C 9.0, the maximum specific activity was 0.12 U mg−1 supernatant protein after 21 h of growth (Fig. 1). The supernatant contained 0.75% yeast extract from the medium, rendering the specific activity relatively low.
FIG. 1.
Extracellular lipase activity during growth at pH60°C 7.6 (A) and pH60°C 9.0 (B). Optical density (O.D.; curve) and specific activity (bars) in units of activity per mg protein in the supernatant which contained 0.75% yeast extract (wt/vol), leading to a high protein value and thus low specific activity. Error bars show standard deviations. A 500-ml amount of anaerobic medium was inoculated with 5% (vol/vol) of a 12-h-old culture of T. lipolytica, the pH of the medium was adjusted by using 0.2 M HCl and 0.2 N NaOH, and the culture was incubated at 60°C. Every time a sample was withdrawn, the pH of the culture was adjusted. After the optical density was measured, the sample was centrifuged and used for protein quantification and lipase assays.
Purification of lipases LipA and LipB.
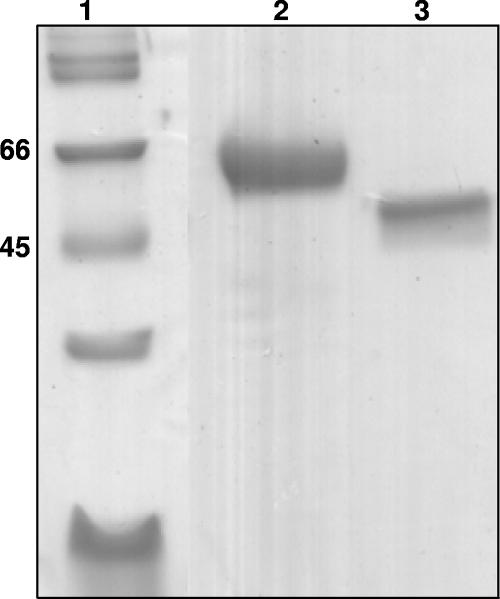
The lipases were purified 140-fold from the culture supernatant using ammonium sulfate precipitation and hydrophobic interaction chromatography (Table 1). Two different lipases were eluted from the octyl Sepharose column using a descending ammonium sulfate gradient. LipA, weakly bound, was eluted at 1.8 M ammonium sulfate, and the second lipase (LipB), more strongly bound, was eluted with 0.5 M ammonium sulfate. SDS-PAGE analysis yielded molecular masses of 50 (LipA) and 57 (LipB) kDa (Fig. 2). Their N-terminal sequences were NGGGATLPLQTSGVLTAGFAP and VKVMATLPADYVAQVIENVKKR, respectively, suggesting that they are two distinct enzymes.
TABLE 1.
Purification of extracellular LipA and LipB from Thermosyntropha lipolytica
| Step, enzyme | Total vol (ml) | Total activity (units) | Total protein (mg) | Sp act (U mg−1) | % Yield | Purification factor |
|---|---|---|---|---|---|---|
| Supernatant | 22,000 | 2,900 | 28,600 | 0.1 | 100 | 1 |
| Holofiber with 10-kDa cutoff | 5,000 | 2,640 | 3,250 | 0.81 | 91 | 8 |
| (NH4)2SO4 precipitation | 800 | 1,580 | 392 | 4.0 | 54.5 | 40 |
| Octyl Sepharose column, LipA | 300 | 660 | 47 | 14 | 23 | 140 |
| Octyl Sepharose column, LipB | 210 | 390 | 29 | 13.4 | 13.4 | 134 |
FIG. 2.
Purified LipA and LipB proteins were analyzed by SDS-PAGE. Lanes: 1, molecular mass marker (kDa); 2, purified LipB from octyl Sepharose column; 3, purified LipA after Q-Sepharose fast-flow chromatography. Gels were stained with GelCode blue stain reagent.
LipA and LipB were cold labile; i.e., both irreversibly lost activity when stored at 0 to 4°C for 24 h or when frozen in various buffers, including phosphate (pH25°C 8.0) buffer. The effect could be minimized by freezing in solutions containing 40% glycerol (vol/vol) and 2 mg/ml bovine serum albumin, leading to residual catalytic activities of 75% and 90% for LipA and LipB, respectively.
Effects of pH and temperature on enzyme activity and stability.
