Abstract
Two transposon-insertional mutants of Listeria monocytogenes showing smaller viable surface-attached cell populations after disinfection with N,N-didecyl-N,N-dimethylammonium chloride were identified. In both mutants, transposon Tn917-lac was found to be inserted into the same gene, lmo1462, which is homologous to the essential Escherichia coli era gene. Both L. monocytogenes lmo1462-disrupted mutants displayed lower growth rates, as was also shown for several E. coli era mutants, and the lmo1462 gene was able to complement the growth defect of an E. coli era mutant. We showed that the disruption of lmo1462 decreased the ability of L. monocytogenes cells to adhere to stainless steel. Our results suggest that this era-like gene is involved in adhesion and contributes to the presence of L. monocytogenes on surfaces.
The food production environment has been widely recognized as a possible source of food contamination by Listeria monocytogenes (18). Indeed, this pathogen is frequently found after cleaning and disinfecting procedures (4, 5). It has been shown that strains from lineage II (serotypes 1/2a and 1/2c) adhere to an inert surface to a greater extent than lineage I strains (serotypes 4b and 1/2b) but only after growth in a diluted growth medium better mimicking natural conditions (10). This difference between lineages was also observed by Borucki et al. (2) when Listeria was grown in modified Welshimer's broth. In addition, strains from genetic lineage II are more frequently found in food-processing environments than strains from lineage I (13, 19, 28). Thus, adhesion potential may be a contributing factor leading to the presence of L. monocytogenes in food-processing plants.
It is likely that increased biocide resistance of biofilms (this term designates the bacterial communities formed after growth of adherent cells) also contributes to the presence of L. monocytogenes in food-processing facilities. Multiple mechanisms of biofilm resistance have been proposed (1, 6, 9, 20, 22). With most of the growth media used, such as brain heart infusion or tryptone soy broth, the growth of L. monocytogenes on surfaces leads to communities distributed as single cells (16, 17). As observed for thick and dense biofilms, these adherent cells are more resistant to antimicrobials than their planktonic counterparts (8, 11), indicating that mechanisms other than poor antimicrobial penetration or limitation of metabolic substrates are involved in the decreased susceptibility of adherent cells to biocides.
The presence of L. monocytogenes after hygiene operations may be because surface-attached cell populations are highly adherent and/or highly resistant to disinfectant. In order to investigate any genetic basis for the persistence of adherent L. monocytogenes after treatment with disinfectant, a bank of transposon-insertional mutants was screened.
Selection of L. monocytogenes mutants with small viable surface-attached cell populations after disinfection.
A total of 3,367 random Tn917-lac (7) insertional mutants of L. monocytogenes EGD (serotype 1/2a) were each inoculated into 100 μl TSBYE (tryptone soy broth enriched with 0.6% yeast extract; Difco laboratories) supplemented with 5 μg ml−1 erythromycin in the wells of sterile 96-well polystyrene microtiter plates (Nunc). Each plate included three wells inoculated with the L. monocytogenes parental strain and two uninoculated wells as controls. Biofilms were allowed to form at 37°C for 24 h and then at 3°C for a subsequent 6-day period. Mutants with smaller amounts of surviving cells after exposure to biocide were selected by using a method similar to that described by Gilbert et al. (12). Briefly, the planktonic contents were carefully removed from the wells, which were then rinsed with 200 μl sterile distilled water and filled with 100 μl Bardac-22 (N,N-didecyl-N,N-dimethylammonium chloride; Lonza, Levallois Perret, France) at 0.01% (vol/vol). Following incubation for 30 min at room temperature, the disinfectant was replaced by 120 μl neutralizing Tween 80 (30 g liter−1) solution, and after 5 min, the wells were gently washed with 150 μl TSBYE and finally filled with 150 μl TSBYE. Since the number of surviving cells correlates with the time taken to grow after disinfection (12), the plates were incubated at 37°C and the optical density at 600 nm (OD600) was measured after a 9-h incubation by using a microtiter plate reader (Fluostar; BMG LabTechnologies, Champigny sur Marne, France). A total of 266 candidate mutants showing low OD600 values (<70% of that of the wild type) in at least two out of three independent experiments were selected and transferred into new microtiter plates. These were retested as described above except that a higher Bardac-22 concentration (0.03%, vol/vol) was used and the OD600 was measured after 24 h of regrowth. Only two mutants, designated 3A6 and 3A10, which displayed an OD600 value of half that of the wild-type strain, were identified after this second screen.
