Abstract
ZO-1, ZO-2, and ZO-3, which contain three PDZ domains (PDZ1 to -3), are concentrated at tight junctions (TJs) in epithelial cells. TJ strands are mainly composed of two distinct types of four-transmembrane proteins, occludin, and claudins, between which occludin was reported to directly bind to ZO-1/ZO-2/ZO-3. However, in occludin-deficient intestinal epithelial cells, ZO-1/ZO-2/ZO-3 were still recruited to TJs. We then examined the possible interactions between ZO-1/ZO-2/ZO-3 and claudins. ZO-1, ZO-2, and ZO-3 bound to the COOH-terminal YV sequence of claudin-1 to -8 through their PDZ1 domains in vitro. Then, claudin-1 or -2 was transfected into L fibroblasts, which express ZO-1 but not ZO-2 or ZO-3. Claudin-1 and -2 were concentrated at cell–cell borders in an elaborate network pattern, to which endogenous ZO-1 was recruited. When ZO-2 or ZO-3 were further transfected, both were recruited to the claudin-based networks together with endogenous ZO-1. Detailed analyses showed that ZO-2 and ZO-3 are recruited to the claudin-based networks through PDZ2 (ZO-2 or ZO-3)/PDZ2 (endogenous ZO-1) and PDZ1 (ZO-2 or ZO-3)/COOH-terminal YV (claudins) interactions. In good agreement, PDZ1 and PDZ2 domains of ZO-1/ZO-2/ZO-3 were also recruited to claudin-based TJs, when introduced into cultured epithelial cells. The possible molecular architecture of TJ plaque structures is discussed.
Keywords: claudin, tight junction, ZO-1, ZO-2, ZO-3
Membrane-associated guanylate kinase-like homologues (MAGUKs) are thought to play a general role in creating and maintaining specialized membrane domains in various types of cells (for reviews, see Woods and Bryant 1993; Anderson 1996; Sheng 1996). These molecules contain several PDZ domains, one SH3 domain, and one guanylate kinase-like (GUK) domain. Among these domains, PDZ domains bind to COOH-terminal ends of various proteins, especially integral membrane proteins, most of which end in valine. Thus, MAGUKs can cross-link multiple integral membrane proteins at the cytoplasmic surface of plasma membranes to establish specialized membrane domains. For example, at the post-synaptic membranes, the COOH termini of Shaker-type K+ channel (-ETDV) and NMDA receptor (-ESDV) were reported to be specifically recognized and cross-linked by the PDZ domains of PSD-95 family members that belong to MAGUKs (Kim et al. 1995; Kornau et al. 1995; Niethammer et al. 1996). The list of MAGUKs is growing rapidly.
Tight junctions (TJs) are one of the specialized domains of plasma membranes (for reviews see Schneeberger and Lynch 1992; Cereijido et al. 1998; Yap et al. 1998; Tsukita and Furuse 1999). TJs occur at the most apical part of lateral membranes of simple epithelial cells, and are considered to be involved in barrier and fence functions; TJs create a primary barrier to the diffusion of solutes through the paracellular pathway, and also function as a fence (a diffusion barrier within plasma membranes) to create and maintain apical and basolateral membrane domains. On freeze-fracture electron microscopy, TJs appear as a continuous, anastomosing network of intramembranous particle strands (TJ strands) (Staehelin 1974). In TJs, individual TJ strands are associated laterally with other TJ strands in apposing membranes of adjacent cells to form paired strands, where the extracellular space is completely obliterated. These TJ strands are further associated with actin filaments through their plaque structures underlying plasma membranes (Madara 1987; Keon et al. 1996). To date, in the TJ plaque structures, three distinct MAGUKs called ZO-1, ZO-2, and ZO-3 have been identified (Mitic and Anderson 1998).
ZO-1 with a molecular mass of 220 kD was first identified as an antigen for a monoclonal antibody raised against the junction-enriched fraction from the liver (Stevenson et al. 1986). Then, ZO-2 was identified as a 160-kD protein that was coimmunoprecipitated with ZO-1 from cell lysates (Gumbiner et al. 1991). A phosphorylated 130-kD protein was also found in the ZO-1 immunoprecipitate (Balda et al. 1993) and is now called ZO-3. Immunofluorescence as well as electron microscopic analyses revealed that ZO-1, ZO-2, and ZO-3 are exclusively concentrated at the cytoplasmic surface of TJs in the immediate vicinity of plasma membranes, although our knowledge of ZO-3 is still fragmentary as compared with ZO-1 and ZO-2. Cloning and sequencing cDNAs encoding these molecules showed that all have three PDZ domains (PDZ1 to -3), one SH3 domain, and one GUK domain in this order from their NH2 termini, indicating that ZO-1, ZO-2, and ZO-3 belong to MAGUKs (Itoh et al. 1993; Willott et al. 1993; Jesaitis and Goodenough 1994; Haskins et al. 1998).