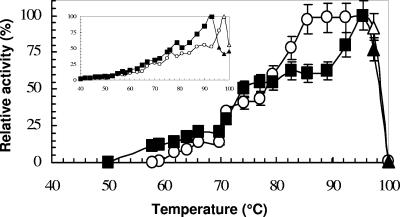
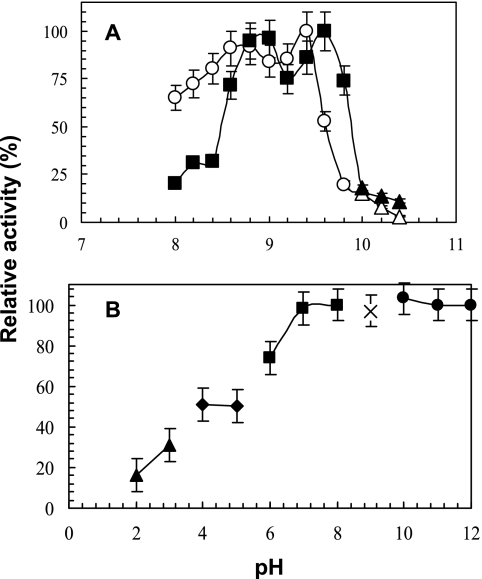
The maximal observed activities for LipA and LipB were at 96°C with the 20-min p-NPL assays (Fig. 3). Because p-NPL is relatively unstable at 96°C and a pH of ≥10.0, the true substrate, trioleate, was used for determining the pH ranges and optima at the optimum temperature of 96°C. A broad pH25°C range for activity was observed for both enzymes. They exhibited a biphasic curve with intermediate maxima of around 8.7 (LipA) and 8.9 (LipB) besides the pH optima of around pH25°C 9.4 (LipA) and 9.6 (LipB) (Fig. 4A). At room temperature, both enzymes were unstable when incubated at acidic pHs (below 7.0) but were stable for 36 h at alkaline pHs, retaining 100% activity at pH25°C 7.0 to 12.0 (Fig. 4B).
FIG. 3.
Effect of temperature on LipA (○) and LipB (▪) activities. The lipase assay was conducted as mentioned in Materials and Methods. The substrate used for the assay was p-NPL. A similar temperature profile but lower specific activity was also obtained when p-NPL was replaced with p-NPP. The insert is the temperature profile when only TAPS buffer was used at pH 9.0. A true substrate, trioleate, was used at temperatures of ≥98°C, and the liberated fatty acids were quantified by using an NEFA C kit. The 100% relative activities were 11.8 ± 0.5 U mg−1 and 13.0 ± 0.6 U mg−1 for LipA and LipB, respectively. The activity values were corrected for control values obtained without enzyme addition. Error bars show standard deviations.
FIG. 4.
Effect of pH25°C on activity of LipA (○) and LipB (▪) (A) and on stability of LipA (B). The 100% relative activities were 12.4 ± 0.5 U mg−1 and 12.8 U mg−1 ± 0.3 U mg−1 for LipA and LipB, respectively. Because the p-NPL substrate was not stable at pH25°C 10 or above, triolein was used at the high pH (panel A, ▴); the liberated fatty acids were measured by using an NEFA C kit. (B) LipA and LipB showed nearly identical profiles for stability; thus, LipB was omitted for clarity. For the stability experiments, 100 mM of the following buffers were used: glycine-HCl (▴), sodium acetate buffer (⧫ and ◊), MES buffer (▪), TAPS buffer (×), and CAPS buffer (•). Results are expressed as percentage of maximal activity. The lipase assays were conducted as described in Materials and Methods.
LipA and LipB showed maximum thermostability at temperature and pH optima in the presence of 0.5 to 2 M (0 to 2 M were tested) ammonium sulfate. They retained 50% activity after incubation at 100°C for 6 h (LipA) and 2 h (LipB). The temperatures for a 24-h half life were 74.1°C (LipA) and 76.5°C (LipB) (graphs not shown).
Substrate specificity and positional specificity.
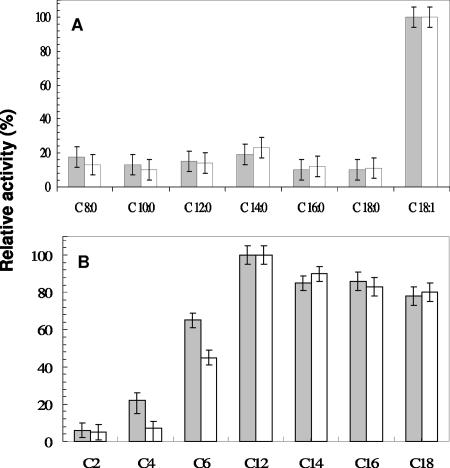
With the use of triglycerides as substrates, both LipA and LipB exhibited no significant activity with tributyrin (C4) and tricaproin (C6) but intermediate high activity toward glycerides with long-chain fatty acids (C12 to C18) and an activity that was about fivefold higher with triolein, i.e., a triglyceride containing unsaturated C18 fatty acids, than with glycerides containing saturated fatty acids (Fig. 5). Among the p-NP esters, similar specificities were observed. The highest activity was observed with the saturated C12 ester; unfortunately, the p-NP oleic acid ester was not available. This substrate specificity identifies LipA and LipB as true lipases. The hydrolytic products of triolein after up to 8 h of incubation with LipA or LipB were increasing amounts of 2-monoolein, indicating that the enzymes specifically hydrolyze the sn-1,3 position of triolein (detailed data not shown).
