Abstract
In the bacterial degradation of polycyclic aromatic hydrocarbons (PAHs), salicylate hydroxylases catalyze essential reactions at the junction between the so-called upper and lower catabolic pathways. Unlike the salicylate 1-hydroxylase from pseudomonads, which is a well-characterized flavoprotein, the enzyme found in sphingomonads appears to be a three-component Fe-S protein complex, which so far has not been characterized. Here, the salicylate 1-hydroxylase from Sphingomonas sp. strain CHY-1 was purified, and its biochemical and catalytic properties were characterized. The oxygenase component, designated PhnII, exhibited an α3β3 heterohexameric structure and contained one Rieske-type [2Fe-2S] cluster and one mononuclear iron per α subunit. In the presence of purified reductase (PhnA4) and ferredoxin (PhnA3) components, PhnII catalyzed the hydroxylation of salicylate to catechol with a maximal specific activity of 0.89 U/mg and showed an apparent Km for salicylate of 1.1 ± 0.2 μM. The hydroxylase exhibited similar activity levels with methylsalicylates and low activity with salicylate analogues bearing additional hydroxyl or electron-withdrawing substituents. PhnII converted anthranilate to 2-aminophenol and exhibited a relatively low affinity for this substrate (Km, 28 ± 6 μM). 1-Hydroxy-2-naphthoate, which is an intermediate in phenanthrene degradation, was not hydroxylated by PhnII, but it induced a high rate of uncoupled oxidation of NADH. It also exerted strong competitive inhibition of salicylate hydroxylation, with a Ki of 0.68 μM. The properties of this three-component hydroxylase are compared with those of analogous bacterial hydroxylases and are discussed in light of our current knowledge of PAH degradation by sphingomonads.
Bacterial isolates belonging to the sphingomonad group have been shown to degrade a wide range of organic pollutants, including recalcitrant compounds such as chlorinated xenobiotics (16), dioxins (26), and polycyclic aromatic hydrocarbons (PAHs) (9, 18, 20, 32). The remarkable capabilities of this group of bacteria suggested that they might be useful for bioremediation purposes and prompted studies on the identification of the catabolic enzymes involved. Independent genetic analyses of different PAH-degrading strains revealed that they contain a unique set of catabolic genes, often carried on a megaplasmid, which is probably responsible for their versatile metabolism (15, 20, 23). Extending over a 40-kb DNA region, the main catabolic gene cluster includes xyl genes involved in the catabolism of monoaromatics interspersed with genes predicted to function in the degradation of polycyclic substrates (designated bph, nah, ahd, or phn). This unique gene arrangement, which is remarkably conserved among strains of various origins, contrasts with that found in other degraders, such as pseudomonads. In Sphingobium yanoikuyae B1, the biodegradation of naphthalene is thought to proceed through a pathway similar to that found in Pseudomonas species, with a first set of enzymes converting naphthalene to salicylate and a second set responsible for the conversion of salicylate to Krebs cycle intermediates (15). In this strain, as in other sphingomonads, many of the gene products involved in this pathway have been identified based on sequence similarity with known Pseudomonas enzymes, but no counterpart was found for NahG, which catalyzes the monohydroxylation of salicylate to catechol (30). In addition, the function of many genes in the catabolism of other PAHs remains to be elucidated.
Peculiar to sphingomonads is the occurrence of multiple copies of genes predicted to encode the terminal component of Rieske-type oxygenases (21, 23). Genes coding for two cognate electron carriers are generally found adjacent to such oxygenase-encoding genes in other bacteria degrading aromatic hydrocarbons, but this is not the case in sphingomonads. Rieske-type oxygenases are known to catalyze the dihydroxylation of various aromatic compounds and often initiate the oxidative biodegradation of pollutants. They constitute a large family of two- or three-component metalloenzymes whose catalytic component is generally a heteromeric α3β3 hexamer containing one Rieske-type [2Fe-2S] cluster and one nonheme iron atom per α subunit. The enzyme responsible for the initial attack on PAHs has recently been identified in Sphingomonas sp. strain CHY-1, a strain able to grow on chrysene as a sole carbon source (9, 25). The oxygenase, designated PhnI, is functionally associated with an NAD(P)H oxidoreductase (PhnA4) and a ferredoxin (PhnA3) in a three-component enzyme complex able to oxidize a wide range of two- to five-ring PAHs (13). Analysis of a knockout mutant demonstrated that PhnI was absolutely required for growth on PAHs, indicating that no other oxygenase could replace PhnI in the initial dihydroxylation of PAHs (9). Another oxygenase, designated PhnII, which catalyzes the C-1 hydroxylation of salicylate to catechol in strain CHY-1 has been identified. When overproduced in recombinant form in Escherichia coli, PhnII required the coexpression of the ancillary proteins PhnA4 and PhnA3 for full activity, indicating that it is a three-component enzyme which shares electron carriers with PhnI (9). PhnII resembles salicylate 5-hydroxylase from Ralstonia sp. strain U2, which oxidizes salicylate to gentisate (31), but it differs from salicylate 1-hydroxylases found in pseudomonads, which are monomeric flavoproteins (29). Interestingly, PhnII is also related to anthranilate dioxygenase from Burkholderia cepacia DBO1, a three-component enzyme that exclusively catalyzes dioxygenation reactions (6). The occurrence of three-component salicylate 1-hydroxylases was first reported by Pinyakong et al. in Sphingobium sp. strain P2, a phenanthrene-degrading strain (22). This strain synthesized three isoenzymes homologous to PhnII which showed different specificities for substituted salicylates. Sequence comparisons using BLAST indicated that PhnII showed substantial similarity with one hydroxylase from strain P2 (83 and 71% identity for the α and β subunits, respectively) but only moderate similarity with the two other P2 enzymes (<51 and <45% identity). Such comparisons of protein sequences did not reveal any clue that would explain the differences in selectivity between these enzymes. In S. yanoikuyae B1, indirect evidence indicated that an analogous hydroxylase catalyzed both salicylate hydroxylation and conversion of 1-hydroxy-2-naphthoate to 1,2-dihydroxynaphthalene (7). The latter reaction is a key step in the biodegradation of phenanthrene by gram-negative bacteria.