Identification of the Tn917-lac insertion target in mutants 3A6 and 3A10.
Southern hybridization confirmed that each mutant carried a single copy of the transposon (data not shown), and sequencing of the DNA flanking the transposon insertion site revealed that Tn917-lac was inserted into the same gene, lmo1462, between nucleotide (nt) 683 and nt 684 in 3A6 and between nt 817 and nt 818 in 3A10. A predicted Rho-independent transcription terminator was located 79 bp downstream from lmo1462, which is the last member of a six-gene operon. The transposon insertion resulted in the deletion of the C-terminal 74 and 28 amino acid (aa) residues of the 301-residue-long lmo1462-encoded product in 3A6 and 3A10, respectively. However, the possibility that these truncated versions of the Lmo1462 protein may retain partial activity in the mutants cannot be excluded. Sequence analysis showed that Lmo1462 is 40% identical and 63% similar across its entire sequence (region of homology: aa 8 to 296) to the Escherichia coli Era protein, a GTPase with RNA-binding activity that has been implicated in a wide array of cellular functions, including DNA replication, protein translation, metabolism, and cell cycle regulation (3, 25, 26). BlastP analysis also indicated that Lmo1462 displayed high homology (66% identity and 81% similarity) to Bex, the Bacillus subtilis Era homolog. Interestingly, bex was identified as a gene whose expression is induced when cells are within a biofilm as opposed to a planktonic state (27).
Complementation of an E. coli era mutant by L. monocytogenes lmo1462.
In order to determine whether lmo1462 from L. monocytogenes was the functional homolog of the E. coli era gene, we tested its ability to complement the E. coli era mutant HT120, for which era expression is dependent on induction by tetracycline because of the rnc40::ΔTn10 mutation (3). For complementation experiments, an lmo1462-containing plasmid, designated pTV1462, was constructed. Briefly, a DNA fragment containing the chromosomal lmo1462 gene, as well as 162 nt upstream of the predicted translational start codon and 102 nt downstream of the translational stop codon, was amplified and ligated to an origin of replication [repA(Ts)] and a kanamycin resistance gene (aphA3) from pTV32-OK (7), creating pTV1462. To construct the control vector pTV, a fragment containing the repA(Ts) and aphA3 genetic elements was circularized by self-ligation. The HT120 strain was then transformed with pTV or pTV1462, and growth was examined at 25°C on LB agar plates in the absence of tetracycline. HT120 containing pTV formed microcolonies that were visible only after 6 days, whereas HT120 containing pTV1462 formed colonies after 1 day, as did a wild-type strain (data not shown). The ability of lmo1462 to complement the growth defect of the E. coli era mutant thus indicated that it is a functional homolog of E. coli era.
Adhesion of mutants 3A6 and 3A10 to inert surfaces.
