Abstract
The growth characteristics of five bacteria, Brevibacterium aurantiacum 1-16-58, Corynebacterium casei DPC 5298T, Corynebacterium variabile DPC 5310, Microbacterium gubbeenense DPC 5286T, and Staphylococcus saprophyticus 4E61, all of which were isolated from the surface of smear cheese, were studied in complex and chemically defined media. All of the coryneforms, except M. gubbeenense, grew in 12% salt, while B. aurantiacum and S. saprophyticus grew in 15% salt. All five bacteria assimilated lactate in a semisynthetic medium, and none of the coryneform bacteria assimilated lactose. Glucose assimilation was poor, except by S. saprophyticus and C. casei. Five to seven amino acids were assimilated by the coryneforms and 12 by S. saprophyticus. Glutamate, phenylalanine, and proline were utilized by all five bacteria, whereas utilization of serine, threonine, aspartate, histidine, alanine, arginine, leucine, isoleucine, and glycine depended on the organism. Growth of C. casei restarted after addition of glutamate, proline, serine, and lactate at the end of the exponential phase, indicating that these amino acids and lactate can be used as energy sources. Pantothenic acid was essential for the growth of C. casei and M. gubbeenense. Omission of biotin reduced the growth of B. aurantiacum, C. casei, and M. gubbeenense. All of the bacteria contained lactate dehydrogenase activity (with both pyruvate and lactate as substrates) and glutamate pyruvate transaminase activity but not urease activity.
Bacterial surface-ripened cheeses are characterized by the development of a viscous, red-orange smear on the surface composed of yeasts, mainly Debaryomyces hansenii and Geotrichum candidum, and gram-positive bacteria such as coryneforms and staphylococci (10). Because of its tolerance to high salt concentrations and low pH, D. hansenii develops first on the cheese, metabolizing lactate to CO2 and H2O and forming alkaline metabolites, such as ammonia, that lead to deacidification of the cheese surface, enabling the growth of the salt-tolerant but less-acid-tolerant bacteria.
Until recently, Brevibacterium linens was considered to be the major component of the surface microflora, and most of the research on smear cheese bacteria has focused on physiological and growth characteristics either in liquid cultures (13, 14, 31, 32), on experimental cheeses in mixed cultures with D. hansenii (25-27), or on cheese agar (30). During the last few years, the microflora of several smear cheeses has been extensively investigated and categorically identified using combinations of phenotypic and genotypic techniques (9, 15, 29, 33, 42). These studies have resulted in the identification of several new species of bacteria, including Arthrobacter arilaitensis and Arthrobacter bergerei (21), Agrococcus casei (6), Corynebacterium casei (7), Corynebacterium mooreparkense (8) (subsequently shown to be Corynebacterium variabile [18]), Microbacterium gubbeenense (8), Staphylococcus equorum subsp. linens (36), Staphylococcus succinus subsp. casei (35), and species not previously identified on smear cheeses such as Brevibacterium aurantiacum (38), Staphylococcus sciuri (5), and Staphylococcus saprophyticus (22, 33). Recently, S. saprophyticus has been shown to dominate the surface of Gubbeen cheese early in ripening but is then replaced by corynebacteria, particularly C. casei (38).
Strains of B. linens and D. hansenii from commercial laboratories are generally inoculated onto the cheese surface early in ripening, but their subsequent recovery during ripening is very low (39) or absent (34), indicating that an adventitious flora develops on smear cheeses. The factors that allow this microflora to develop have not been studied to any great extent. Early work by Purko et al. (37) showed that pantothenic acid production by yeasts stimulated the growth of B. linens. During the early days of cheese ripening, lactose is rapidly transformed into lactate by the large numbers of lactic acid bacteria present in the cheese. The yeasts metabolize lactate, and it is likely that lactate dehydrogenase is the first step in the pathway. As cheese ripening progresses, other C sources such as amino acids from protein hydrolysis appear. Whether lactate and amino acids are used by the smear bacteria as C sources has not been determined.
Metabolism of amino acids is one of the key determinants of flavor formation in cheese. The first step in their metabolism is a transaminase reaction in which the amino group of the amino acid is transferred to an α-ketoacid, resulting in the formation of the corresponding amino and keto acids (reviewed in references 41 and 44). α-Ketoglutarate (αKG) appears to be the important amino group acceptor, since addition of exogenous αKG to cheese increased the rate of flavor formation. It is produced from glutamate by either glutamate amino acid transferase or glutamate dehydrogenase (GDH) and appears to be a rate-limiting reaction in cheese flavor development.
The aim of the present study was to investigate some of the growth characteristics of B. aurantiacum, C. variabile, S. saprophyticus, and two recently described new species, C. casei and M. gubbeenense (7, 8), isolated from smear cheese. Particular attention was given to their ability to metabolize lactose, lactate, and amino acids and to the likely enzymes involved. Nothing is known about the vitamin requirements of these organisms, and this aspect was also studied.
MATERIALS AND METHODS
Strains.
The bacteria used were Corynebacterium casei DPC 5298T, Corynebacterium variabile DPC 5310, Microbacterium gubbeenense DPC 5286T, Brevibacterium aurantiacum 1-16-58, and Staphylococcus saprophyticus 4E61, from the culture collection of the Moorepark Food Research Centre, Moorepark, Fermoy, Ireland. They were originally isolated from various batches of Gubbeen cheese (7, 33). The bacteria were precultured in Trypticase soy broth (TSB; Difco). Before inoculation, a sufficient volume of the TSB preculture was centrifuged at 20,000 × g for 5 min, washed twice with sterile water, and resuspended in sterile water to obtain a suspension with an optical density at 600 nm (OD600) of 1. Two milliliters of this suspension was used to inoculate 100 ml of the relevant medium in a 300-ml flask. During growth, samples were taken periodically (2- to 4-h intervals) for OD measurements. Supernatants after centrifugation at 20,000 × g for 5 min were frozen at −20°C until analyzed. All cultures were grown at 200 rpm at 30°C for 24 to 90 h, depending on the organism and the growth medium used. All experiments were done in duplicate, and comparable results were found for each replicate.