Recent analyses gradually unraveled the molecular interactions around ZO-1, ZO-2, and ZO-3 in the TJ plaque structures. ZO-1 and ZO-2 bind directly to actin filaments at their COOH-terminal regions, suggesting that these molecules function as cross-linkers between TJ strands and actin filaments (Itoh et al. 1997, Itoh et al. 1999; Fanning et al. 1998). ZO-2 has been reported to associate with ZO-1 by the PDZ2/PDZ2 interaction (Fanning et al. 1998; Itoh et al. 1999). ZO-3 has also been reported to associate with ZO-1, but not with ZO-2, although the domains responsible for ZO-3/ZO-1 interaction remain unidentified (Haskins et al. 1998). The important question is how ZO-1, ZO-2, and ZO-3 are recruited to the cytoplasmic surface of TJ strands, i.e., the identities of the membrane binding partner(s) for these MAGUKs.
Occludin with a molecular mass of ∼65 kD was first identified as a component of the TJ strand per se (Furuse et al. 1993; Ando-Akatsuka et al. 1996). This molecule has four-transmembrane domains with both NH2 and COOH termini in the cytoplasm and the COOH-terminal peptide of 150 amino acids of its cytoplasmic tail was reported to directly bind to ZO-1 and ZO-2, indicating that occludin is a membrane-binding partner for ZO-1 and ZO-2 (Furuse et al. 1994; Itoh et al. 1999). ZO-3 was also shown to directly bind to the cytoplasmic domain of occludin (Haskins et al. 1998). However, when occludin-deficient ES cells were produced and differentiated into epithelium-like cells (visceral endoderm cells), they bore well-developed TJs, to which ZO-1 was still recruited (Saitou et al. 1998). Most recently, other four-transmembrane domain proteins, claudins, have been shown to constitute the backbone of TJ strands (Furuse et al. 1998a,Furuse et al. 1998b). Claudins constitute a new gene family, the “claudin” family, which includes at least 16 members (Morita et al. 1999; Tsukita and Furuse 1999; Simon et al. 1999). Interestingly, most of the claudin family members end in YV at their COOH termini, which are good candidates for the binding partners for PDZ domains (see Fig. 5 B).
Figure 5.
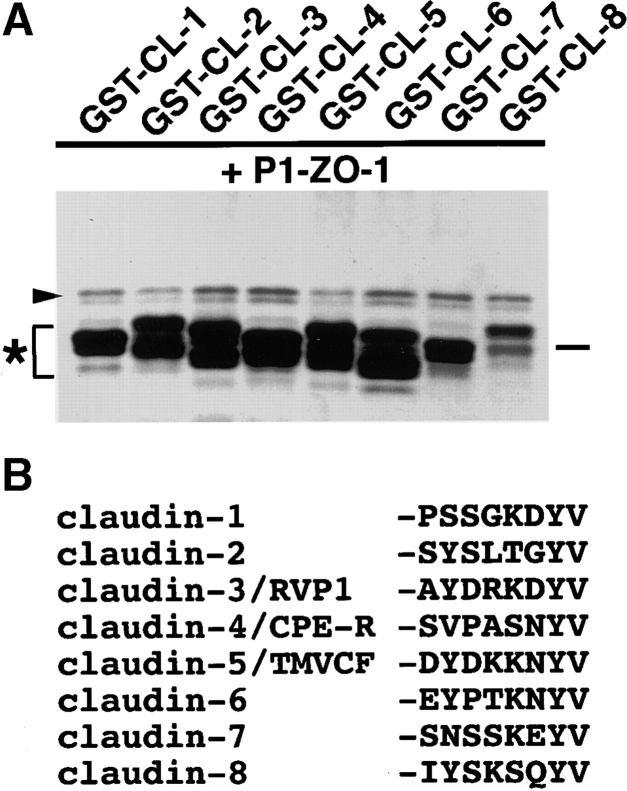
Association of PDZ1 domains of ZO-1 (P1-ZO-1) with the COOH-terminal YV sequence of claudin-1 to -8 (GST-CL-1 to -8). (A) In vitro binding analyses similar to Fig. 4 were performed. P1-ZO-1 bound to all of GST-CL-1 to -8 (arrowhead). Some of the GST fusion proteins (asterisk) were partly degraded. Bar indicates a molecular mass of 31 kD. (B) COOH-terminal amino acid sequences of claudin-1 to -8. All end in YV.
In this study, we first found that not only ZO-1 but also ZO-2 and ZO-3 were still recruited to TJs in intestinal epithelial cells of occludin-deficient mice. We then examined the possible interaction between ZO-1/ZO-2/ZO-3 and the YV sequence of the COOH termini of claudins. In vitro as well as in vivo analyses revealed that PDZ1 domains of ZO-1, ZO-2, and ZO-3 have binding affinities to the COOH-terminal YV of claudins. These findings suggested a new molecular architecture model for the TJ plaque structures.
Materials and Methods
Antibodies and Cells
Mouse anti–ZO-1 mAbs (T8-754 and T7-713), rabbit anti–ZO-2 pAb (pAb62), guinea pig anti–claudin-1 pAb, rat anti–claudin-1 mAb, and rat anti–claudin-2 mAb were described previously (Itoh et al. 1991, Itoh et al. 1993, Itoh et al. 1999; Furuse et al. 1999). The mAbs, T8-754 and T7-713, recognize the NH2- and COOH-terminal halves of ZO-1, respectively. Anti–ZO-3 pAb was raised in rabbits using GST fusion protein with COOH-terminal region (amino acids [aa] 758–904) of mouse ZO-3 as an antigen. Mouse anti–c-myc tag and anti-His tag mAbs were purchased from Sigma Chemical Co.