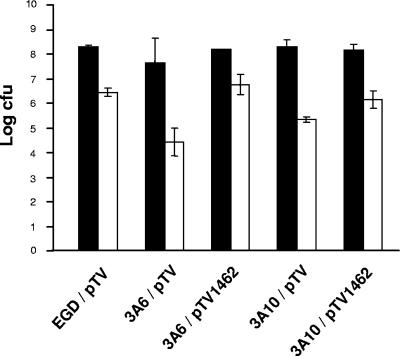
The screening of the bank of transposon mutants exposed to disinfectant did not allow us to determine whether the small residual cell populations observed for the mutants 3A6 and 3A10 were due to either reduced adhesion or increased susceptibility to Bardac-22 (to both). In order to investigate the adhesion ability of the mutants, populations of adherent and nonadherent cells cultured on stainless steel slides were assessed using the selected mutant strains and compared to the wild type. Biofilms were prepared as follows. Bacterial cells taken from tryptone soy agar (TSA) slopes were washed twice in 9 ml of physiological saline, and concentrations of the suspensions were adjusted with TSBYE to 5 × 107 to 5 × 108 CFU ml−1 (OD600 of 0.15 in 1.5-cm-diameter tubes). One-hundred-microliter inocula were deposited on clean, sterile, stainless steel slides (25 by 40 mm) in a 50-cm-diameter petri dish which was placed in a 120-cm-diameter petri dish containing 25 ml of water. The slides were incubated at 30°C for 24 h and then at 3°C for 6 days for biofilm formation (colonized surface area of approximately 1 cm2). Nonadherent (planktonic) cells were collected in 10-ml filter-sterilized ultrapure water poured on the slides, and adherent bacteria were detached from the slides by swabbing as described previously (23). The counts of planktonic and adherent cells were assessed on TSA incubated at 30°C for 24 h. The sum of surface-attached and planktonic populations was referred to as the “total population.” No significant differences (P = 0.29, as determined by variance analysis) were observed in the total populations of the different strains tested (Fig. 1). However, significantly smaller adherent populations were observed for mutants 3A6 (5 × 104 CFU) and 3A10 (2 × 105 CFU) than for the wild type (5 × 106 CFU) (P < 0.0001, as determined by variance analysis), suggesting that adhesion was less efficient in the absence of an intact era gene. The mutants were then complemented with lmo1462 carried by plasmid pTV1462. A wild-type copy of the era-like gene was able to restore adhesion, since the complemented mutants (3A6/pTV1462 and 3A10/pTV1462) had adherent populations significantly larger than those of the control noncomplemented strains (Fig. 1) (P < 0.0001, as determined by variance analysis). In order to investigate whether mutants 3A6 and 3A10 had a higher susceptibility to biocide than the parental strain, these two mutants and the wild-type strain grown either as planktonic cells or as biofilms on stainless steel slides were subjected to Bardac-22 and the CFU that survived this treatment were counted. The analysis of the survival curves obtained did not reveal any significant differences in susceptibility to the disinfectant between the mutant and the wild-type strains (data not shown). As the survival curves obtained from microtiter plates could not be interpreted because of poor reproducibility (data not shown), the hypothesis of a decreased susceptibility of mutants when attached to microtiter plates cannot be excluded. Since the adhesion phenotype of the mutants was clear and considered an interesting result on its own, the susceptibility to Bardac-22 was not further investigated in the present study.
FIG. 1.
Total (solid bars) and adherent (open bars) cell populations of the wild-type strain (EGD/pTV), mutant strains (3A6/pTV and 3A10/pTV), and the lmo1462-complemented mutant strains (3A6/pTV1462 and 3A10/pTV1462). One-hundred-microliter inocula containing 5 × 107 CFU of each strain were deposited on clean, sterile, stainless steel slides which were placed at 30°C for 24 h and then at 3°C for 6 days for biofilm formation. Nonadherent (planktonic) and adherent bacterial cells were counted. The sum of adherent and planktonic CFU is referred to as the “total population.” Bars indicate the grand means of mean values obtained from one triplicate and two single independent experiments (n = 3) in the case of 3A6, from three triplicate independent experiments (n = 3) in the case of 3A10, and from four independent triplicate experiments (n = 4) in the case of the wild-type strain. Error bars represent standard deviations (no error bar is shown for 3A6/pTV1462 total population as all values were identical).
Microscopic observations of adherent mutant cells.