FIG. 5.
The activities of purified LipA (open bars) and LipB (grey bars) toward substrates with different acyl chain lengths were determined under standard conditions. (A) Activities with 5 mM triglycerides. The 100% relative activities represent 0.64 ± 0.03 U mg−1 (LipA) and 0.79 ± 0.04 U mg−1 (LipB) using trioleic acid as the substrate. (B) Activities with 3 mM p-nitrophenyl esters. The 100% activities represent 12.4 ± 0.5 U mg−1 (Lip A) and 13.0 ± 0.6 U mg−1 (LipB) with p-NPL as the substrate. Error bars show standard deviations.
Effects of metal ions, PMSF, and EDTA on the enzymes' activities.
The catalytic activities of LipA and LipB of T. lipolytica were neither enhanced nor inhibited by the presence of 1 mM to 10 mM CaCl2 (Table 2), but a 15% decrease in activity occurred at 25 mM CaCl2. LipA (pH25°C 9.4) and LipB (pH25°C 9.6) showed various degrees of inhibition by the metal ions tested (Table 2), and the inhibition was greater as the concentration of the ion increased. Manganese (in the form of MnCl2) at concentrations between 0.1 and 2 mM increased the activities of LipA and LipB by threefold in comparison to the activities found in assays conducted in the absence of metal ions or in the presence of calcium (0.1 to 10 mM). The effect of manganese was confirmed by carrying out lipase assays using triolein as the substrate and quantifying the liberated fatty acids. In the presence of 5 mM EDTA, nearly half of the activity of both LipA and LipB was lost, and the activity was completely diminished in the presence of 10 mM EDTA. These results indicate that LipA and LipB might require a specific metal for activity.
TABLE 2.
Effects of various metal ions and inhibitors on lipase activity
| Metal ion or inhibitor | Relative remaining activity (%) of enzyme at indicated concn of ion or inhibitor
|
|||
|---|---|---|---|---|
| LipA
|
LipB
|
|||
| 1 mM | 10 mM | 1 mM | 10 mM | |
| Caa | 100 ± 5 | 100 ± 5 | 100 ± 5 | 100 ± 5 |
| Mg | 95 ± 5 | 65 ± 5 | 97 ± 5 | 73 ± 5 |
| Fea | 12 ± 5 | 0 | 20 ± 5 | 0 |
| Mna,b | 340 ± 5 | 270 ± 15 | 315 ± 15 | 250 ± 15 |
| K | 65 ± 5 | 30 ± 10 | 85 ± 5 | 70 ± 10 |
| Zn | 50 ± 15 | 44 ± 15 | 62 ± 15 | 36 ± 15 |
| Na | 78 ± 5 | 70 ± 5 | 83 ± 5 | 80 ± 5 |
| Cs | 77 ± 5 | 37 ± 10 | 72 ± 5 | 44 ± 5 |
| Cu | 45 ± 10 | 0 | 56 ± 10 | 0 |
| Al | 28 ± 5 | 0 | 30 ± 5 | 0 |
| Ni | 80 ± 5 | 22 ± 5 | 50 ± 5 | 15 ± 5 |
| Co | 60 ± 5 | 60 ± 5 | 68 ± 5 | 49 ± 5 |
| PMSF | 70 ± 15 | 6 ± 5 | 62 ± 15 | 0 |
| EDTA | 60 ± 15 | 11 ± 5 | 81 ± 5 | 0 |
Assays were conducted with triolein and lipase activity quantified by using an NEFA C kit.
The addition of Mn or Mn and Ca together gave the same value.
Subsequent metal analyses (by inductively coupled plasma emission spectrometry) yielded data showing that purified LipA and LipB contained Mn2+ ions instead of the Zn2+ ions found in thermophilic lipases from Geobacillus stearothermophilus. After correction against the values of the dialysis buffer, the enzymes contained 0.92 ± 0.1 (mean ± standard deviation) mol Ca2+ and 0.6 ± 0.06 mol Mn2+ ions per mol of monomeric enzyme. Since the addition of manganese ions resulted in a threefold-greater activation of the enzyme, it is assumed that the native enzyme contains 1 mol Mn2+ ions per mol lipase and that approximately 40% of the Mn2+ ions are removed from the protein during the purification procedure or during dialysis.