Mouse L cells, dog MDCK cells, and their transfectants were cultured in DME supplemented with 10% FCS. C1L and C2L cells were described previously (Furuse et al. 1999).
Cloning of cDNA Encoding Mouse ZO-3
Using the BLAST search program, we found that the sequence of mouse EST clone AA016964 was highly homologous but not identical to the PDZ1 domains of ZO-1 and ZO-2, leading to the isolation of full-length cDNA encoding this sequence. During the course of this isolation, a cDNA encoding dog ZO-3 was reported (Haskins et al. 1998) and its sequence was almost the same as that of AA016964, suggesting that AA016964 encodes part of mouse ZO-3. Then, specific primers were synthesized from the sequence of AA016964, and a partial cDNA of mouse ZO-3 was obtained by RT-PCR, which was used as a probe to screen a mouse lung λ ZAP cDNA library. Four positive clones were obtained and sequenced to clarify the entire open reading frame of mouse ZO-3 cDNA. The full-length mouse ZO-3 cDNA was subcloned into pSK- to produce pSK-ZO-3. The mouse ZO-3 sequence has been submitted to EMBL/GenBank/DDBJ under accession number AF157006.
Constructs and Transfection
To fuse a c-myc epitope tag onto the COOH terminus of each construct, the eukaryotic expression vector pME18S-7myc (Itoh et al. 1997) was used. For production of mammalian expression vectors for c-myc–tagged full-length ZO-2 and ZO-3 (pME18S-ZO-2-7myc and pME18S-ZO-3-7myc, respectively), cDNA fragments encoding the entire open reading frame of mouse ZO-2 and ZO-3 were produced by PCR using pSK-ZO-2 (Itoh et al. 1999) and pSK-ZO-3 as template, respectively, and subcloned into pME18S-7myc.
Various deletion mutant constructs with a c-myc tag at their 3′-end were also produced by PCR using the following primers; sense primer from position 344 containing a StuI site and antisense primer from position 3549 containing a SalI site for ZO-2 lacking PDZ1 (pME18S-ΔPDZ1-ZO-2-7myc), sense primer from position 1208 containing a StuI site and antisense primer from position 3549 containing a SalI site for ZO-2 lacking both PDZ1 and -2 (pME18S-ΔPDZ1,2-ZO-2-7myc), sense primer from position 492 containing an EcoRV site and antisense primer from position 2918 containing an XhoI site for ZO-3 lacking PDZ1 (pME18S-ΔPDZ1-ZO-3-7myc), sense primer from position 993 containing EcoRV site and antisense primer from position 2918 containing an XhoI site for ZO-3 lacking both PDZ1 and -2 (pME18S-ΔPDZ1,2-ZO-3-7myc). For the production of ZO-2 lacking PDZ2 (pME18S-ΔPDZ2-ZO-2-7myc), two fragments were amplified by PCR using the sense primer from position 44 containing an EcoRI site and antisense primer from position 886 containing an ApaI site, and sense primer from position 1208 containing an ApaI site and antisense primer from position 3549 containing a SalI site. Two fragments were simultaneously ligated into pME18S-7myc after digestion with the appropriate restriction enzymes. Similarly, for production of ZO-3 lacking PDZ2 (pME18S-ΔPDZ2-ZO-3-7myc), two fragments were amplified by PCR using the sense primer from position 198 containing an EcoRV site and antisense primer from position 761 containing a SacI site, using the sense primer from position 993 containing a SacI site and antisense primer from position 2918 containing an XhoI site, then simultaneously ligated into pME18S-7myc. For production of claudin-1 lacking its COOH-terminal YV (pCAGCL-1ΔYV), the cDNA fragment was amplified by PCR using the sense primer from position 1 and antisense primer from 626 containing a stop codon. The obtained fragment was blunted with T4 polymerase and ligated into pCAGGSneodelEcoRI (Niwa et al. 1991), which was provided by Dr. J. Miysazaki (Osaka University).
Cells (107) were cotransfected with 2 μg of each expression vector and 0.1 μg of pPGKpuro using Lipofectamine reagent for 5 h in Opti-MEM (GIBCO BRL). After 43 h, cells were replated onto four 9-cm dishes in the presence of 8 μg/ml of puromycin to select stable transfectants. Colonies of puromycin-resistant cells were isolated and screened by either immunoblotting or immunofluorescence with anti–c-myc tag mAb or guinea pig anti–claudin-1 pAb.
To construct GFP-tagged expression vectors, GFP cDNA was excised from pQBI25 and introduced into pCAGGSneodelEcoRI generating pCAG-GFP. The following cDNAs were amplified by PCR and subcloned into SwaI-digested pCAG-GFP to express each PDZ domain tagged with GFP at its COOH-terminal end: PDZ1 (aa 19–113), PDZ2 (aa 181–292), or PDZ3 (aa 423–503) of ZO-1, PDZ1 of ZO-2 (aa 4–100), and PDZ1 of ZO-3 (aa 5–96). These GFP-tagged proteins were introduced into cultured MDCK cells as described previously (Itoh et al. 1999).