Effect of NaCl.
The effect of NaCl (0 to 15%) on the growth of the different bacteria was tested in YT medium, which consisted of (per liter), yeast extract (Difco), 5 g; tryptone (Difco), 5 g; 365 ml of 0.2 M Na2HPO4·H2O and 130 ml of 0.2 M NaH2PO4, and the required amount of salt. The pH was 6.5, and the medium was autoclaved at 121°C for 15 min.
Sugar, lactate, and amino acid assimilation.
A semisynthetic medium (SSM) modified from Katsumata et al. (23) and Liebl et al. (28) was used. It contained the following (per liter): vitamin assay Casamino Acids (Difco), 5 g; (NH4)2SO4, 10 g; urea, 3 g; K2HPO4·3H2O, 1.13 g; MgSO4·7H2O, 0.4 g; FeSO4·7H2O, 2 mg; MnSO4·4H2O, 2 mg; NaCl, 50 g; protocatechuic acid, 15.4 mg; thiamine, 2 mg; biotin, 0.5 mg; pyridoxine, 0.6 mg; pyridoxal, 0.6 mg; pyridoxamine, 0.6 mg; nicotinic acid, 0.65 mg; calcium pantothenate, 1.2 mg; folic acid, 0.5 mg; and riboflavin, 0.5 mg. Glucose, lactose, or sodium lactate was added to the medium as appropriate at a concentration of 10 g/liter. The pH of each medium was adjusted to 6.5 by the addition of 1 M HCl prior to filter sterilization (0.22 μm; PES membrane [Nalgene]).
Utilization of glutamate, proline, serine, or lactate as energy sources.
Further investigations were conducted on C. casei DPC 5298 and M. gubbeenense DPC 5286 to determine whether these bacteria could use glutamate, proline, serine, or lactate as energy sources. The strains were grown in SSM modified by reducing the Casamino Acids to 1.25 g/liter. When the stationary phase was reached, 1 ml of concentrated solutions of sodium glutamate, proline, or serine at 1 g/liter or sodium lactate at 10 g/liter was added and growth was subsequently monitored at 600 nm.
Growth of C. casei in MM.
A minimal medium (MM) for the growth of C. casei was subsequently designed and contained the same vitamins and minerals as SSM, but ammonium sulfate, urea, and the Casamino Acids were replaced with glutamate, proline, serine, glycine, histidine monohydrochloride, alanine, and phenylalanine (each at 200 mg/liter). Lactate (10 g/liter [MM plus lactate]), was added as appropriate. The pH of each medium was adjusted to 6.5 by the addition of 1 M HCl prior to filter sterilization (0.22 μm; PES membrane [Nalgene]).
Vitamin requirements.
To identify the vitamin requirements, the strains were grown in SSM modified by omission of each of the vitamins in turn. The inocula for these were grown in SSM supplemented with 10 g/liter sodium lactate. After centrifugation (15,000 × g for 5 min), the cells were washed twice in sterile water, resuspended, and added to the medium from which one of the vitamins had been omitted. The inoculum used was standardized to an OD600 of 1, and 0.05 ml of this suspension was added to 5 ml of fresh medium in a 30-ml sterile container (Bibby Sterilin) and incubated for 72 h at 30°C at 200 rpm. After this, the OD600 of the subcultures was compared with the OD600 of a positive control without vitamin omissions by Student's t test. These experiments were performed in triplicate.
Enzyme assays.
Cells, grown in 250-ml volumes of filter-sterilized (0.22 μm) SSM in 1-liter Erlenmeyer flasks at 30°C at 200 rpm, were centrifuged for 15 min at 8,000 rpm at 4°C. The pellets were washed once with 0.85% NaCl and centrifuged for 15 min at 8,000 rpm at 4°C. After decanting the supernatant, the pellets were resuspended in 5 to 10 ml of 50 mM phosphate buffer (pH 6.5), transferred to a 10-ml beaker, and kept on ice. Cells of C. casei, C. variabile, and M. gubbeenense were sonicated (Soniprep 150; MSE, London, United Kingdom) at 4°C for 5 min at 1-min intervals with cooling between each sonication to 4°C for 1 min. Cells of B. aurantiacum and S. saprophyticus were broken open in a bead breaker (Biospec Products, Bartlettsville, OK) for 4 and 8 min, respectively, at 2-min intervals with cooling to 4°C for 2 min between each beating. Cell debris was removed by centrifugation at 10,000 × g for 4 min at 4°C. LDH was measured using both pyruvate and lactate as substrates by the procedures of Bergmeyer and Bernt (1, 2). Glutamate dehydrogenase (GDH), glutamate-pyruvate transaminase (GPT), and urease were measured by the procedures of Schmidt (40), Bergmeyer and Bernt (3), and Schlegel and Kaltwasser (39), respectively. All assays were carried out at least in triplicate at 25°C, except that of LDH with lactate as substrate, which was conducted at 30°C.
Analyses.
Growth was monitored by measurement of the OD600, diluting cultures with an OD of >0.5 with uninoculated medium to maintain the linearity of biomass with OD. Growth rates were calculated from linear regression of the slopes of the linear portion of the growth curves relating log OD to time and multiplying by 2.303 to convert to natural logarithms. At least five points were used in each calculation. The pH was also monitored during growth. Glucose, l-lactic acid, and lactose concentrations were determined using enzymatic kits (Sigma for glucose and Boehringer-Mannheim for l-lactate and lactose) following the instructions of the manufacturer. Protein was determined using the Bradford method (Sigma) with bovine serum albumin as the standard. For amino acid determination, samples were deproteinized by mixing equal volumes of 24% (wt/vol) trichloroacetic acid and sample. These were allowed to stand for 10 min before centrifugation at 14,400 × g (Microcentaur; MSE, London, United Kingdom) for 10 min. Supernatants were removed and diluted with 0.2 M sodium citrate buffer (pH 2.2) to give approximately 250 nmol/ml of each amino acid residue. Samples were then diluted 1 in 2 with the internal standard, norleucine, to give a final concentration of 125 mM. Amino acids were quantified using a JEOL JLC-500/V amino acid analyzer (JEOL UK Ltd., Garden City, Herts., United Kingdom) fitted with a JEOL Na+ high-performance cation-exchange column.