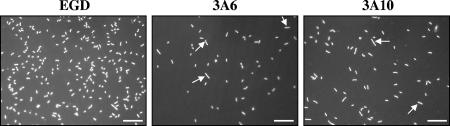
In an attempt to analyze the impact of lmo1462 mutation on adherent-cell morphology, mutant cells attached to polyurethane slides were examined by epifluorescence microscopy. Slides were prepared and incubated as described for the adhesion experiments. Adherent cells were stained with 5 μg ml−1 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) solution for 15 min in darkness, rinsed with 25 ml of peptone solution, and immediately examined in a wet state under an epifluorescence microscope (magnification, ×100) (Axioskop; Zeiss, Le Pecq, France) connected to a charge-coupled device camera (Jaim 50; Adersa, Palaiseau, France) and a digital image acquisition system. A variation in cell length was observed for both mutants (Fig. 2). Fifteen percent of the mutant cells were longer than 2 μm, compared to only 2% for the wild type (175 cells were tested for each strain, with a minimum of five fields examined per strain). It is not likely that a larger amount of elongated cells could be the cause of reduced attachment. Indeed, cells having a larger area in contact with the substratum adhere more strongly, as shown by Gomez-Suarez et al. (15), who compared spherical and rod-shaped microorganisms. Furthermore, when comparing only the numbers of normal-sized attached cells (85% for the mutants and 98% for the wild type), the large differences observed in Fig. 1 between the two mutants and the wild type are nearly unchanged (e.g., log 2.00 versus log 2.06 in the case of 3A6). Microscopic observations of the mutant cells in a planktonic state also revealed larger amounts of elongated cells than for the wild type (data not shown), as was also observed for several E. coli era conditional mutants (14, 21). This result suggested the involvement of lmo1462, either directly or indirectly, in cell division.
FIG. 2.
Morphologies of surface-attached wild-type and lmo1462 mutant cells. Cells of wild-type strain EGD and mutants 3A6 and 3A10 grown on polyurethane slides at 37°C for 24 h and incubated at 3°C for 6 days were stained with DAPI and visualized under an epifluorescence microscope (magnification, ×100). Arrows indicate some of the cells that have an increased size. Scale bars, 10 μm.
Growth characteristics of mutants 3A6 and 3A10.
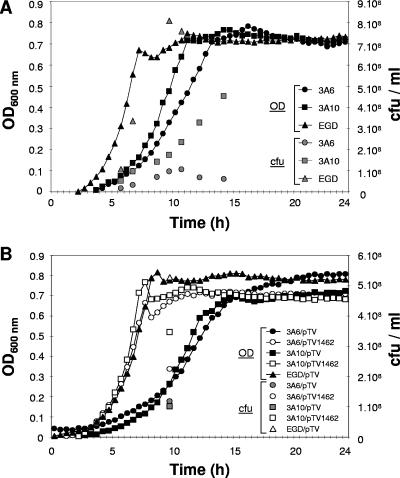
To investigate the physiological effect of the lmo1462 disruption, the growth rates of the mutants and the wild-type strain were determined. Growth was monitored at 30°C for 30 h in 96-well sterile microplates using a microplate reader (Fluostar), each well containing 100-μl TSBYE supplemented with kanamycin when appropriate and an initial bacterial concentration of 107 CFU ml−1. The growth at 30°C was also monitored by CFU counts on TSA plates incubated for 24 h at 30°C. The growth of mutants 3A6 and 3A10 was slower than that of the wild type, as determined both by OD measurements and CFU counts (Fig. 3A). Similarly, mutant strains containing control vector pTV grew more slowly than those containing plasmid pTV1462 (which carried a copy of the lmo1462 gene) or the wild type (Fig. 3B). The growth rates were calculated from OD values obtained from three independent experiments by using Microfit software (version 1.0; Institute of Food Research, Norwich, United Kingdom). The growth rate differences between strains that contained a wild-type copy of the era gene (i.e., the wild-type and complemented mutants) and strains that did not (the noncomplemented mutants) were highly significant. Mutants 3A6 and 3A10 displayed mean growth rates of 0.95 h−1 and 0.89 h−1, respectively, which were both significantly different from that of the wild type (1.49 h−1) (P = 0.0027, as determined by variance analyses; n = 3). Bacterial strains 3A6/pTV1462 and 3A10/pTV1462 had significantly higher mean growth rates (1.41 h−1 and 1.89 h−1, respectively) than 3A6/pTV and 3A10/pTV (0.72 h−1 and 0.81 h−1, respectively) (P = 0.0047 and P = 0.0014, respectively, as determined by mean comparison; n = 3), indicating that the lmo1462-containing plasmid pTV1462 complemented the growth defect of the mutants. These results showed that disruption of lmo1462 was detrimental but not lethal to the cell, as also reported for bex null mutation in B. subtilis (24) and era conditional mutations in E. coli (3).