So far, none of the three-component hydroxylases considered above has been purified, and therefore there are no biochemical and catalytic data on this important class of enzymes. Here, we purified the PhnII enzyme to near homogeneity and investigated the kinetics of the catalytic hydroxylation of salicylate and several analogues. We also found that 1-hydroxy-2-naphthoate is not a substrate but triggers NADH oxidation at a high rate in a totally uncoupled reaction. This reaction might inhibit PAH degradation by sphingomonads when phenanthrene is an abundant substrate.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains of E. coli and Pseudomonas putida and plasmids used for gene overexpression, as well as general culture conditions, have been previously described (9). Plasmid pVEHD11 carrying the phnA1bA2b coding sequence fused to a 5′ extension coding for a His tag was constructed in two steps. An NdeI-BamHI fragment carrying the two genes of interest was isolated from plasmid pDAB11 (9) and subcloned into pET15b (Novagen) to obtain pEHD11. The insert was then extracted from pEHD11 as a XbaI fragment and cloned into pVLT31 (8). A plasmid carrying phnA1bA2b in the right orientation with respect to the Ptac promoter was selected and designated pVEHD11. His-tagged PhnII (ht-PhnII) was overproduced either in E. coli BL21(DE3) carrying pEHD11 or in P. putida KT2442 carrying pVEHD11. Overproduction of ht-PhnA3, ht-PhnA4, and ht-RedB356, which were used as electron carriers for ht-PhnII in oxygenase assays, has been previously described (13).
Large-scale cultures of strain KT2442(pVEHD11) were grown in Luria-Bertani rich medium in a 12-liter fermentor as previously described (13). Expression of the recombinant oxygenase was induced at 25°C with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and each culture was incubated for 20 h before it was harvested by centrifugation. The bacterial pellet was washed with 50 mM Tris-HCl buffer (pH 7.5) and kept frozen until use.
Purification of oxygenase component PhnII.
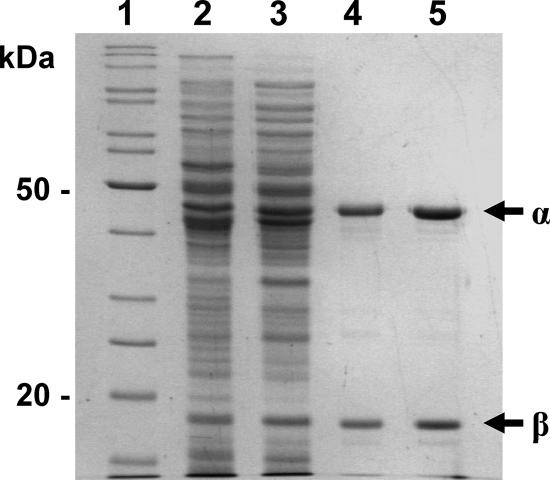
All purification procedures were carried out under argon, using buffers equilibrated for at least 24 h in a glove box maintained under anoxic conditions (O2 concentration, <2 ppm; Jacomex, France). The temperature was kept at 0 to 4°C unless otherwise indicated. A crude extract was prepared by thawing a bacterial pellet (100 g) in 2 volumes of lysis buffer (50 mM Tris-HCl, pH 7.5), followed by lysozyme treatment (0.5 mg/ml) for 15 min at 30°C. The suspension was then subjected to ultrasonication for a total of 10 min, including a 5-min pause, at 75% of the maximal intensity using a Vibra Cell apparatus run in pulse mode with 5 s/pulse (Fisher Bioblock Scientific, Illkirch, France). The lysate was centrifuged at 12,000 × g for 30 min, and the resulting cell extract was diluted twofold with TG buffer (25 mM Tris-HCl [pH 7.5] containing 5% glycerol and 2 mM β-mercaptoethanol) and applied to two 40-ml columns of DEAE-cellulose (DE52; Whatman) equilibrated with TG buffer. After the columns were washed with 4 bed volumes of the same buffer, the oxygenase was eluted as a brown band with buffered 0.3 M NaCl. The eluate (about 50 ml) was immediately applied to a small column (7 ml) of immobilized metal affinity chromatography (IMAC) resin loaded with Co2+ (Talon; BD Biosciences Ozyme, France). The column was washed successively with 8 bed volumes of TG buffer containing 0.5 M NaCl and 5 bed volumes of the same buffer supplemented with 20 mM imidazole and 1 mM salicylate. A brown protein fraction was then eluted with TG buffer containing 0.15 M imidazole and 1 mM salicylate. A portion of the preparation (1.3 ml containing about 40 mg of protein) was applied to a column (1.5 by 60 cm) of Superdex SD200 equilibrated in TG buffer containing 0.1 M NaCl and eluted at a rate of 0.43 ml/min. The purified protein fraction was concentrated on a small column of DEAE-cellulose, eluted in about 1 ml of TG buffer containing 0.3 M NaCl, and frozen as pellets in liquid nitrogen. This preparation, designated ht-PhnII, was judged to be at least 95% pure by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1).
FIG. 1.
SDS-PAGE protein analysis during purification of recombinant ht-PhnII from strain KT2442 carrying pVEHD11. Lane 1, protein markers; lane 2, soluble cell extract; lane 3, DEAE column eluate; lane 4, IMAC column eluate; lane 5, dialyzed purified protein. The arrows indicate the α and β subunits of ht-PhnII.
Purification of other proteins.
The ferredoxin (ht-PhnA3) from strain CHY-1 and the reductase component of the biphenyl dioxygenase from Comamonas testosteroni B-356 (ht-RedB356) were purified as previously described (13). The catechol 2,3-dioxygenase from P. putida mt2 (XylE) was purified using a previously described procedure (11).
Enzyme assays.