RESULTS
Effect of salt.
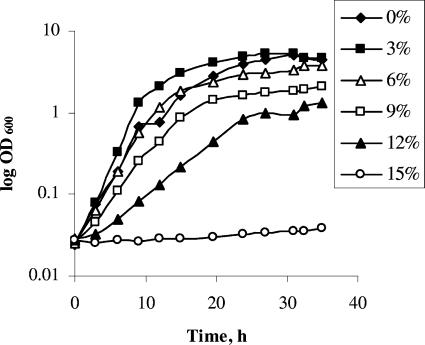
The effect of salt on the growth of C. casei 5298 is shown in Fig. 1. Growth occurred in up to 12% salt but was fastest in 3% salt. Similar growth rates occurred in 0 and 6% salt, while growth and the final cell density were much lower in 9% and, especially, 12% salt, and no growth occurred in 15% salt. Similar results were found for the other three coryneforms, except that growth of B. aurantiacum occurred in 15% salt and M. gubbeenense did not grow in 9% salt (data not shown). Growth of S. saprophyticus occurred at all salt levels but was best in 0% salt (data not shown).
FIG. 1.
Effect of different levels of salt on the growth of C. casei DPC 5298T in YT medium at 30°C and 200 rpm.
Sugar and lactate assimilation.
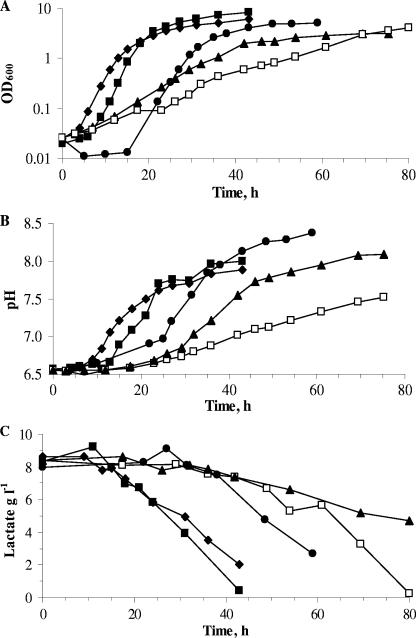
The five strains were able to grow in SSM in the absence of glucose, lactose, or lactate but generally grew faster in the presence of glucose or lactate (Table 1). B. aurantiacum and M. gubbeenense had the slowest growth rates, while C. variabile, C. casei, and S. saprophyticus grew quite rapidly. Lactate was used by all five species (Fig. 2) and did not limit growth because it was not depleted at the end of the exponential phase of growth. This suggested that other substrates were limiting. The pH increased during growth from 6.5 to 7.5 to 8.4 (Fig. 2). Lactose was only used by S. saprophyticus, while glucose was used rapidly by S. saprophyticus and slowly by all of the corynebacteria, except B. aurantiacum, which was unable to use it (data not shown).
TABLE 1.
Growth rates of several bacteria isolated from the surface of smear cheese in SSM and SSM supplemented with glucose, lactose, or lactate
| Organism | Growth rate (h−1) ona:
|
|||
|---|---|---|---|---|
| SSM | SSMG | SSML | SSM + lactate | |
| B. aurantiacum 1-16-58 | 0.09 | 0.09 | 0.11 | 0.13 |
| C. casei DPC 5298T | 0.34 | 0.35 | 0.34 | 0.38 |
| C. variabile DPC 5310 | 0.17 | 0.55 | 0.16 | 0.54 |
| M. gubbeenense DPC 5286T | 0.09 | 0.13 | 0.09 | 0.12 |
| S. saprophyticus 4E61 | 0.28 | 0.57 | 0.51 | 0.35 |
SSMG, SSM supplemented with 10 g liter−1 glucose; SSML, SSM supplemented with 10 g liter−1 lactose; SSM + lactate, SSM supplemented with 10 g liter−1 lactate.
FIG. 2.
Growth (A), pH increase (B), and lactate consumption (C) of C. casei DPC 5298T (♦), S. saprophyticus 4E61 (▪), C. variabile DPC 5310 (•), B. aurantiacum 1-16-58 (□), and M. gubbeenense DPC 5286T (▴), in the semisynthetic medium containing 10 g sodium lactate/liter at 30°C and 200 rpm.
Amino acid utilization.