Lipases are generally members of the serine hydrolases, with serine as an essential residue for catalytic activity. The addition of 10 mM of the serine hydrolase inhibitor PMSF inhibited the activity of LipA and LipB (Table 2), indicating that LipA and LipB are also serine hydrolases.
DISCUSSION
Two highly thermostable enzymes are present.
Very few lipases from thermophiles and none from an anaerobic thermophile have been characterized so far (43). The anaerobic thermophile T. lipolytica produced two extracellular lipases which appear to be encoded by two distinct genes, as indicated by their different biochemical properties and different N-terminal sequences. However, this needs to be verified in the future by sequence analysis of the encoding genes. Attempts to clone the two lipases were unsuccessful. A maximum temperature of 96°C (while using the true substrate triolein instead of the usually used p-NP esters) indicates that these two lipases are the most thermophilic ones so far reported. The only other lipase with a similar high thermal activity and stability was found, surprisingly, in the mesophilic Burkholderia cepacia (39). Its lipase retains 50% activity after 13 h of incubation at 90°C. However, comparing the physical properties of these lipases is quite difficult due to the differences in the assay substrates and procedures. None of the lipases from aerobic thermophiles exhibit the high thermostability of LipA and LipB (27, 36, 50).
Role of metal content in activity and stability of LipA and LipB.
Many bacterial lipases for which structures are available contain a Ca2+-binding site. These include the enzymes from Chromobacterium viscosum (28), Pseudomonas aeruginosa (35), and Burkholderia glumae (basonym Pseudomonas glumae) (BGL) (37). Many lipases have been found to require certain metals for activity and/or to enhance activity and (thermo)stability. The two thermophilic lipases from Geobacillus stearothermophilus (basonym Bacillus stearothermophilus), P1 (55) and L1 (24), contain, besides Ca2+ ions, Zn2+ ions, which is unique among the lipases that have been characterized so far. Zinc is believed to be involved in enhancing the thermostability of these enzymes. In addition, the activity of the thermophilic lipase of Geobacillus thermoleovorans strain ID-1 (30) is enhanced by the addition of calcium and zinc ions. The activities of other lipases, from Pseudomonas sp. (13, 38), Acinetobacter sp. (51), Bacillus licheniformis (25), and Bacillus subtilis 168 (31), are enhanced by calcium. Staphylococcal lipases require calcium for activity and stability (40, 16). Kim et al. showed by means of fluorescence emission kinetics that calcium ions enhance the thermostability of the G. stearothermophilus lipase (27).
The importance of calcium ions and their involvement in stabilizing the tertiary structures of lipases was observed in the crystal structure of the B. glumae lipase (37). Calcium ions function as ligands to a number of adjacent residues at the active site. The loss of the calcium ion through either pH change or mutation to a residue that affects the calcium interactions has been proposed to disrupt the enzyme structure and decrease its thermal stability, as observed in Staphylococcus hyicus (49). However, the addition of calcium ions neither enhanced nor reduced the activity and/or stability of LipA or LipB. Based on the observed stoichiometry, the Ca2+ ion appears to be tightly bound. In contrast, manganese ions specifically enhanced the activities of both LipA and LipB by threefold (Table 2). Based on the metal analysis, the Mn2+ ion does not bind as tightly as the Ca2+ ion. Moreover, the effect of manganese on the thermostability of LipA and LipB; i.e., manganese extended the half-lives of both enzymes by 4 h at 60°C and 75°C, respectively, suggests that LipA and LipB contain a manganese-coordinated domain similar to the Zn-coordinated domain found in G. stearothermophilus lipases, with a similar function of stabilizing the protein structure. The crystal structure of G. stearothermophilus lipase indicated that the zinc-coordinated extra domain makes tight interactions with the loop extended from the C terminus of the lid helix, thus making strong, hydrophobic interactions with its neighboring domains, including the core domain. It has been proposed that these interactions lead to rigid packing of the active site, and they appeared to play an important role in the enzyme catalytic activity and thermostability of the enzyme. This is in agreement with the observed maximal activity at 65°C. The lid apparently requires high temperatures in order to move away and to expose the active site (24). The Mn2+ ion, a slightly larger metal ion than the Zn2+ ion, might have the same effect in LipA and LipB as zinc in other lipases, contributing to the enzyme activity and stability at the elevated temperatures. Other structural features, such as high hydrophobic interactions, higher hydrogen bonding, amino acid substitutions, and high salt bridge content, among others (10, 11, 18, 23, 32, 42, 54), which were observed and studied in thermophilic proteins, could contribute to the thermostability of LipA and LipB, and have not been investigated here due to lack of the encoding gene sequence and crystal structure. However, the most profound effect on thermostability was noticed with the addition of 0.5 to 2 M ammonium sulfate to the enzyme solution prior to assaying activity, suggesting that hydrophobic interactions are important for the integrity of the LipA and LipB protein structures. This conclusion is in agreement with the results of Benjwal and coworkers, who showed that molar concentrations of NaCl or Na2SO4 increased the apparent melting temperature of a lipoprotein by up to 20°C and that these salts decelerated protein unfolding (6). Similar effects have also been reported for other enzymes (1, 20, 52). Ammonium sulfate is a chaotropic agent. It increases the chaos (entropy) in water and thereby increases hydrophobic interactions; the proteins become less flexible, which stabilizes the tertiary structure and lowers domain unfolding.