Immunofluorescence Microscopy
Cells plated on glass coverslips were rinsed in PBS and fixed with 1% formaldehyde in PBS for 15 min at room temperature. Cells were then treated with 0.3% Triton X-100 in PBS for 15 min and washed three times with PBS. After soaking in PBS containing 1% BSA, the samples were treated with primary antibodies for 1 h in a moist chamber. They were then washed three times with PBS, followed by incubation for 30 min with appropriate secondary antibodies. The samples were washed with PBS three times, embedded in 95% glycerol-PBS containing 0.1% para-phenylendiamine and 1% n-propylagate, and examined with a photomicroscope (model Axiophoto; Carl Zeiss). Frozen sections of the intestine of occludin-deficient or control mice were immunofluorescently stained as described previously (Itoh et al. 1997).
In Vitro Binding Assay Using Fusion Proteins
The cDNA fragments encoding the cytoplasmic domain of claudin-1 to -8 and the cytoplasmic domain of claudin-1 lacking its COOH-terminal YV were amplified using specific primers and subcloned into pGEX vector (see Morita et al. 1999). Recombinant NH2-terminal half of ZO-1 (N-ZO-1; aa 1–862), COOH-terminal half of ZO-1 (C-ZO-1; aa 863–1745), 6xHis-tagged NH2-terminal portion of ZO-2 (N-ZO-2; aa 1–938), and 6xHis-tagged COOH-terminal portion of ZO-2 (C-ZO-2; aa 939–1167) were produced in Sf9 cells by baculovirus infection (Itoh et al. 1997, Itoh et al. 1999). For various mutants of ZO-1/ZO-2/ZO-3 carrying 6xHis tag, the following cDNAs were amplified by PCR and subcloned into pET32 (Novagen): PDZ1 of ZO-1 (P1-ZO-1; aa 19–113), PDZ2 of ZO-1 (P2-ZO-1; aa 181–292), PDZ3 of ZO-1 (P3-ZO-1; aa 423–503), PDZ1 of ZO-2 (P1-ZO-2; aa 4–100), PDZ2 of ZO-2 (P2-ZO-2; aa 282–388), and PDZ3 of ZO-2 (P3-ZO-2; aa 491–571), full-length of ZO-3 (aa 1–904), PDZ1 of ZO-3 (P1-ZO-3; aa 5–96), PDZ2 of ZO-3 (P2-ZO-3; aa 189–263), and PDZ3 of ZO-3 (P3-ZO-3; aa 371–449). These recombinant proteins were expressed in E. coli (BL21).
In vitro binding assay was performed basically as previously described (Itoh et al. 1997). In brief, GST fusion proteins with the cytoplasmic domains of claudins expressed in E. coli were purified using glutathione–Sepharose 4B beads (Amersham Pharmacia Biotech Ltd.). After washing with PBS, the beads were incubated with lysate of Sf9 cells or E. coli expressing each recombinant protein of ZO-1, ZO-2, and ZO-3 described above followed by washing with PBS containing 1% Triton X-100. Then, bound proteins were eluted with 300 μl of 50 mM Tris-HCl buffer (pH 8.0) containing 20 mM glutathione. Each eluate was analyzed by SDS-PAGE.
To estimate the dissociation constant between the cytoplasmic domain of claudin-1 and the PDZ1 domains of ZO-1/ZO-2/ZO-3, 200 μl of the glutathione–Sepharose bead slurry containing 40 μg of GST-claudin-1 was incubated with 2 ml of the E. coli lysate containing 0.01–0.5 μg of the PDZ1 domain of ZO-1, ZO-2, or ZO-3. The beads were washed and bound proteins were eluted with 1 ml of 50 mM Tris-HCl buffer (pH 8.0) containing 10 mM glutathione. The amounts of recombinant PDZ1 domains in the cell lysate or in each eluate were estimated by comparing the intensity of their Coomassie brilliant blue–stained bands with those of various amounts of BSA using Adobe Photoshop™ 3.0J histogram, and Scatchard plot of the data was generated. Experiments were repeated three times for each estimation of K d.
Immunoprecipitation
For metabolic labeling of transfectants, cells cultured on 9-cm dishes were washed twice with methionine-free medium supplemented with 2% FCS, and then incubated with 3 ml of the same medium containing 0.2 mCi [35S]methionine for 5 h. After washing with ice-cold PBS three times, cells were lysed in 2 ml of extraction buffer (1% Triton X-100, 0.5% sodium deoxycholate, 0.2% SDS, 150 mM NaCl, 10 mM Hepes [pH 7.4], 2 mM phenylmethylsulfonyl, 2 mg/ml leupeptin). Cell lysates were clarified by centrifugation at 100,000 g for 30 min and incubated with 50 μl of protein G–Sepharose bead slurry (Zymed Laboratories Inc.) coupled with anti–c-myc tag mAb for 3 h. After washing five times with the extraction buffer, immunoprecipitates were eluted from beads with the SDS-PAGE sample buffer.
Gel Electrophoresis and Immunoblotting
One-dimensional SDS-PAGE (10–12.5%) was performed based on the method of Laemmli 1970. Gels were stained with Coomassie brilliant blue R-250 and exposed to imaging plates and analyzed using a BAS2000 (Fuji Film). For immunoblotting, proteins separated by SDS-PAGE were electrophoretically transferred onto nitrocellulose sheets, which were then incubated with primary antibodies. The antibodies were detected with a blotting detection kit (Amersham Pharmacia Biotech Ltd.).