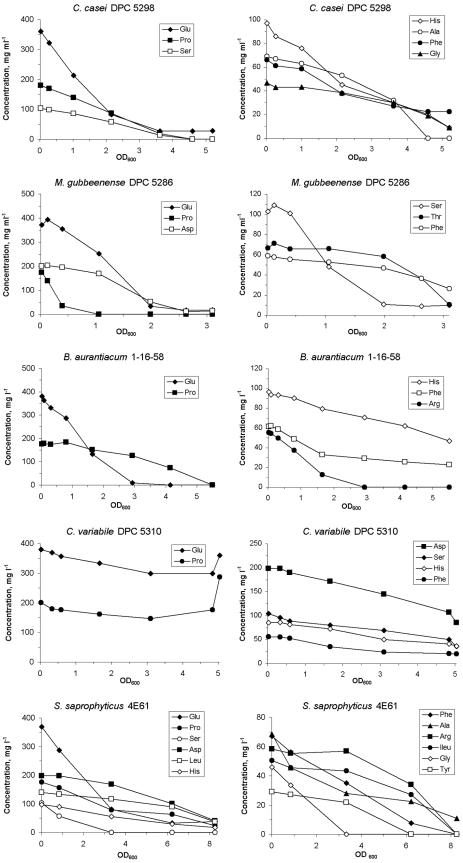
The utilization of amino acids during growth of the five bacteria in SSM containing sodium lactate is shown in Fig. 3. Amino acid depletion correlated with the increase in biomass. B. aurantiacum used 5 amino acids, C. variabile and M. gubbeenense used 6 each, C. casei used 7, and S. saprophyticus used 12. Glutamate, proline, and phenylalanine were used by all organisms; serine was used by all, except B. aurantiacum; and histidine was used by all, except M. gubbeenense. Use of the other amino acids depended on the organism. C. variabile utilized the amino acids to a much lower extent than the four other bacteria, and at the end of growth, increases in glutamate and proline were found. These patterns of utilization were similar in the media containing glucose for C. variabile, C. casei, M. gubbeenense, and S. saprophyticus or lactose for S. saprophyticus (data not shown). In the absence of sugar and lactate, the same amino acids as in SSM plus lactate were utilized by C. casei, M. gubbeenense, and S. saprophyticus, except that C. casei also utilized aspartate and threonine, while M. gubbeenense did not utilize threonine and phenylalanine in SSM. In SSM, B. aurantiacum only utilized glutamate and C. variabile only used aspartate, proline, and glutamate (data not shown). This was likely due to the fact that the latter two bacteria only grew to an OD of ∼0.6 in SSM compared with ∼5 in SSM plus lactate. C. casei and S. saprophyticus grew to an OD of 2.5, while M. gubbeenense grew to an OD of 1 in SSM compared with ODs of ∼5, 6, and 3, respectively, in SSM plus lactate.
FIG. 3.
Amino acid consumption as a function of OD600 during growth of C. casei DPC 5298T, C. variabile DPC 5310, B. aurantiacum 1-16-58, M. gubbeenense DPC 5286T, and S. saprophyticus 4E61 in the SSM containing 10 g sodium lactate/liter at 30°C and 200 rpm.
Utilization of glutamate, proline, serine, or lactate as energy sources.
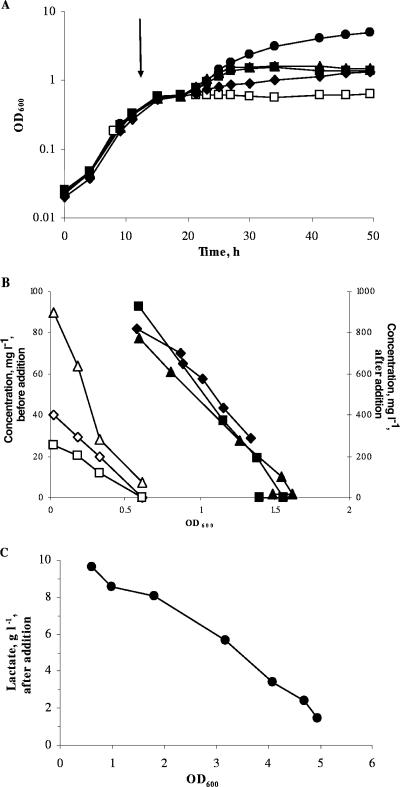
Addition of individual concentrated solutions of glutamate, proline, serine, or lactate at the end of the exponential phase of growth of C. casei resulted in resumption of growth (Fig. 4), with growth rates of 0.12, 0.05, 0.10, and 0.19 h−1, respectively. Plots of consumption of amino acids against biomass were linear, indicating that the amino acids were used as energy sources. It is likely that the lower growth rates obtained after addition of the amino acids compared to the initial one in SSM (0.34 h−1; Table 1) are due to the lower levels of amino acids added at the end of the exponential phase and the fact that they were added individually. Similar results were obtained for M. gubbeenense, except that growth did not occur after the addition of serine (data not shown).
FIG. 4.
Effect of the addition of glutamate (▴), proline (♦), serine (▪), or lactate (•) at the end of the exponential phase on the growth (A) of C. casei DPC 5298 and the level of amino acids (B) and lactate utilization (C) as a function of OD600. □, control culture without addition. The arrow indicates the time at which the additions were made to the medium. Open symbols, concentration of amino acids before addition; closed symbols, concentration of amino acids after addition of the amino acid.
Vitamin requirements.
The effects of omission of individual vitamins from SSM on the growth of C. casei, C. variabile, M. gubbeenense, B. aurantiacum, and S. saprophyticus are shown in Table 2. Calcium panthotenate was essential for growth of C. casei and M. gubbeenense. Omission of biotin reduced the growth of B. aurantiacum, C. casei, and M. gubbeenense, while omission of thiamine reduced the growth of M. gubbeenense and S. saprophyticus. Single omission of these vitamins did not affect the growth of C. variabile. The simultaneous omission of pyridoxine, pyridoxal, and pyridoxamine did not affect the growth of any of the organisms tested (data not shown).
TABLE 2.