Salicylate hydroxylase activity was determined using three different assays. In the coupled assay, the formation of catechol from salicylate was coupled to ring cleavage by catechol 2,3-dioxygenase (XylE). Each reaction mixture contained 0.1 μM ht-PhnII, 1.5 or 3 μM ht-PhnA3, 0.3 μM ht-RedB356, 0.03 μM XylE, 0.2 mM NADH, and 0.5 mM salicylate in 50 mM potassium phosphate buffer (pH 7.0). Reactions were carried out at 30°C in quartz cuvettes and were initiated by injection of a 100-fold-concentrated solution of the protein mixture with a gas-tight syringe. The protein mixture was prepared 30 min before the assays were started and was kept on ice under argon in phosphate buffer containing 1 mM dithiothreitol, 0.05 mM ferrous ammonium sulfate, and 10% glycerol. The absorption at 376 nm was recorded at 0.1-s intervals over 1 min with an HP8452 spectrophotometer (Agilent Technologies, Les Ulis, France). The enzyme activity was calculated from the initial linear portion of the time course, using an absorption coefficient of 34,000 M−1·cm−1 for 2-hydroxymuconate semialdehyde (11). Assuming that each molecule of the latter product resulted from the oxidation of one molecule of catechol, 1 U of activity was defined as the amount of enzyme that converted 1 μmol of salicylate to catechol per min.
The enzyme activity was also measured by recording NADH oxidation at 340 nm and was calculated using an absorbance coefficient of 6,220 M−1·cm−1. The protein mixture used had the same composition as that described above except that XylE was omitted. Alternatively, the oxygenase activity was determined by monitoring O2 uptake using a Clark-type O2 electrode under conditions similar to those previously described (13). The assay mixtures contained NADH at an initial concentration of 0.4 mM. After approximately 60 nmol O2 had been consumed, 300 U of liver catalase (Sigma-Aldrich) was added to estimate the amount of H2O2 formed during the assay. In some cases, 0.6 ml of the reaction mixture was immediately withdrawn and acidified with 0.1% acetic acid. After centrifugation and filtration, samples were analyzed by high-performance liquid chromatography (HPLC) to quantify the catechols formed as described below.
Steady-state kinetics.
Sets of salicylate hydroxylase assays were carried out using substrate concentrations that ranged from 0.5 to 100 μM. The activity in the XylE-coupled assay was measured as described above. All assays were performed in duplicate, and at least 12 concentrations were tested. For titration of the hydroxylase with anthranilate, enzyme activity was measured by monitoring NADH oxidation at 340 nm. Assays were performed for substrate concentrations ranging from 5 to 500 μM. Oxidation rates were calculated using an absorption coefficient increment of 0.874 M−1·cm−1 to take into account the absorbance of anthranilate at 340 nm. Plots of the initial reaction rate versus substrate concentration were fitted to the Michaelis-Menten equation using the curve fit option of Kaleidagraph (Synergy Software). Only curve fits showing correlation coefficients greater than 0.98 were considered.
Chemical identification and quantification of the enzymatic products.
The oxidation products generated by PhnII were analyzed by gas chromatography-mass spectrometry (GC-MS). Samples were derivatized with bis(trimethysilyl)trifluoroacetamide-trimethylchlorosilane (99:1) from Supelco (Sigma-Aldrich) prior to analysis using an HP6890 gas chromatograph coupled to an HP5973 mass spectrometer (Agilent Technologies). The operating conditions were the same as those previously described (12), and mass spectrum acquisition was carried out in the scan mode.
Concentrations of catechol and methylcatechols formed by salicylate 1-hydroxylase were determined by HPLC using a Kontron system equipped with an F430 UV detector. Samples (0.1 ml) were injected onto a C8 reverse-phase column (4 by 150-mm; Zorbax; Agilent Technologies) run at 0.8 ml/min. The column was eluted with a linear gradient of 0 to 50% acetonitrile in water over 15 min. Detection was carried out at 276 nm. Calibration curves were generated by injecting known amounts of 3-methylcatechol, 4-methylcatechol, or unsubstituted catechol.
Determination of the iron content of proteins.
To extract iron from proteins, samples (150 μl) were treated with 2.5 N HCl for 30 min at 95°C and then diluted with 135 μl of ultrapure water. Iron was assayed in triplicate as a complex with bathophenanthroline by measuring absorbance at 536 nm (2). A calibration curve was generated by diluting a standard solution of ferric nitrate containing 1 g/liter Fe (Merck).
Protein analyses.
Routine protein determination was performed using the Bradford assay (4) or a bicinchoninic acid reagent kit (Pierce) with bovine serum albumin as a standard. A modified microbiuret assay was used for measuring the protein contents of purified preparations of ht-PhnA3 and ht-PhnII (19). SDS-PAGE on mini-slab gels was performed as previously described (14). The molecular mass of purified ht-PhnII was determined by size exclusion chromatography on a Superdex SD200 column (1.5 by 60 cm; Amersham Biosciences). The column was run at a flow rate of 0.2 ml/min and calibrated with the following protein markers: ferritin (443 kDa), catalase (240 kDa), aldolase (150 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), and myoglobin (17 kDa), all obtained from Sigma-Aldrich.
EPR analysis.
The concentration of purified ht-PhnII was adjusted to 30 μM in 25 mM Tris-HCl (pH 7.5) containing 5% glycerol, and the ht-PhnII was reduced with 1 mM dithionite. The electron paramagnetic resonance (EPR) spectrum was recorded at 4 K as previously described (13).
RESULTS
Purification and biochemical properties of salicylate 1-hydroxylase from strain CHY-1.