Substrate specificity and true lipases.
The substrate specificity preferences of LipA and LipB are different from those of other described thermophilic lipases. Whereas LipA and LipB both had similar preferences for long-chain fatty acid esters (C12 to C18 p-NP esters) and, especially, toward the unsaturated ones present in triolein (C18:1 as glycerol ester), other described thermophilic lipases have maximal activity with shorter, acryl esters, and several could be called lipase esterases and esterases instead of true lipases. For example, the thermophilic enzyme from the aerobic strain G. thermoleovorans ID-1 showed maximum activity toward tricaproin (C6) and p-nitrophenyl caproate (C6) (9), that from G. stearothermophilus P1 toward p-nitrophenyl caprate (C10) and tricaprylin (C8) (50), that from G. stearothermophilus L1 towards p-nitrophenyl caprylate (C8) and tripropionin (C3) (26), and that from Geobacillus thermocatenulatus toward p-nitrophenyl butyrate and tributyrin (C4) (48).
LipA and LipB hydrolyze specifically the ester bonds at positions 1 and 3 of triglycerides. This specificity is similar to that exhibited by lipases from, e.g., Bacillus sp. strain THL027 (12), G. thermocatenulatus (47), B. subtilis 168 (31), and some gram-negative bacteria (29, 34).
Most of the lipases that are utilized commercially are from Pseudomonas species, presumably because these lipases were the first to be isolated, cloned, and characterized. Their three-dimensional structures were also among the earliest to be revealed. Moreover, most of these lipases have undergone a variety of molecular modifications yielding effective and efficient enzymes with desirable characteristics (17, 22). However, the properties of LipA and LipB should make these enzymes of great interest for specific industrial applications. The described properties at high temperatures and activities at alkaline pHs extend the diversity of lipases and indicate that extremophilic bacteria are a rich source of biotechnologically interesting enzymes, including lipases in particular.
Footnotes
Published ahead of print on 12 October 2007.
REFERENCES
- 1.Arakawa, T., and M. Tokunaga. 2004. Electrostatic and hydrophobic interactions play a major role in the stability and refolding of halophilic proteins. Protein Pept. Lett. 11:125-132. [DOI] [PubMed] [Google Scholar]
- 2.Arpigny, J. L., and K.-E. Jaeger. 1999. Bacterial lipolytic enzymes: classification and properties. Biochemistry 343:177-183. [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. Seidman, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates, New York, NY.