Effect of single omission of vitamins on growth of several bacteria isolated from the surface of smear cheese
| Vitamin omitted | Growth (OD600) of straina:
|
||||
|---|---|---|---|---|---|
| B. aurantiacum 1-16-58 | C. casei DPC 5298T | C. variabile DPC 5310 | M. gubbeenense DPC 5286T | S. saprophyticus 4E61 | |
| None | 1.8 ± 0.05a | 2.2 ± 0.27 | 1.9 ± 0.25 | 1.8 ± 0.03 | 2.4 ± 0.19 |
| Riboflavin | 1.7 ± 0.07 | 2.1 ± 0.30 | 1.7 ± 0.35 | 1.7 ± 0.10 | 2.1 ± 0.23 |
| Folic acid | 1.8 ± 0.03 | 2.3 ± 0.13 | 1.8 ± 0.16 | 1.7 ± 0.07 | 2.0 ± 0.07 |
| Calcium panthotenate | 1.7 ± 0.04 | 0.16 ± 0.004* | 1.8 ± 0.16 | 0.01 ± 0.001* | 2.1 ± 0.15 |
| Nicotinic acid | 1.7 ± 0.04 | 2.2 ± 0.06 | 1.8 ± 0.17 | 1.8 ± 0.10 | 2.1 ± 0.11 |
| Pyridoxamine | 1.8 ± 0.02 | 1.8 ± 0.16 | 1.7 ± 0.04 | 1.9 ± 0.14 | 2.2 ± 0.28 |
| Pyridoxal HCl | 1.8 ± 0.04 | 2.1 ± 0.32 | 1.8 ± 0.04 | 1.8 ± 0.07 | 2.3 ± 0.33 |
| Pyridoxine | 1.8 ± 0.02 | 2.4 ± 0.15 | 2.1 ± 0.33 | 1.8 ± 0.02 | 2.4 ± 0.12 |
| Biotin | 1.1 ± 0.02* | 1.4 ± 0.11* | 1.8 ± 0.02 | 1.3 ± 0.04* | 1.9 ± 0.09 |
| Thiamine | 1.8 ± 0.04 | 2.3 ± 0.31 | 1.8 ± 0.03 | 0.8 ± 0.02* | 1.2 ± 0.02* |
Growth was measured as the OD600 after 72 h of incubation at 30°C. Each value is the average ± standard deviation. *, mean significantly different from control at a 95% confidence interval.
Growth of C. casei in MM.
C. casei grew in MM and MM plus lactate, each of which contained only 7 amino acids as N sources (Fig. 5). Growth in MM plus lactate was faster (0.31 h−1) than in MM (0.24 h−1). In both media, all amino acids were utilized, but only data for glutamate, proline, serine, and alanine are shown in Fig. 5. Glutamate was the most rapidly used amino acid, but it was not rate limiting. The rate of utilization of all the amino acids was slower in the presence of lactate, indicating that lactate was being used as an energy source. Lactate was also linearly depleted during growth in MM plus lactate (data not shown). M. gubbeenense did not grow in this medium.
FIG. 5.
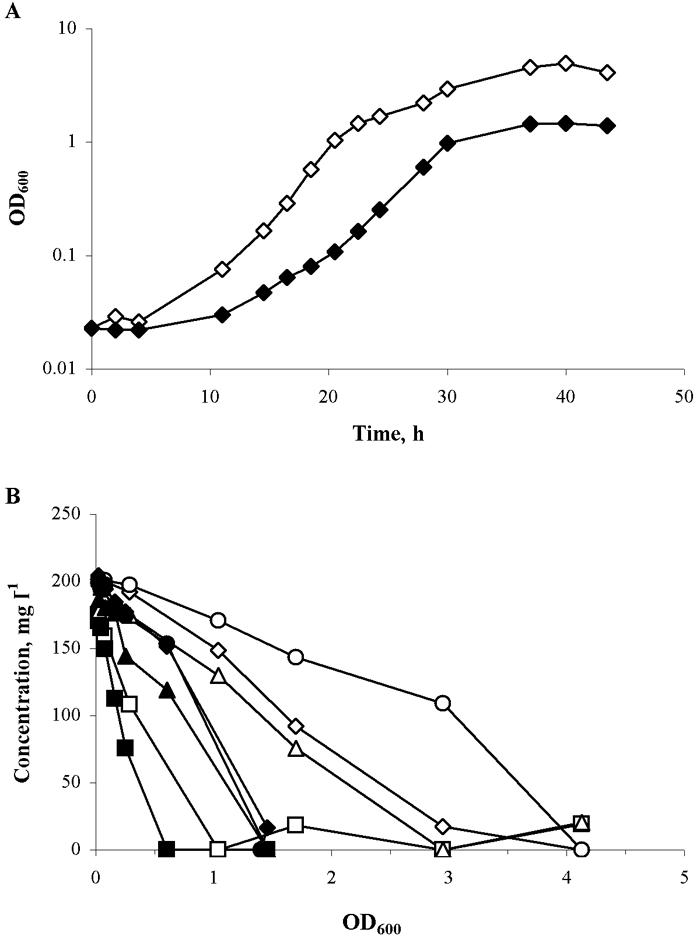
Growth (A) and amino acid utilization as a function of OD600 (B) of C. casei DPC 5298 in MM with (open symbols) or without (closed symbols) lactate. ▪ and □, glutamate; ▴ and ▵, proline; ♦ and ⋄, serine; • and ○, alanine.
Enzyme activities.
Cells of each organism grown in SSM plus lactate contained GPT and LDH activities with both pyruvate and lactate as substrates (Table 3). None of them contained urease activity, and only S. saprophyticus had GDH activity. The differences in activities of LDH, using pyruvate and lactate as substrates, probably reflect the different conditions used in the respective assays.
TABLE 3.
Enzyme activities in cell-free extracts of the strains used in this studya
| Strain | Enzyme activity (U/mg protein)
|
|||
|---|---|---|---|---|
| LDH
|
GPT | GDH | ||
| Pyruvate as substrate | Lactate as substrate | |||
| M. gubbeenense DPC 5286 | 0.19 ± 0.031 | 0.121 ± 0.0029 | 0.155 ± 0.0484 | NDb |
| C. casei DPC 5298 | 0.99 ± 0.057 | 0.022 ± 0.0011 | 0.097 ± 0.0238 | ND |
| C. variabile DPC 5310 | 0.11 ± 0.005 | 0.046 ± 0.0024 | 0.177 ± 0.0157 | ND |
| B. aurantiacum 1-16-58 | ND | 0.065 ± 0.0007 | 0.210 ± 0.0095 | ND |
| S. saprophyticus 4E61 | 0.12 ± 0.013 | 0.010 ± 0.0003 | 0.042 ± 0.0198 | 0.065 ± 0.0084 |
Urease activity was not detected in any strain.
ND, not detected.