The structural genes encoding the hydroxylase were cloned into pET15b in order to produce the enzyme with a His tag fused at the N terminus of the large subunit. When the plasmid was introduced into an appropriate E. coli overexpression strain, two polypeptides of the expected sizes were detected in the cell extract by SDS-PAGE, but the enzyme was essentially inactive. Transfer of the genes into plasmid pVLT31 under control of the Ptac promoter resulted in an efficient expression system producing active enzyme in both E. coli and P. putida. The latter organism was preferred as a host to overproduce the recombinant enzyme because it consistently provided better purification yields than E. coli. The hydroxylase was purified under anoxic conditions using a three-step procedure as described in Materials and Methods, which yielded a protein preparation that was judged to be more than 95% pure based on SDS-PAGE (Fig. 1). This enzyme consisted of two polypeptides with apparent Mr of 47,000 and 21,000. We observed that the enzyme activity fell dramatically upon IMAC, most likely because the enzyme lost a good part of its active site Fe(II) ion. This loss of activity could be prevented by supplementing buffers with 1 mM salicylate, while ethanol or isopropanol had no protective effect. After the last purification step, maximal activity was obtained by incubating the protein preparation for 1 h at 4°C with 0.1 mM ferrous ammonium sulfate. The purified enzyme exhibited a specific activity of 0.89 U/mg in a coupled assay in which the catechol formed by the hydroxylase was oxidized to 2-hydroxymuconic acid semialdehyde by catechol dioxygenase (see Materials and Methods). The molecular mass of the hydroxylase as deduced from gel filtration experiments was 190 kDa, which is consistent with an α3β3 hexameric structure. The enzyme exhibited a broad absorbance maximum at 456 nm with a shoulder around 530 nm, which is typical for proteins containing Rieske-type [2Fe-2S] clusters. The occurrence of such clusters in the protein was confirmed by EPR analysis of a dithionite-reduced sample, which gave a rhombic signal centered around a g value of 1.91 (data not shown). The iron content of the enzyme was calculated to be 8.52 ± 0.21 Fe atoms/mol based on chemical assay, which is close to the theoretical value for an α3β3 hexamer containing one [2Fe-2S] cluster and one nonheme iron per α subunit. Overall, the salicylate hydroxylase from strain CHY-1 exhibited biochemical properties similar to those of classical Rieske-type dioxygenases that catalyze the dihydroxylation of various aromatic hydrocarbons (5).
Catalytic properties and substrate specificity.
The activity of the purified ht-PhnII oxygenase required ancillary proteins PhnA4 and PhnA3 to transfer electrons derived from NADH oxidation to the enzyme active site. The dependence of the enzyme complex activity on the concentration of each electron carrier was studied in assays in which the molar ratio of the carrier to the oxygenase component was independently varied. While half-saturation was reached at a reductase-to-oxygenase ratio close to 0.5, a much higher relative concentration of ferredoxin ht-PhnA3 was needed to obtain maximum activity. At the highest protein ratio tested (about a 200-fold molar excess of ferredoxin), the enzyme activity was not fully saturated. Nevertheless, in the assays described here this ratio was set at 30 or below in order to prevent excessive substrate-independent NADH oxidation by the enzyme complex. Also, the reductase component of the biphenyl dioxygenase from C. testosteroni, ht-RedB356, was substituted for PhnA4 in most assays because the reductase from strain CHY-1 proved to be unstable (13). No loss of hydroxylase activity was observed as a result of this substitution.
The substrate specificity of the hydroxylase with various salicylate analogues was first determined by NADH oxidation assays (Table 1). At the end of each assay, the product formed was extracted with ethyl acetate and analyzed by GC-MS. The enzyme showed high levels of activity with all the methylsalicylates tested, with a preference for 4-methylsalicylate. Analogues bearing a substituent with a high electron-withdrawing effect, like 5-chloro- and 5-nitrosalicylate, were poor substrates. 2,4- and 2,6-dihydroxybenzoates were converted to products identified as trihydroxybenzenes by GC-MS, but gentisate (2,5-dihydroxybenzoate) gave no detectable product. These three compounds yielded relatively small amounts of product (if any) which did not correlate with the high NADH oxidation rates observed, indicating that the catalytic reaction was uncoupled. Anthranilate also gave rise to substantial NADH oxidation while producing little 2-aminophenol as the only detectable product. When 1-hydroxy-2-naphthoate and 2-hydroxy-1-naphthoate were provided as substrates, the NADH oxidation was the highest but no product was formed, suggesting that the reaction was fully uncoupled. To further investigate the catalytic properties of strain CHY-1 salicylate 1-hydroxylase, enzyme reactions were monitored by amperometric measurement of O2 uptake.
TABLE 1.
Substrate specificity of ht-PhnII in the NADH oxidation assay and GC-MS analysis of the products formed
| Substratea | Enzyme activityb
|
Product detected by GC-MSc
|
|||
|---|---|---|---|---|---|
| μmol NADH· min−1·mg−1 | % | Retention time (min) | Molecular mass (Da) | Identity | |
| Salicylate | 0.578 | 100 | 8.87 | 254 | Catechol |
| 3-Methylsalicylate | 0.44 | 76 | 9.86 | 268 | 3-Methylcatechol |
| 4-Methylsalicylate | 0.625 | 108 | 9.74 | 268 | 4-Methylcatechol |
| 5-Methylsalicylate | 0.396 | 68.5 | 9.74 | 268 | 4-Methylcatechol |
| 5-Chlorosalicylate | 0.267 | 46 | 10.79 | 288 | 4-Chlorocatechol |
| 5-Nitrosalicylate | 0.184 | 32 | 13.39 | 299 | 4-Nitrocatechol |
| Anthranilate | 0.39 | 67 | 10.21 | 253 | 2-Aminophenol |
| 3,5-Dichlorosalicylate | NDd | ND | 12.14 | 322 | 3,5-Dichlorocatechol |
| 2,4-Dihydroxybenzoate | 0.425 | 91 | 11.98 | 342 | Trihydroxybenzene |
| 2,6-Dihydroxybenzoate | 0.327 | 70 | 11.34 | 342 | Trihydroxybenzene |
| Gentisate | 0.462 | 99 | No product | ||
| 1-Hydroxy-2-naphthoate | 0.64 | 137 | No product | ||
| 2-Hydroxy-1-naphthoate | 0.75 | 160 | No product | ||
The initial substrate concentration was 0.5 mM except for 1-hydroxy-2-naphthoate(0.25 mM), 2-hydroxy-1-naphthoate (0.25 mM), and 5-nitrosalicylate (0.125 mM).