Results
Recruitment of ZO-1, ZO-2, and ZO-3 to TJs of Intestinal Epithelial Cells in Occludin-deficient Mice
We previously produced occludin-deficient embryonic stem cells, and differentiated them into epithelium-like cells (visceral endoderms) (Saitou et al. 1998). Unexpectedly, these cells bore well-developed TJs, where ZO-1 was still concentrated. In this study, we first examined whether ZO-2 and ZO-3 as well as ZO-1 are recruited to TJs of highly polarized epithelial cells such as intestinal epithelial cells in occludin-deficient mice, which were born normally (Saitou, M., unpublished data). Frozen sections of occludin-deficient intestinal epithelial cells were double stained with anti-occludin mAb/anti–claudin-3 pAb, anti–claudin-3 pAb/anti–ZO-1 mAb, anti–ZO-1 mAb/anti–ZO-2 pAb, or anti–ZO-1 mAb/anti–ZO-3 pAb. As shown in Fig. 1, not only ZO-1 but also ZO-2 and ZO-3 were targeted to and concentrated at TJs of these well-polarized occludin-deficient epithelial cells. These findings suggested the possible interactions between claudins and ZO-1/ZO-2/ZO-3, prompting us to in vitro binding analyses.
Figure 1.
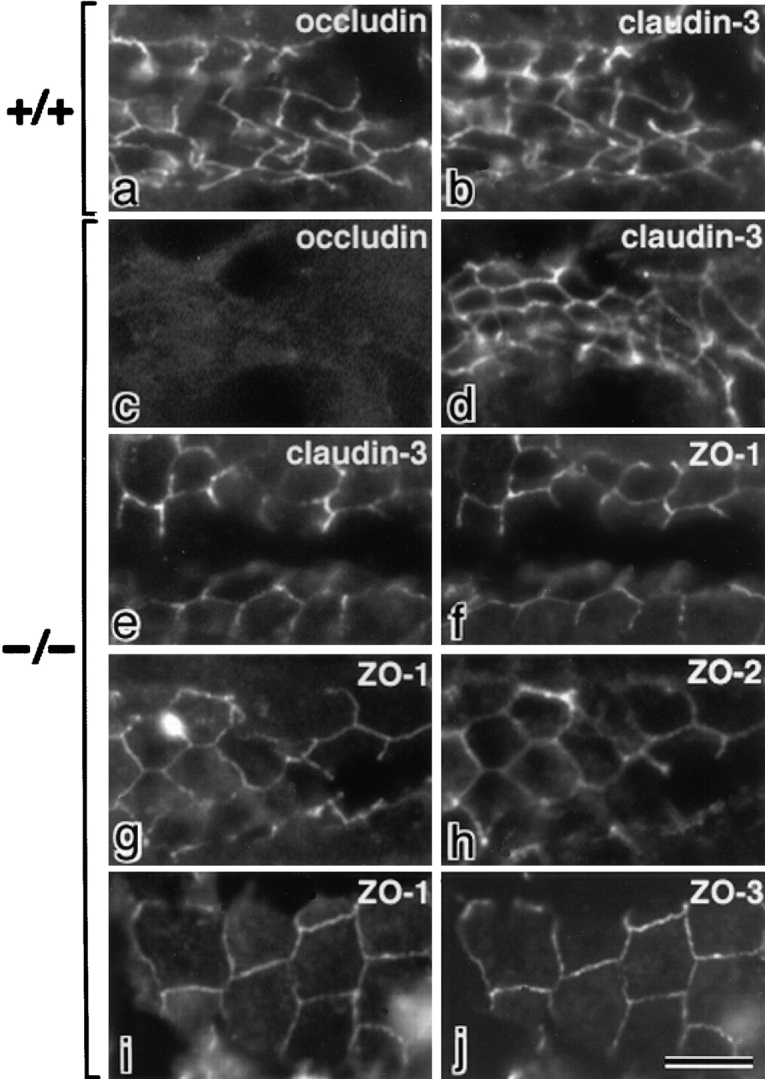
Recruitment of ZO-1/ZO-2/ZO-3 to TJs of intestinal epithelial cells in occludin-deficient mice. Frozen sections of intestinal epithelial cells in wild-type (a and b) and occludin-deficient mice (c–j) were double stained with rat anti–occludin mAb (a and c)/rabbit anti–claudin-3 pAb (b and d), rabbit anti–claudin-3 pAb (e)/mouse anti–ZO-1 mAb (f), mouse anti–ZO-1 mAb (g)/rabbit anti–ZO-2 pAb (h), or mouse anti–ZO-1 mAb (i)/rabbit anti–ZO-3 pAb (j). In occludin-deficient intestinal epithelial cells, not only ZO-1 but also ZO-2 and ZO-3 were concentrated at the claudin-3–positive TJs. Bar, 10 μm.