DISCUSSION
The two main sources of carbon available to microorganisms at the beginning of cheese ripening are lactose and lactate. However, lactose is rapidly transformed to lactate during the first days of ripening by the lactic acid bacteria present on the cheese surface. Therefore, after a few days of ripening, lactate is the most important C source on the cheese surface. All five bacteria studied assimilated lactate, but only S. saprophyticus utilized lactose. It is generally believed that yeasts are responsible for metabolizing lactate on the surface of smear cheese. The present data suggest that the bacteria also play a role in this regard. LDH activity with lactate as the substrate was present in all five bacteria, suggesting that transformation of lactate to pyruvate is the first step in lactate utilization. Malic enzyme may also be involved, as has been found in C. glutamicum (19).
The four coryneform bacteria studied represented three different genera. Despite this, their amino acid utilization patterns were fairly similar. All four species utilized glutamate, proline, phenylalanine, and, except for B. aurantiacum, serine. Utilization of the other amino acids depended on the organism. The results for B. aurantiacum agree with those of Famelart et al. (13), who showed that B. linens ATCC 9175, recently reclassified as B. aurantiacum (17), rapidly consumed phenylalanine, arginine, proline, glutamate, histidine, and tyrosine in a medium containing Casamino Acids and sodium lactate. Tyrosine was not consumed by the strain of B. aurantiacum used in the present study. In C. variabile, utilization of glutamate and proline was much slower than in the other four organisms and C. variabile also appeared to be able to synthesize glutamate and proline because both amino acids increased in the medium during the stationary phase of growth. Further work is necessary to try to understand this phenomenon. These organisms are moderately halophilic (Fig. 1), and glutamate and proline may be important in maintaining the osmotic balance in the cells (12, 24).
GPT activity was detected in all five organisms, while GDH activity was only detected in S. saprophyticus. GPT catalyzes the transamination of pyruvate from glutamate, resulting in formation of alanine from pyruvate and αKG from glutamate, from which energy can be generated through the tricarboxylic acid cycle. αKG is also important in the catabolism of amino acids for flavor formation by the lactic acid bacteria in cheese (for reviews, see references 41 and 44), and this may also be its role in the smear bacteria. LDH activity with lactate as substrate was also detected in all organisms (Table 3) and is the likely source of pyruvate for the GPT activity. None of the organisms, except C. casei and S. saprophyticus, used alanine, implying that GPT activity is also a possible source of alanine in these organisms.
C. casei and M. gubbeenense used serine, which can be converted to pyruvate and NH3 by both l-serine dehydratase and cystathionine β-lyase, both of which have been characterized in C. glutamicum (34). An l-serine dehydratase has also been characterized in B. aurantiacum ATCC 9175 (20).
Urea and (NH4)2SO4 were also present in the SSM used. Whether these compounds were involved in N metabolism was not determined, but the fact that urease activity was not detected in any organism (Table 3) suggests that urea is not used by these organisms and that it could be omitted from the medium. This conclusion is supported by the fact that C. casei, at least, was able to grow in a defined medium containing seven amino acids as the only sources of C and N. Further investigations are necessary to identify which of these amino acids are essential for growth.
To our knowledge, this is the first time that the vitamin requirements of B. aurantiacum, C. casei, and M. gubbeenense have been studied. Omission of biotin and thiamine from the medium significantly reduced growth of M. gubbeenense. It has been previously shown that pantothenic acid is essential for growth of B. linens and that this vitamin is produced by D. hansenii (37). The present study showed that pantothenic acid was also required by C. casei and M. gubbeenense. There was no significant effect of omission of vitamins on the growth of C. variabile, suggesting that this organism has no vitamin requirements. The growth of S. saprophyticus was only limited when thiamine was omitted from the medium. This is in agreement with Cove and Holland (11), who found that strains of S. saprophyticus isolated from human skin required thiamine for growth.
Coryneform bacteria are considered to be halotolerant microorganisms (16), and the present results show that they grow quite well in the presence of 6% salt. Concentrations of >9% salt retard growth, and only S. saprophyticus and B. aurantiacum grew (slowly) in the presence of 15% salt. The four coryneforms grew best in low (3%) levels of salt, but the effect was not great. Similar results have been reported for various strains of B. linens, Corynebacterium ammoniagenes, and Corynebacterium flavescens (4, 31) but, in contrast to the present findings, not C. variabile (31). We have no explanation for this contradictory result. The enhancing effect of salt on growth may be due in part to the requirement of Na+ gradients to drive transport processes across the cell membrane (43).
Extrapolation of the growth characteristics of these organisms in liquid cultures to their growth on the surface of smear cheese may be risky since the composition of the cheese matrix is far more complex than the medium utilized in this study and competition for the substrates and/or negative or positive interactions may occur between organisms. However, coryneform bacteria dominate the later stages of the ripening process (9, 34, 39). At this point, the level of lactate would be low and that of the free amino acids high, as a result of proteolysis by the bacteria and yeast on the surface. The fact that these organisms are salt tolerant and able to grow using only amino acids as energy sources could explain their prevalence on the cheese surface at the later stages of ripening.
Acknowledgments
J.M. thanks Teagasc for awarding him a Walsh Fellowship, during which the research in this paper was undertaken.
Footnotes
Published ahead of print on 5 October 2007.