FIG. 3.
Effect of disruption of lmo1462 on growth of L. monocytogenes at 30°C in TSBYE. The growth of the wild-type EGD strain and mutants 3A6 and 3A10 (A) and the growth of the mutants containing either plasmid pTV1462 (carrying the lmo1462 gene) or control vector pTV (B) were measured by monitoring the OD600 and determining the number of CFU at various time points. Data are presented for one representative experiment out of three independent experiments. For more clarity, only the CFU data corresponding to the enumeration performed at 9 h 30 min are presented in panel B.
Concluding remarks.
In this study, we identified two lmo1462-disrupted mutants that displayed a decreased number of surface-attached cells on stainless steel compared to the number of surface-attached wild-type cells, indicating that their capacity to adhere to surfaces was reduced. These results suggest that the L. monocytogenes era-like gene lmo1462 is involved in adhesion. To our knowledge, this is the first report of an L. monocytogenes gene whose mutation decreased the adhesion of this pathogen to inert surfaces.
Acknowledgments
We thank A. Brisabois (AFSSA-LERQAP, Maisons-Alfort, France) for providing L. monocytogenes serovar 1/2a strain EGD, D. L. Court (NCI/FCRDC, Frederick, MD) for providing E. coli HT120 containing the rnc40::ΔTn10 mutation, and A. Bleiweis (University of Florida, Gainesville, FL) for sending plasmid pTV32-OK carrying transposon Tn917-lac. Special thanks to J. Verran for English correction and A.-M. Leconte for help with organization.
Footnotes
Published ahead of print on 5 October 2007.
REFERENCES
- 1.Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. [DOI] [PubMed] [Google Scholar]
- 2.Borucki, M. K., J. D. Peppin, D. White, F. Loge, and D. R. Call. 2003. Variation in biofilm formation among strains of Listeria monocytogenes. Appl. Environ. Microbiol. 69:7336-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Britton, R. A., B. S. Powell, S. Dasgupta, Q. Sun, W. Margolin, J. R. Lupski, and D. L. Court. 1998. Cell cycle arrest in Era GTPase mutants: a potential growth rate-regulated checkpoint in Escherichia coli. Mol. Microbiol. 27:739-750. [DOI] [PubMed] [Google Scholar]
- 4.Chasseignaux, E., P. Gerault, M. T. Toquin, G. Salvat, P. Colin, and G. Ermel. 2002. Ecology of Listeria monocytogenes in the environment of raw poultry meat and raw pork meat processing plants. FEMS Microbiol. Lett. 210:271-275. [DOI] [PubMed] [Google Scholar]
- 5.Chasseignaux, E., M. T. Toquin, C. Ragimbeau, G. Salvat, P. Colin, and G. Ermel. 2001. Molecular epidemiology of Listeria monocytogenes isolates collected from the environment, raw meat and raw products in two poultry- and pork-processing plants. J. Appl. Microbiol. 91:888-899. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Cvitkovitch, D. G., J. A. Gutierrez, J. Behari, P. J. Youngman, J. E. Wetz, P. J. Crowley, J. D. Hillman, L. J. Brady, and A. S. Bleiweis. 2000. Tn917-lac mutagenesis of Streptococcus mutans to identify environmentally regulated genes. FEMS Microbiol. Lett. 182:149-154. [DOI] [PubMed] [Google Scholar]
- 8.Das, J. R., M. Bhakoo, M. V. Jones, and P. Gilbert. 1998. Changes in the biocide susceptibility of Staphylococcus epidermidis and Escherichia coli cells associated with rapid attachment to plastic surfaces. J. Appl. Microbiol. 84:852-858. [DOI] [PubMed] [Google Scholar]
- 9.Davies, D. 2003. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2:114-122. [DOI] [PubMed] [Google Scholar]
- 10.Folsom, J. P., G. R. Siragusa, and J. F. Frank. 2006. Formation of biofilm at different nutrient levels by various genotypes of Listeria monocytogenes. J. Food Prot. 69:826-834. [DOI] [PubMed] [Google Scholar]