- 4.Balcão, V., M. Paiva, and F. X. Malcata. 1996. Bioreactors with immobilized lipases: state of the art. Enzyme Microb. Technol. 18:392-416. [DOI] [PubMed] [Google Scholar]
- 5.Bell, P. J. L., N. Nevalainen, H. W. Morgan, and P. L. Bergquist. 1999. Rapid cloning of thermoalkalophilic lipases from Bacillus sp. using PCR. Biotechnol. Lett. 21:1003-1006. [Google Scholar]
- 6.Benjwal, S., S. Jayaraman, and O. Gursky. 2005. Electrostatic effects on the stability of discoidal high-density lipoproteins. Biochemistry 44:10218-10226. [DOI] [PubMed] [Google Scholar]
- 7.Bornscheuer, U. T., C. Bessler, R. Srinivas, and S. H. Krishna. 2002. Optimizing lipases and related enzymes for efficient application. Trends Biotechnol. 20:433-437. [DOI] [PubMed] [Google Scholar]
- 8.Brzozowski, A. M., U. Derewenda, Z. S. Derewenda, G. G. Dodson, D. M. Lawson, J. P. Turkenburg, F. Bjorkling, B. Huge-Jensen, S. A. Patkar, and L. Thim. 1991. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature 351:491-494. [DOI] [PubMed] [Google Scholar]
- 9.Cho, A. R., S. K. Yoo, and E. J. Kim. 2000. Cloning, sequencing and expression in Escherichia coli of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol. Lett. 186:235-238. [DOI] [PubMed] [Google Scholar]
- 10.Criswell, A. R., E. Bae, B. Stec, J. Konisky, and G. N. Phillips. 2003. Structures of thermophilic and mesophilic adenylate kinases from the genus Methanococcus. J. Mol. Biol. 330:1087-1099. [DOI] [PubMed] [Google Scholar]
- 11.Dello Russo, A., R. Rullo, G. Nitti, M. Masullo, and V. Bocchini. 1997. Iron superoxide dismutase from the archaeon Sulfolobus solfataricus: average hydrophobicity and amino acid weight are involved in the adaptation of proteins to extreme environments. Biochim. Biophys. Acta 1343:23-30. [DOI] [PubMed] [Google Scholar]
- 12.Dharmsthiti, S., and S. Luchai. 1999. Production, purification and characterization of thermophilic lipase from Bacillus sp. THL027. FEMS Microbiol. Lett. 179:241-246. [DOI] [PubMed] [Google Scholar]
- 13.Dong, H., S. Gao, S. Han, and S. Cao. 1999. Purification and characterization of a Pseudomonas sp. lipase and its properties in non-aqueous media. Biotechnol. Appl. Biochem. 30:251-256. [PubMed] [Google Scholar]
- 14.Ferrato, F., F. Carriere, L. Sarda, and R. Verger. 1997. A critical reevaluation of the phenomenon of interfacial activation. Methods Enzymol. 286:327-347. [DOI] [PubMed] [Google Scholar]
- 15.Freier, D., C. P. Mothershed, and J. Wiegel. 1988. Characterization of Clostridium thermocellum JW-20. Appl. Environ. Microbiol. 54:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gotz, F., H. M. Verheij, and R. Rosenstein. 1998. Staphylococcal lipases: molecular characterization, secretion, and processing. Chem. Phys. Lipids 93:15-25. [DOI] [PubMed] [Google Scholar]
- 17.Gupta, R., N. Gupta, and P. Rathi. 2004. Bacterial lipases: an overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 64:763-781. [DOI] [PubMed] [Google Scholar]
- 18.Haney, P., J. Konisky, K. K. Koretke, Z. Luthey-Schulten, and P. G. Wolynes. 1997. Structural basis for thermostability and identification of potential active site residues for adenylate kinases from the archael genus Methanococcus. Proteins Struct. Funct. Genet. 28:117-130. [DOI] [PubMed] [Google Scholar]
- 19.Hjorth, A., F. Carriere, C. Cudrey, H. Woldike, E. Boel, D. M. Lawson, F. Ferrato, C. Cambillau, G. G. Dodson, L. Thim, and R. Verger. 1993. A structural domain (the lid) found in pancreatic lipases is absent in the guinea pig (phospho) lipase. Biochemistry 32:4702-4707. [DOI] [PubMed] [Google Scholar]
- 20.Hutcheon, G. W., N. Vasisht, and A. Bolhuis. 2005. Characterisation of a highly stable alpha-amylase from the halophilic archaeon Haloarcula hispanica. Extremophiles 9:487-495. [DOI] [PubMed] [Google Scholar]
- 21.Jaeger, K.-E., B. W. Dijkstra, and M. T. Reetz. 1999. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu. Rev. Microbiol. 53:315-351. [DOI] [PubMed] [Google Scholar]
- 22.Jaeger, K. E., and M. T. Reetz. 1998. Microbial lipases form versatile tools for biotechnology. Trends Biotechnol. 16:396-403. [DOI] [PubMed] [Google Scholar]