In Vitro Binding of ZO-1, ZO-2, and ZO-3 to the COOH Termini of Claudins
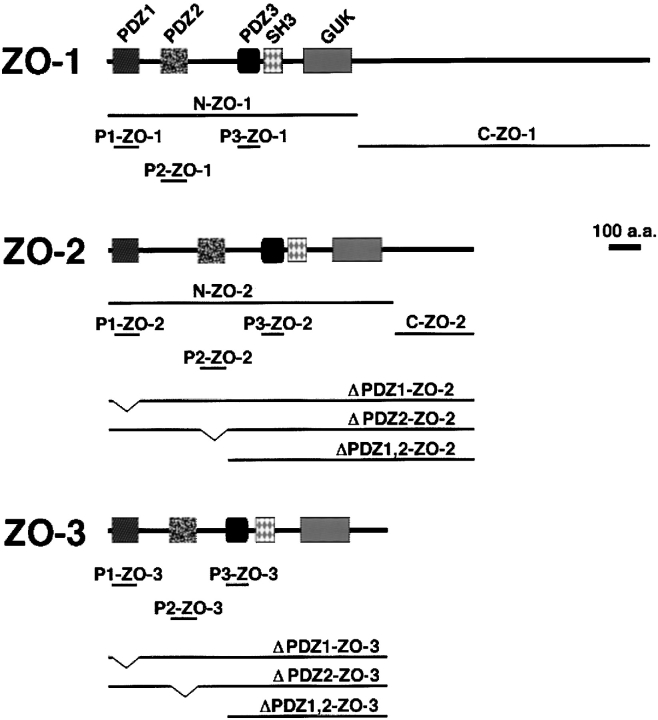
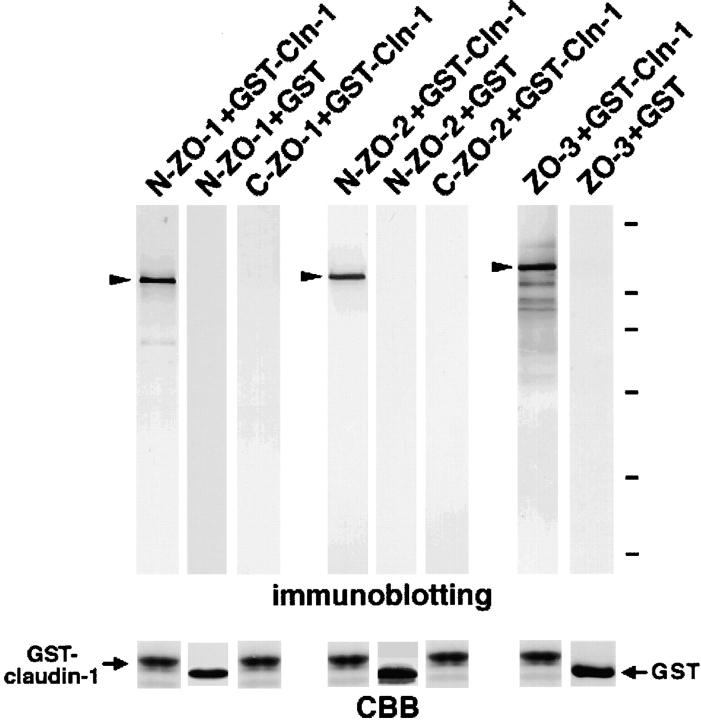
As shown in Fig. 2, various constructs for ZO-1, ZO-2, and ZO-3 were used in this study for in vitro biding assays and in vivo colocalization analyses. First, we produced recombinant N-ZO-1, C-ZO-1, N-ZO-2 (6xHis tag), and C-ZO-2 (6xHis tag) in Sf9 cells by baculovirus infection (Itoh et al. 1997, Itoh et al. 1999), and full-length ZO-3 in E. coli, and their cell lysates containing the same amount of recombinant proteins were mixed with beads conjugated with the GST-fusion protein with the cytoplasmic domain of claudin-1. Bound proteins were then eluted from beads and each eluate was subjected to SDS-PAGE followed by immunoblotting with corresponding antibodies. As shown in Fig. 3N-ZO-1, N-ZO-2, and full-length ZO-3, but not C-ZO-1 or C-ZO-2, appeared to specifically bind to the GST-fusion protein with the cytoplasmic domain of claudin-1.
Figure 2.
Structures of ZO-1, ZO-2, and ZO-3 and their deletion mutants. ZO-1, ZO-2, and ZO-3 contain three PDZ domains (PDZ1, PDZ2, PDZ3), one SH3 domain (SH3) and one guanylate kinase-like domain (GUK) in this order from their NH2 termini. ZO-1 and ZO-2 were divided into NH2-terminal (N-ZO-1, N-ZO-2) and COOH-terminal regions (C-ZO-1, C-ZO-2). When these constructs were expressed in Sf9 cells by baculovirus infection, N- and C-ZO-2, but not N- or C-ZO-1, were tagged with 6xHis. When the other constructs were expressed in E. coli or mammalian cells, they were tagged with 6xHis or c-myc, respectively.
Figure 3.