The values are means calculated from duplicate assays. The standard deviations are less than 5%.
Products were extracted with ethyl acetate after 2 min of reaction and analyzed as trimethylsilyl derivatives.
ND, not determined. The absorbance of the oxidation product at 340 nm precluded accurate estimation of the activity.
Uncoupled reactions catalyzed by salicylate 1-hydroxylase.
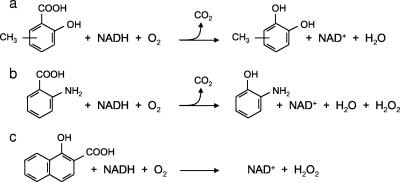
Substrate-dependent oxygen uptake by CHY-1 hydroxylase and hydrogen peroxide formation during the assays were measured for several salicylate analogues (Table 2). When possible, catechol products were quantified in order to determine the coupling efficiency between oxygen consumption and catechol formation. Fairly good coupling was observed upon enzymatic oxidation of salicylate and methylsalicylates, although a small proportion of the O2 consumed was reduced to H2O2. As the enzyme also formed hydrogen peroxide in the absence of substrate (data not shown), we assumed that this low activity might have resulted from autooxidation of the ht-PhnA3 ferredoxin, as previously observed with a similar enzyme system (13). When anthranilate and nitrosalicylate were used as substrates, a greater proportion of the oxygen consumed was converted to peroxide. The two hydroxynaphthoate molecules gave rise to high rates of oxygen uptake with no product formation. HPLC measurement at the end of the reaction revealed that the substrate concentration had not significantly decreased. These results indicated that both hydroxynaphthoates triggered a highly uncoupled enzymatic reaction, as confirmed by the high proportion of oxygen recovered as hydrogen peroxide. Some of the reactions catalyzed by the hydroxylase from strain CHY-1 are illustrated in Fig. 2.
TABLE 2.
Substrate-dependent O2 uptake, product formation, and H2O2 release in PhnII-catalyzed reactions
| Substrate | O2 uptake (μmol/min/mg)a | H2O2/O2 ratio | Amt of catechol produced (μmol/min/mg)b |
|---|---|---|---|
| Salicylate | 0.757 ± 0.007 | 0.144 | 0.64 ± 0.04 |
| 3-Methylsalicylate | 0.711 ± 0.010 | 0.23 | 0.50 ± 0.01 |
| 5-Methylsalicylate | 0.659 ± 0.005 | 0.19 | 0.61 ± 0.01 |
| 5-Chlorosalicylate | 0.630 ± 0.025 | 0.19 | ND |
| 5-Nitrosalicylate | 0.416 ± 0.005 | 0.38 | ND |
| Anthranilate | 0.630 ± 0.02 | 0.42 | ND |
| 1-Hydroxy-2-naphthoate | 3.07 ± 0.01 | 0.70 | No product |
| 2-Hydroxy-1-naphthoate | 8.56 ± 0.01 | 0.65 | No product |
Maximum velocities were calculated from the initial O2 uptake rate determined during the first minute of the time course. The PhnA3/PhnII molar ratio was 30.
Determined by HPLC quantification after about 3 min. ND, not determined (no appropriate standard was available to quantify the relevant products).
FIG. 2.
Reactions catalyzed by the three-component hydroxylase from strain CHY-1. The scheme shows reaction products formed from (a) methylsalicylate, (b) anthranilate, and (c) the pseudosubstrate 1-hydroxy-2-naphthoate.
Steady-state kinetics.
Steady-state rates of salicylate hydroxylation by the purified enzyme were measured at substrate concentrations ranging from 0.5 to 100 μM. The XylE-coupled assay was used to monitor the reaction because it directly measured the rate of product formation, whereas other assays might have introduced a bias due to the imperfect coupling of the reaction. The enzyme titration gave data that could be fitted by the Michaelis function, from which an apparent Km of 1.10 ± 0.2 μM was calculated. This value remained constant when the ht-PhnA3/ht-PhnII molar ratio was varied. On the other hand, the apparent turnover number of the hydroxylase was found to depend on this ratio and to vary from one enzyme preparation to another, probably as a function of the iron content of the enzyme. The catalytic velocity ranged from 0.73 ± 0.04 s−1 at a ratio of 15 for one preparation to 2.9 s−1 at a ratio of 30 for another preparation. Inhibition of the hydroxylase activity was observed at high salicylate concentrations, reaching 30% with 0.5 mM salicylate and 51% with 1 mM salicylate. The kinetic parameters of the enzyme with anthranilate were also examined at substrate concentrations that ranged from 5 to 400 μM. Steady-state rates were determined using NADH oxidation assays. PhnII exhibited an almost 30-fold-higher Km (28 ± 6 μM) for this substrate than for salicylate, whereas its Vmax values, which were considered the NADH oxidation rates, were very similar for the two substrates. However, since NADH oxidation was poorly coupled to anthranilate hydroxylation (Table 2), the actual rate of substrate hydroxylation was lower in the case of anthranilate.
Inhibition of salicylate hydroxylation by 1-hydroxy-2-naphthoate.
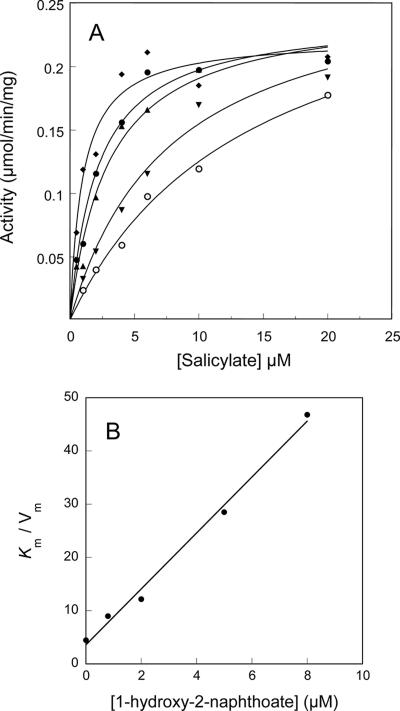
Bacterial degradation of phenanthrene leads to the intermediate metabolite 1-hydroxy-2-naphthoate, which in gram-negative species like Pseudomonas species is thought to be converted to 1,2-dihydroxynaphthalene by the same enzyme that hydroxylates salicylate (1). Since 1-hydroxy-2-naphthoate was not a substrate of the hydroxylase studied here and caused uncoupling of the reaction, we examined whether this compound inhibited salicylate hydroxylation in vitro. To this end, we obtained salicylate titration curves with various concentrations of 1-hydroxy-2-naphthoate (Fig. 3A). The results showed that the hydroxylation reaction was strongly inhibited through a mechanism that appeared to be competitive. A plot of Km/Vmax values versus inhibitor concentration gave a straight line, which allowed us to calculate an inhibition constant of 0.68 μM (Fig. 3B).