- 23.Jaenicke, R., and G. Bohm. 1998. The stability of proteins in extreme environments. Curr. Opin. Struct. Biol. 8:738-748. [DOI] [PubMed] [Google Scholar]
- 24.Jeong, S. T., H. K. Kim, S. J. Kim, S. W. Chi, J. G. Pan, T. K. Oh, and S. E. Ryu. 2002. Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase. J. Biol. Chem. 277:17041-17047. [DOI] [PubMed] [Google Scholar]
- 25.Khyami-Horani, H. 1996. Thermotolerant strain of Bacillus licheniformis producing lipase. World J. Microbiol. Biotechnol. 12:399-401. [DOI] [PubMed] [Google Scholar]
- 26.Kim, H. K., S. Y. Park, J. K. Lee, and T. K. Oh. 1998. Gene cloning and characterization of thermostable lipase from Bacillus stearothermophilus L1. Biosci. Biotechnol. Biochem. 62:66-71. [DOI] [PubMed] [Google Scholar]
- 27.Kim, M. H., H. K. Kim, J. K. Lee, S. Y. Park, and T. K. Oh. 2000. Thermostable lipase of Bacillus stearothermophilus: high-level production, purification, and calcium-dependent thermostability. Biosci. Biotechnol. Biochem. 64:280-286. [DOI] [PubMed] [Google Scholar]
- 28.Lang, D., B. Hofmann. L. Haalck. H. J. Hecht. F. Spener, R. D. Schmid, and D. Schomburg. 1996. Crystal structure of a bacterial lipase from Chromobacterium viscosum ATCC 6918 refined at 1.6 Å resolution. J. Mol. Biol. 259:704-717. [DOI] [PubMed] [Google Scholar]
- 29.Lee, P., G. H. Chung, and J. S. Rhee. 1993. Purification and characterization of Pseudomonas fluorescens SIK-W1 lipase expressed in Escherichia coli. Biochim. Biophys. Acta 1169:156-164. [DOI] [PubMed] [Google Scholar]
- 30.Lee, W., Y. S. Koh, K. J. Kim, B. C. Kim, H. J. Choi, D. S. Kim, M. T. Suhartono, and Y. R. Pyun. 1999. Isolation and characterization of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol. Lett. 179:393-400. [DOI] [PubMed] [Google Scholar]
- 31.Lesuisse, E., K. Schanck, and C. Colson. 1993. Purification and preliminary characterization of the extracellular lipase of Bacillus subtilis 168, an extremely basic pH-tolerant enzyme. Eur. J. Biochem. 216:155-160. [DOI] [PubMed] [Google Scholar]
- 32.Macedo-Ribeiro, S., B. Darimont, R. Sterner, and R. Huber. 1996. Small structural changes account for the high thermostability of 1[4Fe-4S] ferredoxin from the hyperthermophilic bacterium Thermotoga maritima. Structure 4:1291-1301. [DOI] [PubMed] [Google Scholar]
- 33.Martinez, C., P. De Geus, M. Lauwereys, G. Matthyssens, and C. Cambillau. 1992. Fusarium solani cutinase is a lipolytic enzyme with a catalytic serine accessible to solvent. Nature 356:615-618. [DOI] [PubMed] [Google Scholar]
- 34.Matsumae, H., and T. Shibatani. 1994. Purification and characterization of lipase from Serratia marcescens Sr41 8000 responsible for asymmetric hydrolysis of 3-phenylglysidic acid esters. J. Ferment. Bioeng. 77:152-158. [Google Scholar]
- 35.Nardini, M., D. A. Lang, K. E. Jaeger, and B. W. Dijkstra. 2000. Crystal structure of Pseudomonas aeruginosa lipase in the open conformation. The prototype for family I.1 of bacterial lipases. J. Biol. Chem. 275:31219-31225. [DOI] [PubMed] [Google Scholar]
- 36.Nawani, N., and J. Kaur. 2000. Purification, characterization and thermostability of lipase from a thermophilic Bacillus sp. J33. Mol. Cell. Biochem. 206:91-96. [DOI] [PubMed] [Google Scholar]
- 37.Noble, M. E., A. Cleasby, L. N. Johnson, M. R. Egmond, and L. G. Frenken. 1993. The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate. FEBS Lett. 331:123-128. [DOI] [PubMed] [Google Scholar]
- 38.Rashid, N., Y. Shimada, S. Ezaki, H. Atomi, and T. Imanaka. 2001. Low-temperature lipase from psychrotrophic Pseudomonas sp. strain KB700A. Appl. Environ. Microbiol. 67:4064-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rathi, P., S. Bradoo, R. K. Saxena, and R. Gupta. 2000. A hyper-thermostable, alkaline lipase from Pseudomonas sp. with the property of thermal activation. Biotechnol. Lett. 22:495-498. [Google Scholar]
- 40.Rosenstein, R., and F. Götz. 2000. Staphylococcal lipases: biochemical and molecular characterization. Biochimie 82:1005-1014. [DOI] [PubMed] [Google Scholar]