Association of N-ZO-1, N-ZO-2, and full-length ZO-3 with the cytoplasmic domain of claudin-1 in vitro. The GST fusion protein with the cytoplasmic domain of claudin-1 (GST-Cln-1) or GST protein (GST), which was bound to glutathione–Sepharose beads, was incubated with the lysate of Sf9 cells expressing N-ZO-1, C-ZO-1, 6xHis-N-ZO-2, or 6xHis-C-ZO-2, or with the lysate of E. coli expressing 6xHis-tagged full-length ZO-3. In each lysate, the amount of recombinant protein was adjusted to be the same. After washing, the proteins associated with GST fusion protein or GST were eluted from the beads with a buffer containing glutathione. The eluates from N-ZO-1–, C-ZO-1–, 6xHis-N-ZO-2–, 6xHis-C-ZO-2–, or 6xHis-ZO-3–incubated beads were separated by SDS-PAGE followed by immunoblotting with anti–ZO-1 mAb T8754, anti–ZO-1 mAb T7713, anti-His tag mAb, anti-His tag mAb, or anti-His tag mAb (immunoblotting). The amount of GST–claudin-1 (GST-claudin-1) and GST (GST) in each eluate was determined by Coomassie brilliant blue staining (CBB). As indicated by arrowheads, N-ZO-1, N-ZO-2, full-length ZO-3, but not C-ZO-1 or C-ZO-2, showed binding affinity to the cytoplasmic domain of claudin-1. Bars indicate molecular masses of 200, 116, 97, 66, 45, and 31 kD, respectively, from the top.
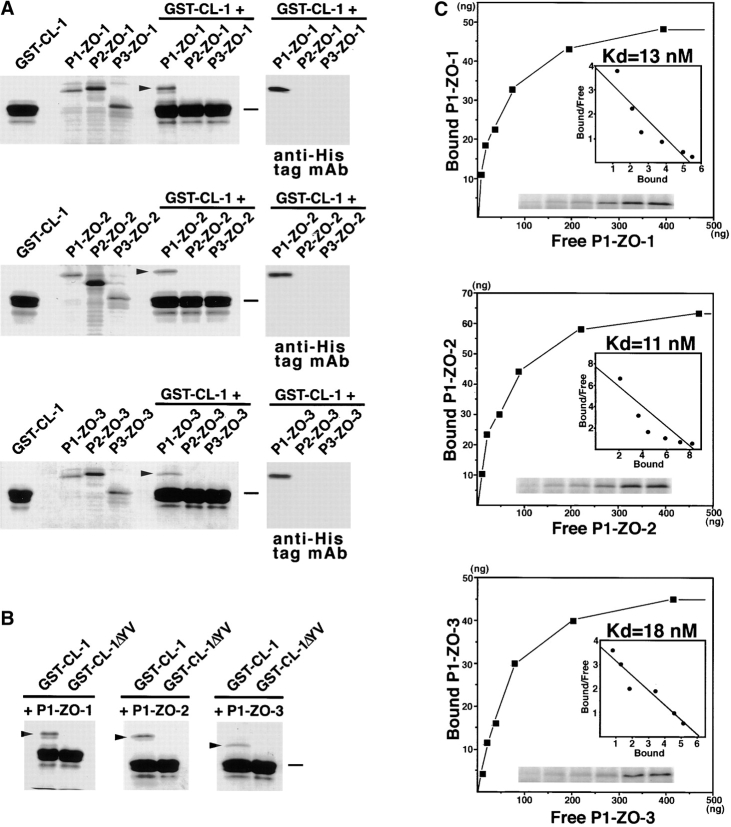
Next, using similar in vitro binding assays, we narrowed down the domains responsible for the ZO-1/ZO-2/ZO-3–claudin-1 interaction. Because preliminary experiments showed that PDZ domains of ZO-1/ZO-2/ZO-3, but not SH3 or GUK domains, are involved in this interaction, we produced recombinant 6xHis-tagged PDZ1, -2, and -3 domains of ZO-1, ZO-2, and ZO-3 in E. coli, and performed the in vitro binding analyses with the GST fusion protein with the cytoplasmic domain of claudin-1. In these analyses, the direct binding was evaluated by Coomassie brilliant blue staining of each eluate. As a result, among nine recombinant PDZ domains, only PDZ1 domains of ZO-1, ZO-2, and ZO-3 were specifically associated with the GST–claudin-1 fusion protein (Fig. 4 A). Interestingly, these PDZ1 domains lost their binding affinity to the cytoplasmic domain of claudin-1, when the YV sequence was deleted from the COOH terminus of claudin-1 (Fig. 4 B). We then estimated the dissociation constant between the cytoplasmic domain of claudin-1 and PDZ1 domains of ZO-1/ZO-2/ZO-3 (Fig. 4 C). For this purpose, various concentrations of these PDZ1 domains were incubated with beads bearing the GST fusion protein with the cytoplasmic domain of claudin-1 or GST (for control). The binding constant was estimated by subtracting the binding to the GST-conjugated Sepharose beads (Itoh et al. 1997). For each PDZ1 domain the binding was saturable, and Scatchard analysis revealed a single class of affinity binding sites with K d's of ∼13, 11, and 18 nM for the PDZ1 domain of ZO-1, ZO-2, and ZO-3, respectively.
Figure 4.