FIG. 3.
Inhibition of salicylate hydroxylation by 1-hydroxy-2-naphthoate. (A) Steady-state rates of salicylate hydroxylation measured in the presence of 1-hydroxy-2-naphthoate at the following concentrations: 0 μM (⧫), 0.8 μM (•), 2 μM (▴), 5 μM (▾), and 8 μM (○). Activities were measured using the coupled assay with an enzyme complex containing ht-PhnII and ht-PhnA3 at a molar ratio of 1:15. Michaelis curve fits were adjusted to the data points as described in Materials and Methods. (B) Plot of Km/Vmax versus inhibitor concentration.
DISCUSSION
In this paper, we describe the first characterization of a three-component salicylate 1-hydroxylase, an enzyme that appears to be present only in PAH-degrading strains of the sphingomonad group. The oxygenase component of the enzyme exhibited biochemical properties and a cofactor content very similar to those of typical ring-hydroxylating dioxygenases catalyzing the dihydroxylation of various aromatic substrates (5). In addition, the PhnII protein shows significant sequence similarity with anthranilate dioxygenase from B. cepacia DBO1 (52 and 38% identity for the α and β subunits, respectively [6]), which converts anthranilate to catechol. Nevertheless, PhnII exclusively catalyzes monohydroxylations, as exemplified by the conversion of anthranilate to 2-aminophenol. Phylogenetic analyses have previously shown that the oxygenase components of anthranilate dioxygenase from strain DBO1 and of salicylate 1-hydroxylases from sphingomonads (including PhnII) form a distinct group of enzymes, together with halobenzoate dioxygenases and salicylate 5-hydroxylase (6). Curiously, this group includes enzymes that function only as dioxygenases on the one hand or as monohydroxylases on the other, all of which use substituted benzoates as substrates. This observation raises questions about the structural features that determine whether an enzyme functions as a mono- or dioxygenase. Such a question might be addressed in experiments where a monohydroxylase is modified to acquire the ability to catalyze dihydroxylations through site-directed mutagenesis of residues predicted to be at the enzyme active site.
In in vitro assays, the catalytic activity of the hydroxylase was highly dependent on the molar ratio of the ferredoxin (PhnA3) and oxygenase (PhnII) components, and half-saturation of the activity occurred for an approximately 200-fold excess of PhnA3. Similar observations were made with other three-component oxygenases, including the naphthalene dioxygenase (PhnI) from strain CHY-1 (13). However, in the latter case, half-saturation was achieved with a 14-fold excess of PhnA3, suggesting that the ferredoxin has different affinities for the PhnI and PhnII oxygenases. This difference might have physiological significance inasmuch as the two oxygenases might compete for the same ferredoxin in vivo. Indeed, a single ferredoxin-encoding gene homologous to phnA3 was identified in the catabolic cluster responsible for aromatic hydrocarbon degradation in three sphingomonads (15, 20, 23). In vivo, the ferredoxin requirement is certainly not as high as it is in vitro, because the high protein concentration of the cytosolic compartment probably favors ferredoxin oxygenase interactions.
The three-component salicylate hydroxylase from strain CHY-1 has a low apparent Michaelis constant for its substrate similar to those reported for counterparts of the flavoprotein type found in Pseudomonas species (1.72 to 1.9 μM) (1, 29). The specific activities of the two types of enzymes are also on the same order (0.89 U/mg versus 0.27 U/mg for the P. putida enzyme [1]), although direct comparison is difficult because the in vitro activity of the CHY-1 enzyme is strongly dependent on the ferredoxin/oxygenase molar ratio. ht-PhnII appeared to be almost as active on methylsalicylates as on salicylate. This result is consistent with our finding that the aromatic ring-hydroxylating dioxygenase responsible for the initial attack on PAHs in CHY-1 can oxidize several methyl- and dimethylnaphthalenes (unpublished results), which are then predicted to be further oxidized to methylsalicylates. ht-PhnII also hydroxylated salicylates with chlorine, nitro, and hydroxyl substituents, although at lower rates. Broad substrate specificities were also observed for the two flavoprotein hydroxylases from Pseudomonas stutzeri AN10 (3) and the three-component hydroxylases from Sphingobium sp. strain P2 (22). However, enzyme activity appeared to be dependent on the nature of the substituent group, with the order of preference as follows: CH3 > Cl > NO3 > OH. This can be clearly seen by comparing the hydroxylase activities of ht-PhnII with salicylate analogues bearing one of these groups in the C-5 position (Table 1). In this respect, 5-hydroxysalicylate or gentisate was not oxidized by ht-PhnII, whereas the isomers bearing an hydroxyl in the C-4 or C-6 position were hydroxylated. This finding indicates that a polar substituent at the C-5 position of the ring prevents productive interaction of the substrate at the enzyme active site, possibly because the substrate binding pocket has a hydrophobic character that hinders access of gentisate. Anthranilate was a poor substrate for ht-PhnII, as indicated by the uncoupling of the hydroxylation reaction and the relatively high Km of the enzyme for this substrate. In comparison, the Km of anthranilate dioxygenase for anthranilate has been estimated to be about 1 μM (10).