- 41.Rua Luisa, M., C. Schmidt-Dannert, S. Wahl, A. Sprauer, and R. D. Schmid. 1997. Thermoalkalophilic lipase of Bacillus thermocatenulatus large-scale production, purification and properties: aggregation behaviour and its effect on activity. J. Biotechnol. 56:89-102. [DOI] [PubMed] [Google Scholar]
- 42.Russell, R. J., J. M. Ferguson, D. W. Hough, M. J. Danson, and G. L. Taylor. 1997. The crystal structure of citrate synthase from the hyperthermophilic archaeon Pyrococcus furiosus at 1.9 A resolution. Biochemistry 36:9983-9994. [DOI] [PubMed] [Google Scholar]
- 43.Salameh, M., and J. Wiegel. 2007. Lipases from extremophiles and potential for industrial applications. Adv. Appl. Microbiol. 61:253-283. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook. J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Sarda, L., and P. Desnuelle. 1958. Action of pancreatic lipase on emulsified esters. Biochim. Biophys. Acta 30:513-521. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt-Dannert, C. 1999. Recombinant microbial lipases for biotechnological applications. Bioorg. Med. Chem. 7:2123-2130. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt-Dannert, C., H. Sztajer, W. Stocklein, U. Menge, and R. D. Schmid. 1994. Screening, purification and properties of a thermophilic lipase from Bacillus thermocatenulatus. Biochim. Biophys. Acta 1214:43-53. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt-Dannert, C., M. L. Rua, H. Atomi, and R. D. Schmid. 1996. Thermoalkalophilic lipase of Bacillus thermocatenulatus. I. Molecular cloning, nucleotide sequence, purification and some properties. Biochim. Biophys. Acta 1301:105-114. [DOI] [PubMed] [Google Scholar]
- 49.Simons, J. W., M. D. van Kampen, I. Ubarretxena-Belandia, R. C. Cox, C. M. Alves dos Santos, M. R. Egmond, and H. M. Verheij. 1999. Identification of a calcium binding site in Staphylococcus hyicus lipase: generation of calcium-independent variants. Biochemistry 38:2-10. [DOI] [PubMed] [Google Scholar]
- 50.Sinchaikul, S., B. Sookkheo, S. Phutrakul, F. M. Pan, and S. T. Chen. 2001. Optimization of a thermostable lipase from Bacillus stearothermophilus P1: overexpression, purification, and characterization. Protein Expr. Purif. 22:388-398. [DOI] [PubMed] [Google Scholar]
- 51.Snellman, E. A., E. R. Sullivan, and R. R. Colwell. 2002. Purification and properties of the extracellular lipase, LipA, of Acinetobacter sp. RAG-1. Eur. J. Biochem. 269:5771-5779. [DOI] [PubMed] [Google Scholar]
- 52.Spencer, D. S., K. Xu, M. T. Logan, and H. X. Zhou. 2005. Effects of pH, salt, and macromolecular crowding on the stability of FK506-binding protein: an integrated experimental and theoretical study. J. Mol. Biol. 351:219-232. [DOI] [PubMed] [Google Scholar]
- 53.Svetlitshnyi, V., F. Rainey, and J. Wiegel. 1996. Thermosyntropha lipolytica gen. nov., sp. nov., a lipolytic, anaerobic, alkalitolerant, thermophilic bacterium utilizing short and long chain fatty acids in syntrophic coculture with a methanogenic archaeum. Int. J. Syst. Bacteriol. 46:1131-1137. [DOI] [PubMed] [Google Scholar]
- 54.Tanner, J. J., R. M. Hecht, and K. L. Krause. 1996. Determinants of enzyme thermostability observed in the molecular structure of Thermus aquaticus d-glyceraldehyde-3-phosphate dehydrogenase at 2.5 Å resolution. Biochemistry 35:2597-2609. [DOI] [PubMed] [Google Scholar]
- 55.Tyndall, J. D., S. Sinchaikul, L. A. Fothergill-Gilmore, P. Taylor, and M. D. Walkinshaw. 2002. Crystal structure of a thermostable lipase from Bacillus stearothermophilus P1. J. Mol. Biol. 323:859-869. [DOI] [PubMed] [Google Scholar]
- 56.Verger, R., and G. H. De Haas. 1976. Interfacial enzyme kinetics of lipolysis. Annu. Rev. Biophys. Bioeng. 5:77-117. [DOI] [PubMed] [Google Scholar]
- 57.Villeneuve, P., J. M. Muderhwa, J. Graille, and M. J. Haas. 2000. Customizing lipases for biocatalysis: a survey of chemical, physical and molecular biological approaches. J. Mol. Catal. B 9:113-148. [Google Scholar]
- 58.Wiegel, J. 1998. Anaerobic alkalithermophiles, a novel group of extremophiles. Extremophiles 2:257-267. [DOI] [PubMed] [Google Scholar]
- 59.Winkler, U. K., and M. Stuckmann. 1979. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J. Bacteriol. 138:663-670. [DOI] [PMC free article] [PubMed] [Google Scholar]