REFERENCES
- 1.Bergmeyer, H. U., and E. Bernt. 1974. Lactate dehydrogenase UV assay with pyruvate and NADH, p. 574-579. In H. U. Bergmeyer (ed.), Methods of enzymatic analysis, 2nd English ed., vol. 2. Academic Press, New York, NY. [Google Scholar]
- 2.Bergmeyer, H. U., and E. Bernt. 1974. Lactate dehydrogenase colorimetric assay with L lactate, NAD, phenazine methosulphate and INT, p. 579-582. In H. U. Bergmeyer (ed.), Methods of enzymatic analysis, 2nd English ed., vol. 2. Academic Press, New York, NY. [Google Scholar]
- 3.Bergmeyer, H. U., and E. Bernt. 1974. Glutamate-pyruvate transaminase UV assay, manual method, p. 752-758. In H. U. Bergmeyer (ed.), Methods of enzymatic analysis, 2nd English ed., vol. 2. Academic Press, New York, NY. [Google Scholar]
- 4.Bernard, T., M. Jebbar, Y. Rassouli, S. Himdi-Kabbab, J. Hamelin, and C. Blanco. 1993. Ectoine accumulation and osmotic regulation in Brevibacterium linens. J. Gen. Microbiol. 139:129-136. [Google Scholar]
- 5.Bockelmann, W., and T. Hoppe-Seyler. 2001. The surface flora of bacterial smear-ripened cheeses from cow's and goat's milk. Int. J. Food Microbiol. 11:307-314. [Google Scholar]
- 6.Bora, N., M. Vancanneyt, R. Gelsomino, J. Swings, N. Brennan, T. M. Cogan, S. Larpin, N. Desmasures, F. E. Lechner, R. M. Kroppenstedt, A. C. Ward, and M. Goodfellow. 2007. Agrococcus casei sp. nov., isolated from the surfaces of smear-ripened cheeses. Int. J. Syst. Evol. Microbiol. 57:92-97. [DOI] [PubMed] [Google Scholar]
- 7.Brennan, N. M., R. Brown, M. Goodfellow, A. C. Ward, T. P. Beresford, P. J. Simpson, P. F. Fox, and T. M. Cogan. 2001. Corynebacterium mooreparkense sp. nov. and Corynebacterium casei sp. nov., isolated from the surface of a smear-ripened cheese. Int. J. Syst. Evol. Microbiol. 51:843-852. [DOI] [PubMed] [Google Scholar]
- 8.Brennan, N. M., R. Brown, M. Goodfellow, A. C. Ward, T. P. Beresford, M. Vancanneyt, T. M. Cogan, and P. F. Fox. 2001. Microbacterium gubbeenense sp. nov., from the surface of a smear-ripened cheese. Int. J. Syst. Evol. Microbiol. 51:1969-1976. [DOI] [PubMed] [Google Scholar]
- 9.Brennan, N. M., A. C. Ward, T. P. Beresford, P. F. Fox, M. Goodfellow, and T. M. Cogan. 2002. Biodiversity of the bacterial flora on the surface of a smear cheese. Appl. Environ. Microbiol. 68:820-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corsetti, A., J. Rossi, and M. Gobbetti. 2001. Interactions between yeasts and bacteria in the smear surface-ripened cheeses. Int. J. Food Microbiol. 69:1-10. [DOI] [PubMed] [Google Scholar]
- 11.Cove, J. H., and K. T. Holland. 1990. The vitamin requirements of staphylococci isolated from human skin. J. Appl. Bacteriol. 49:29-37. [DOI] [PubMed] [Google Scholar]
- 12.Csonka, L. N. 1989. Physiology and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53:121-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Famelart, M.-H., C. Bouillanne, A. Kobilinsky, and M. Desmazeaud. 1987. Studies on Brevibacterium linens metabolism in a fermentor. Appl. Microbiol. Biotechnol. 26:378-382. [Google Scholar]
- 14.Famelart, M.-H., A. Kobilinsky, C. Bouillanne, and M. Desmazeaud. 1987. Influence of temperature, pH and dissolved oxygen on growth of Brevibacterium linens in a fermentor. Appl. Microbiol. Biotechnol. 25:442-448. [Google Scholar]
- 15.Feurer, C., T. Vallaeys, G. Corrieu, and F. Irlinger. 2004. Does smearing inoculum reflect the bacterial composition of the smear at the end of the ripening of a French soft, red-smear cheese? J. Dairy Sci. 87:3189-3197. [DOI] [PubMed] [Google Scholar]
- 16.Frings, E., H. J. Kunte, and E. A. Galinsky. 1993. Compatible solutes in representatives of the genera Brevibacterium and Corynebacterium: occurrence of tetrahydropyrimidines and glutamine. FEMS Microbiol. Lett. 109:25-32. [Google Scholar]
- 17.Gavrish, E. Y., V. I. Krauzova, N. V. Potekhina, S. G. Karasev, E. G. Plotnikova, O. V. Altyntseva, L. A. Korosteleva, and L. I. Evtushenko. 2005. Three new species of brevibacteria, Brevibacterium antiquum sp. nov., Brevibacterium aurantiacum sp. nov. and Brevibacterium permense sp. nov. Microbiology 73:176-183. [PubMed] [Google Scholar]
- 18.Gelsomino, R., M. Vancanneyt, C. Snauwaert, K. Vandemeulebroecke, B. Hoste, T. M. Cogan, and J. Swings. 2005. Corynebacterium mooreparkense, a later heterotypic synonym of Corynebacterium variabile. Int. J. Syst. Evol. Microbiol. 55:1129-1131. [DOI] [PubMed] [Google Scholar]
- 19.Gourdon, P., M. F. Baucher, N. D. Lindley, and A. Guyonvarch. 2000. Cloning of the malic enzyme gene from Corynebacterium glutamicum and role of the enzyme in lactate metabolism. Appl. Environ. Microbiol. 66:2981-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamouy, D., and M. J. Desmazeaud. 1985. Characterization of an L-serine dehydratase activity in permeabilized cells of Brevibacterium linens. Lait 65:103-121. [Google Scholar]
- 21.Irlinger, F., F. Bimet, J. Delettre, M. Lefevre, and P. A. D. Grimont. 2005. Two new coryneform species isolated from the surface of cheeses are species of the genus Arthrobacter: Arthrobacter bergerei sp. nov. and Arthrobacter arilaitensis sp. nov. Int. J. Syst. Bacteriol. 55:457-462. [DOI] [PubMed] [Google Scholar]