- 11.Frank, J. F., and R. Koffi. 1990. Surface-adherent growth of Listeria monocytogenes is associated with increased resistance to surfactant sanitizers and heat. J. Food Prot. 53:550-554. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert, P., J. R. Das, M. V. Jones, and D. G. Allison. 2001. Assessment of resistance towards biocides following the attachment of micro-organisms to, and growth on, surfaces. J. Appl. Microbiol. 91:248-254. [DOI] [PubMed] [Google Scholar]
- 13.Giovannacci, I., C. Ragimbeau, S. Queguiner, G. Salvat, J. L. Vendeuvre, V. Carlier, and G. Ermel. 1999. Listeria monocytogenes in pork slaughtering and cutting plants. Use of RAPD, PFGE and PCR-REA for tracing and molecular epidemiology. Int. J. Food Microbiol. 53:127-140. [DOI] [PubMed] [Google Scholar]
- 14.Gollop, N., and P. E. March. 1991. A GTP-binding protein (Era) has an essential role in growth rate and cell cycle control in Escherichia coli. J. Bacteriol. 173:2265-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez-Suarez, C., H. J. Busscher, and H. C. van der Mei. 2001. Analysis of bacterial detachment from substratum surfaces by the passage of air-liquid interfaces. Appl. Environ. Microbiol. 67:2531-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herald, J., and A. Zottola. 1988. Attachment of Listeria monocytogenes to stainless steel surfaces at various temperatures and pH values. J. Food Sci. 53:1549-1552. [Google Scholar]
- 17.Kalmokoff, M. L., J. W. Austin, X. D. Wan, G. Sanders, S. Banerjee, and J. M. Farber. 2001. Adsorption, attachment and biofilm formation among isolates of Listeria monocytogenes using model conditions. J. Appl. Microbiol. 91:725-734. [DOI] [PubMed] [Google Scholar]
- 18.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 19.Kerouanton, A. 1999. Etude de la variabilité génétique des souches de Listeria monocytogenes en fonction de leur origine dans différentes filieres de l'industrie agroalimentaire. Ph.D. thesis. Université de Bretagne Occidentale, Brest, France.
- 20.Lewis, K. 2005. Persister cells and the riddle of biofilm survival. Biochemistry (Moscow) 70:267-274. [DOI] [PubMed] [Google Scholar]
- 21.Lu, Q., and M. Inouye. 1998. The gene for 16S rRNA methyltransferase (ksgA) functions as a multicopy suppressor for a cold-sensitive mutant of era, an essential RAS-like GTP-binding protein in Escherichia coli. J. Bacteriol. 180:5243-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 23.Midelet, G., and B. Carpentier. 2002. Transfer of microorganisms, including Listeria monocytogenes, from various materials to beef. Appl. Environ. Microbiol. 68:4015-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minkovsky, N., A. Zarimani, V. K. Chary, B. H. Johnstone, B. S. Powell, P. D. Torrance, D. L. Court, R. W. Simons, and P. J. Piggot. 2002. Bex, the Bacillus subtilis homolog of the essential Escherichia coli GTPase Era, is required for normal cell division and spore formation. J. Bacteriol. 184:6389-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pillutla, R. C., J. Ahnn, and M. Inouye. 1996. Deletion of the putative effector region of Era, an essential GTP-binding protein in Escherichia coli, causes a dominant-negative phenotype. FEMS Microbiol. Lett. 143:47-55. [DOI] [PubMed] [Google Scholar]
- 26.Sayed, A., S. Matsuyama, and M. Inouye. 1999. Era, an essential Escherichia coli small G-protein, binds to the 30S ribosomal subunit. Biochem. Biophys. Res. Commun. 264:51-54. [DOI] [PubMed] [Google Scholar]
- 27.Stanley, N. R., R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 185:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thevenot, D., M. L. Delignette-Muller, S. Christieans, and C. Vernozy-Rozand. 2005. Prevalence of Listeria monocytogenes in 13 dried sausage processing plants and their products. Int. J. Food Microbiol. 102:85-94. [DOI] [PubMed] [Google Scholar]