The hydroxylation of anthranilate, like that of other poor substrates, gave rise to significant uncoupling of the reaction, resulting in the formation of hydrogen peroxide. Such a side reaction has been observed for enzymes with similar structures that function as dioxygenases (13, 17) and seems to be a consequence of the catalytic mechanism of this type of enzyme (27, 28). Indeed, it has been observed that in naphthalene dioxygenase, substrate binding to the active site increases the affinity of the enzyme for oxygen (28). Such a mechanism might explain the high rate of substrate-induced reduction of oxygen to peroxide catalyzed by PhnII in the presence of pseudosubstrates, such as hydroxynaphthoate. Interestingly, salicylate hydroxylases of the flavoenzyme type, which have a different catalytic mechanism, were also found to catalyze uncoupled reactions when they were incubated with certain salicylate analogues, such as benzoate (24).
Unlike the flavoprotein hydroxylase from P. putida (1), the enzyme from CHY-1 was unable to hydroxylate 1-hydroxy-2-naphthoate, a metabolite that is produced in the catabolic pathway of phenanthrene. Moreover, we obtained evidence that this metabolite causes inhibition of salicylate hydroxylation, which might be deleterious to bacterial cells growing on PAHs. Indeed, salicylate is an intermediate in the degradation of several PAHs, including naphthalene, anthracene, phenanthrene, and fluorene, and its hydroxylation might be a bottleneck if it is subjected to metabolic inhibition. Such inhibition could be circumvented in vivo if an enzyme oxidizes 1-hydroxy-2-naphthoate and the concentration is maintained at a level low enough to prevent inhibition of PhnII or if another salicylate hydroxylase functionally substitutes for PhnII. In the phenanthrene-degrading organism Sphingobium sp. strain P2, three isoenzymes have been shown to catalyze salicylate hydroxylation (22), and counterparts were identified in two other sphingomonads from an analysis of catabolic genes (15, 23). In S. yanoikuyae B1, the hydroxylase encoded by bphA1cA2c, which shares high sequence similarity with PhnII (the α and β subunits show 79 and 66% identity, respectively), has been proposed to catalyze hydroxylation of both salicylate and 1-hydroxy-2-naphthoate, as deduced from analysis of a bphA1c knockout mutant strain (7). This mutant was not able to grow on salicylate and naphthalene and accumulated 1-hydroxy-2-naphthoate when it was incubated with phenanthrene. However, the ability of the enzyme to catalyze the conversion of 1-hydroxy-2-naphthoate to 1,2-dihydroxynaphthalene has not been clearly demonstrated. Besides, the latter study indicated that the bphA1c product is required for hydroxylation of salicylate, implying that this enzyme could not be replaced by either of the two other isoenzymes present in the bacterium. Hence, the physiological function and raison d'être of multiple copies of three-component hydroxylases in the catabolism of PAHs by sphingomonads remain to be elucidated. Also, further work is needed to identify unambiguously the enzyme in sphingomonads responsible for the hydroxylation of 1-hydroxy-2-naphthoate, which is an essential step in phenanthrene catabolism.
Acknowledgments
We thank Jacques Gaillard for recording EPR spectra and John Willison for helpful discussions and critical reading of the manuscript.
This work was supported by grants from the Centre National de la Recherche Scientifique, the Commisariat à l'Energie Atomique, and Université Joseph Fourier to UMR5249.
Footnotes
Published ahead of print on 28 September 2007.
REFERENCES
- 1.Balashova, N. V., A. Stolz, H. J. Knackmuss, I. A. Kosheleva, A. V. Naumov, and A. M. Boronin. 2001. Purification and characterization of a salicylate hydroxylase involved in 1-hydroxy-2-naphthoic acid hydroxylation from the naphthalene- and phenanthrene-degrading bacterial strain Pseudomonas putida BS202-P1. Biodegradation 12:179-188. [DOI] [PubMed] [Google Scholar]
- 2.Blair, D., and H. Diehl. 1961. Bathophenanthroline disulphonic acid and bathocuproine disulphonic acid, water soluble reagents for iron and copper. Talanta 7:163-174. [Google Scholar]
- 3.Bosch, R., E. R. Moore, E. Garcia-Valdes, and D. H. Pieper. 1999. NahW, a novel, inducible salicylate hydroxylase involved in mineralization of naphthalene by Pseudomonas stutzeri AN10. J. Bacteriol. 181:2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Butler, C. S., and J. R. Mason. 1997. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv. Microb. Physiol. 38:47-84. [DOI] [PubMed] [Google Scholar]
- 6.Chang, H.-K., P. Mohseni, and G. J. Zylstra. 2003. Characterization and regulation of the genes for a novel anthranilate 1,2-dioxygenase from Burkholderia cepacia DBO1. J. Bacteriol. 185:5871-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, O., K. Y. Choi, G. J. Zylstra, Y. S. Kim, S. K. Kim, J. H. Lee, H. Y. Sohn, G. S. Kwon, Y. M. Kim, and E. Kim. 2005. Catabolic role of a three-component salicylate oxygenase from Sphingomonas yanoikuyae B1 in polycyclic aromatic hydrocarbon degradation. Biochem. Biophys. Res. Commun. 327:656-662. [DOI] [PubMed] [Google Scholar]
- 8.de Lorenzo, V., L. Eltis, B. Kessler, and K. N. Timmis. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17-24. [DOI] [PubMed] [Google Scholar]