- 22.Irlinger, F., A. Morvan, N. El Solh, and J.-L. Bergere. 1997. Taxonomic characterisation of coagulase-negative staphylococci in ripening flora from traditional French cheeses. Syst. Appl. Microbiol. 20:319-328. [Google Scholar]
- 23.Katsumata, R., A. Ozaki, T. Oka, and A. Furuya. 1984. Protoplast transformation of glutamate-producing bacteria with plasmid DNA. J. Bacteriol. 159:306-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawahara, Y., T. Oshumi, Y. Yoshihara, and S. Ikeda. 1989. Proline in the osmoregulation of Brevibacterium lactofermentum. Agric. Biol. Chem. 53:2475-2479. [Google Scholar]
- 25.Leclercq-Perlat, M.-N., A. Oumer, J.-L. Bergere, H.-E. Spinnler, and G. Corrieu. 1999. Growth of Debaryomyces hansenii on a bacterial surface-ripened soft cheese. J. Dairy Sci. 66:271-281. [Google Scholar]
- 26.Leclercq-Perlat, M.-N., A. Oumer, J.-L. Bergere, H.-E. Spinnler, and G. Corrieu. 2000. Behavior of Brevibacterium linens and Debaryomyces hansenii as ripening flora in controlled production of smear soft cheese from reconstituted milk: growth and substrate consumption dairy foods. J. Dairy Sci. 83:1665-1673. [DOI] [PubMed] [Google Scholar]
- 27.Leclercq-Perlat, M.-N., A. Oumer, J.-L. Bergere, H.-E. Spinnler, and G. Corrieu. 2000. Behavior of Brevibacterium linens and Debaryomyces hansenii as ripening flora in controlled production of smear soft cheese from reconstituted milk: protein degradation. J. Dairy Sci. 83:1674-1683. [DOI] [PubMed] [Google Scholar]
- 28.Liebl, W., R. Klamer, and K.-H. Schleifer. 1989. Requirement of chelating compounds for the growth of Corynebacterium glutamicum in synthetic media. Microbiol. Biotechnol. 32:205-210. [Google Scholar]
- 29.Maoz, A., R. Mayr, and S. Scherer. 2003. Temporal stability and biodiversity of two complex antilisterial cheese-ripening microbial consortia. Appl. Environ. Microbiol. 69:4012-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masoud, W., and M. Jakobsen. 2003. Surface ripened cheeses: the effects of Debaryomyces hansenii, NaCl and pH on the intensity of pigmentation produced by Brevibacterium linens and Corynebacterium flavescens. Int. Dairy J. 13:231-237. [Google Scholar]
- 31.Masoud, W., and M. Jakobsen. 2005. The combined effects of pH, NaCl and temperature on growth of cheese ripening cultures of Debaryomyces hansenii and coryneform bacteria. Int. Dairy J. 15:69-77. [Google Scholar]
- 32.Mimura, H., S. Nagata, and T. Matsumoto. 1994. Concentrations and compositions of internal free amino acids in a halotolerant Brevibacterium sp. in response to salt stress. Biosci. Biotechnol. Biochem. 58:1873-1874. [Google Scholar]
- 33.Mounier, J., R. Gelsomino, S. Goerges, M. Vancanneyt, K. Vandemeulebroecke, B. Hoste, S. Scherer, J. Swings, G. F. Fitzgerald, and T. M. Cogan. 2005. The surface microflora of four smear-ripened cheeses. Appl. Environ. Microbiol. 71:6489-6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Netzer, R., P. Peters-Wendisch, L. Eggeling, and H. Sahm. 2004. Cometabolism of a nongrowth substrate: l-serine utilization by Corynebacterium glutamicum. Appl. Environ. Microbiol. 70:7148-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Place, R. B., D. Hiestaind, S. Burri, and M. Teuber. 2002. Staphylococcus succinus subsp. casei subsp. nov., a dominant isolate from a surface ripened cheese. Syst. Appl. Microbiol. 25:353-359. [DOI] [PubMed] [Google Scholar]
- 36.Place, R. B., D. Hiestaind, H. R. Gallmann, and M. Teuber. 2003. Staphylococcus equorum subsp. linens, subsp. nov., a starter culture component for surface ripened semi-hard cheeses. Syst. Appl. Microbiol. 26:30-37. [DOI] [PubMed] [Google Scholar]
- 37.Purko, M., W. O. Nelson, and W. A. Wood. 1951. The associative action between certain yeast and Bacterium linens. J. Dairy Sci. 34:699-705. [Google Scholar]
- 38.Rea, M. C., S. Gorges, R. Gelsomino, N. M. Brennan, J. Mounier, M. Vancanneyt, S. Scherer, J. Swings, and T. M. Cogan. 2007. Stability of the biodiversity of the surface consortia of Gubbeen, a red-smear cheese. J. Dairy Sci. 90:2200-2210. [DOI] [PubMed] [Google Scholar]
- 39.Schlegel, H. G., and H. Kaltwasser. 1974. Urease, p. 1080-1085. In H. U. Bergmeyer (ed.), Methods of enzymatic analysis, 2nd English ed., vol. 2. Academic Press, New York, N.Y. [Google Scholar]
- 40.Schmidt, E. 1974. Glutamate dehydrogenase, p. 650-656. In H. U. Bergmeyer (ed.), Methods of enzymatic analysis, 2nd English ed., vol. 2. Academic Press, New York, NY. [Google Scholar]
- 41.Smit, G., B. A. Smit, and W. J. M. Engels. 2005. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 29:591-610. [DOI] [PubMed] [Google Scholar]
- 42.Valdés-Stauber, N., S. Scherer, and H. Seiler. 1997. Identification of yeasts and coryneform bacteria from the surface microflora of brick cheeses. Int. J. Food Microbiol. 34:115-129. [DOI] [PubMed] [Google Scholar]
- 43.Ventosa, A., J. J. Nieto, and A. Oren. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62:504-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yvon, M., and L. Rijnen. 2001. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 11:185-201. [Google Scholar]