- 9.Demaneche, S., C. Meyer, J. Micoud, M. Louwagie, J. C. Willison, and Y. Jouanneau. 2004. Identification and functional analysis of two aromatic ring-hydroxylating dioxygenases from a Sphingomonas strain degrading various polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70:6714-6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eby, D. M., Z. M. Beharry, E. D. Coulter, D. M. Kurtz, Jr., and E. L. Neidle. 2001. Characterization and evolution of anthranilate 1,2-dioxygenase from Acinetobacter sp. strain ADP1. J. Bacteriol. 183:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugo, N., J. Armengaud, J. Gaillard, K. N. Timmis, and Y. Jouanneau. 1998. A novel [2Fe-2S] ferredoxin from Pseudomonas putida mt2 promotes the reductive reactivation of catechol 2,3-dioxygenase. J. Biol. Chem. 273:9622-9629. [DOI] [PubMed] [Google Scholar]
- 12.Jouanneau, Y., and C. Meyer. 2006. Purification and characterization of an arene cis-dihydrodiol dehydrogenase endowed with broad substrate specificity toward PAH dihydrodiols. Appl. Environ. Microbiol. 72:4726-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jouanneau, Y., C. Meyer, J. Jakoncic, V. Stojanoff, and J. Gaillard. 2006. Characterization of a naphthalene dioxygenase endowed with an exceptionally broad substrate specificity toward polycyclic aromatic hydrocarbons. Biochemistry 45:12380-12391. [DOI] [PubMed] [Google Scholar]
- 14.Jouanneau, Y., C. Meyer, I. Naud, and W. Klipp. 1995. Characterization of an fdxN mutant of Rhodobacter capsulatus indicates that ferredoxin I serves as electron donor to nitrogenase. Biochim. Biophys. Acta 1232:33-42. [DOI] [PubMed] [Google Scholar]
- 15.Kim, E., and G. J. Zylstra. 1999. Functional analysis of genes involved in biphenyl, naphthalene, phenanthrene, and m-xylene degradation by Sphingomonas yanoikuyae B1. J. Ind. Microbiol. Biotechnol. 23:294-302. [DOI] [PubMed] [Google Scholar]
- 16.Kumari, R., S. Subudhi, M. Suar, G. Dhingra, V. Raina, C. Dogra, S. Lal, J. R. van der Meer, C. Holliger, and R. Lal. 2002. Cloning and characterization of lin genes responsible for the degradation of hexachlorocyclohexane isomers by Sphingomonas paucimobilis strain B90. Appl. Environ. Microbiol. 68:6021-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, K. 1999. Benzene-induced uncoupling of naphthalene dioxygenase activity and enzyme inactivation by production of hydrogen peroxide. J. Bacteriol. 181:2719-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leys, N. M. E. J., A. Ryngaert, L. Bastiaens, W. Verstraete, E. M. Top, and D. Springael. 2004. Occurrence and phylogenetic diversity of Sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70:1944-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelley, J. W., C. W. Garner, and G. H. Little. 1978. A simple rapid buiret method for the estimation of protein in samples containing thiols. Anal. Biochem. 86:341-343. [DOI] [PubMed] [Google Scholar]
- 20.Pinyakong, O., H. Habe, and T. Omori. 2003. The unique aromatic catabolic genes in sphingomonads degrading polycyclic aromatic hydrocarbons (PAHs). J. Gen. Appl. Microbiol. 49:1-19. [DOI] [PubMed] [Google Scholar]
- 21.Pinyakong, O., H. Habe, N. Supaka, P. Pinpanichkarn, K. Juntongjin, T. Yoshida, K. Furihata, H. Nojiri, H. Yamane, and T. Omori. 2000. Identification of novel metabolites in the degradation of phenanthrene by Sphingomonas sp. strain P2. FEMS Microbiol. Lett. 191:115-121. [DOI] [PubMed] [Google Scholar]
- 22.Pinyakong, O., H. Habe, T. Yoshida, H. Nojiri, and T. Omori. 2003. Identification of three novel salicylate 1-hydroxylases involved in the phenanthrene degradation of Sphingobium sp. strain P2. Biochem. Biophys. Res. Commun. 301:350-357. [DOI] [PubMed] [Google Scholar]
- 23.Romine, M. F., L. C. Stillwell, K. K. Wong, S. J. Thurston, E. C. Sisk, C. Sensen, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White-Stevens, R. H., and H. Kamin. 1972. Studies of a flavoprotein, salicylate hydroxylase. I. Preparation, properties, and the uncoupling of oxygen reduction from hydroxylation. J. Biol. Chem. 247:2358-2370. [PubMed] [Google Scholar]
- 25.Willison, J. C. 2004. Isolation and characterization of a novel sphingomonad capable of growth with chrysene as sole carbon and energy source. FEMS Microbiol. Lett. 241:143-150. [DOI] [PubMed] [Google Scholar]
- 26.Wittich, R. M., H. Wilkes, V. Sinnwell, W. Francke, and P. Fortnagel. 1992. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl. Environ. Microbiol. 58:1005-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolfe, M. D., and J. D. Lipscomb. 2003. Hydrogen peroxide-coupled cis-diol formation catalyzed by naphthalene 1,2-dioxygenase. J. Biol. Chem. 278:829-835. [DOI] [PubMed] [Google Scholar]
- 28.Wolfe, M. D., J. V. Parales, D. T. Gibson, and J. D. Lipscomb. 2001. Single turnover chemistry and regulation of O2 activation by the oxygenase component of naphthalene 1,2-dioxygenase. J. Biol. Chem. 276:1945-1953. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto, S., M. Katagiri, H. Maeno, and O. Hayaishi. 1965. Salicylate hydroxylase, a monooxygenase requiring flavin adenine dinucleotide. I. Purification and general properties. J. Biol. Chem. 240:3408-3413. [PubMed] [Google Scholar]
- 30.You, I. S., D. Ghosal, and I. C. Gunsalus. 1991. Nucleotide sequence analysis of the Pseudomonas putida PpG7 salicylate hydroxylase gene (nahG) and its 3′-flanking region. Biochemistry 30:1635-1641. [DOI] [PubMed] [Google Scholar]
- 31.Zhou, N. Y., J. Al-Dulayymi, M. S. Baird, and P. A. Williams. 2002. Salicylate 5-hydroxylase from Ralstonia sp. strain U2: a monooxygenase with close relationships to and shared electron transport proteins with naphthalene dioxygenase. J. Bacteriol. 184:1547-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zylstra, G. J., and E. Kim. 1997. Aromatic hydrocarbon degradation by Sphingomonas yanoikuyae B1. J. Ind. Microbiol. Biotechnol. 19:408-414. [DOI] [PubMed] [Google Scholar]