Association of PDZ1 domains of ZO-1, ZO-2, and ZO-3 with the COOH-terminal YV sequence of claudin-1. All SDS-PAGE gels were stained with Coomassie brilliant blue. (A) The GST fusion protein with the cytoplasmic domain of claudin-1 (GST-CL-1), which was bound to glutathione–Sepharose beads, was incubated with the lysate of E. coli expressing 6xHis-PDZ1 domains (P1-ZO-1, P1-ZO-2, P1-ZO-3), 6xHis-PDZ2 domains (P2-ZO-1, P2-ZO-2, P2-ZO-3), and 6xHis-PDZ3 domains (P3-ZO-1, P3-ZO-2, P3-ZO-3) of ZO-1, ZO-2, and ZO-3. After washing, the proteins associated with GST fusion proteins were eluted from the beads with a buffer containing glutathione. The eluates from PDZ domain-incubated beads were separated by SDS-PAGE followed by Coomassie brilliant blue staining or by immunoblotting with anti-His tag mAb (anti-His tag mAb). Among the 9 PDZ domains, only P1-ZO-1, P1-ZO-2, and P1-ZO-3 were associated with the cytoplasmic domain of claudin-1 (arrowheads). (B) Similar binding assay was performed between P1-ZO-1/P1-ZO-2/P1-ZO-3 and GST fusion protein with the COOH-terminal YV-deleted cytoplasmic domain of claudin-1 (GST-CL-1ΔYV). P1-ZO-1, P1-ZO-2, and P1-ZO-3 bound to GST-CL-1 (arrowheads), but not to GST-CL-1ΔYV. Bars indicate a molecular mass of 31 kD. (C) Quantitative analysis of the binding between PDZ1 domains of ZO-1/ZO-2/ZO-3 and the cytoplasmic domain of claudin-1. Glutathione–Sepharose bead slurry containing GST-claudin-1 was incubated with E. coli lysate containing 0.01–0.5 μg of the PDZ1 domain of ZO-1, ZO-2, or ZO-3 (from the top). The amounts of the PDZ1 domain of ZO-1, ZO-2, or ZO-3 in the E. coli lysate and in each eluate (inset) were estimated as described in Materials and Methods. Each point represents the mean value of triplicate determinations. The binding was saturable, and Scatchard analysis (inset) indicated that the K d values for the PDZ1 domains of ZO-1 (P1-ZO-1), ZO-2 (P1-ZO-2), and ZO-3 (P1-ZO-3) were ∼13, 11, and 18 nM, respectively.
Finally, with the similar binding assays, we examined the binding ability of the PDZ1 domain of ZO-1 to the cytoplasmic domains of claudin-1 to -8 (Fig. 5). Since some GST fusion proteins with the cytoplasmic domains of claudins were easily degraded, it was difficult to estimate the amount of GST fusion proteins precisely, but it is clear that the PDZ1 domain of ZO-1 directly bound to the cytoplasmic domain of claudin-1 to -8. PDZ1 domains of ZO-2 and ZO-3 also showed binding affinity to claudin-1 to -8 (data not shown).
Recruitment of Endogenous ZO-1 to Claudin-based Networks in L Transfectants
In vitro binding analyses indicated that the PDZ1 domains of ZO-1, ZO-2, and ZO-3 are directly associated with the COOH termini of claudins. To examine these bindings in vivo, the immunoprecipitation experiments with epithelial cells would be one of the most appropriate approaches, but this method does not work in the case of claudins; the claudin molecules are polymerized into strands which are highly insoluble in detergents. The transfection experiments with epithelial cells would also be another appropriate approach, but the data obtained are not easily interpreted, since multiple species of claudins as well as ZO-1/ZO-2/ZO-3 are endogenously expressed in epithelial cells. To avoid these technical difficulties, we used L transfectants expressing various types of claudin. As shown previously, parental L cells lack either TJs or the expression of claudins and occludin (Furuse et al. 1998b). When claudin-1 or -2 was singly transfected into L cells (C1L and C2L cells, respectively), introduced claudins were concentrated at cell–cell borders as planes in an elaborate network pattern at the immunofluorescence microscopic level, where they reconstituted well-developed networks of TJ strands at the electron microscopic level (Furuse et al. 1998b, Furuse et al. 1999). Using these C1L and C2L cells, we evaluated the interactions of various constructs of ZO-1/ZO-2/ZO-3 with claudins by examining whether they were recruited to the claudin-based networks.
As shown in Fig. 6, ZO-1, but not ZO-2 or ZO-3, was expressed endogenously in parental L cells (and also in C1L and C2L cells). This endogenous ZO-1 did not show characteristic concentration in L cells (Fig. 7, a and b). In contrast, in both C1L and C2L cells in which claudin-1 and claudin-2 were concentrated at cell–cell borders in an elaborate network, respectively, endogenous ZO-1 was colocalized with claudins (Fig. 7, c–f). Close inspection revealed that ZO-1 was also concentrated as elaborate networks, which almost overlapped with those based on claudin (Fig. 7, c–f, insets). When claudin-1 mutant lacking its COOH-terminal YV was transfected into L cells (C1ΔYVL cells), introduced claudin-1 mutant was again concentrated at cell–cell borders as elaborate networks (Fig. 7 g). Interestingly, however, ZO-1 was not recruited to these claudin-based networks in C1ΔYVL cells (Fig. 7 h). These findings, together with the in vitro binding data suggested that ZO-1 also directly binds to the COOH-terminal YV of claudins through its PDZ1 domain inside cells.
Figure 6.
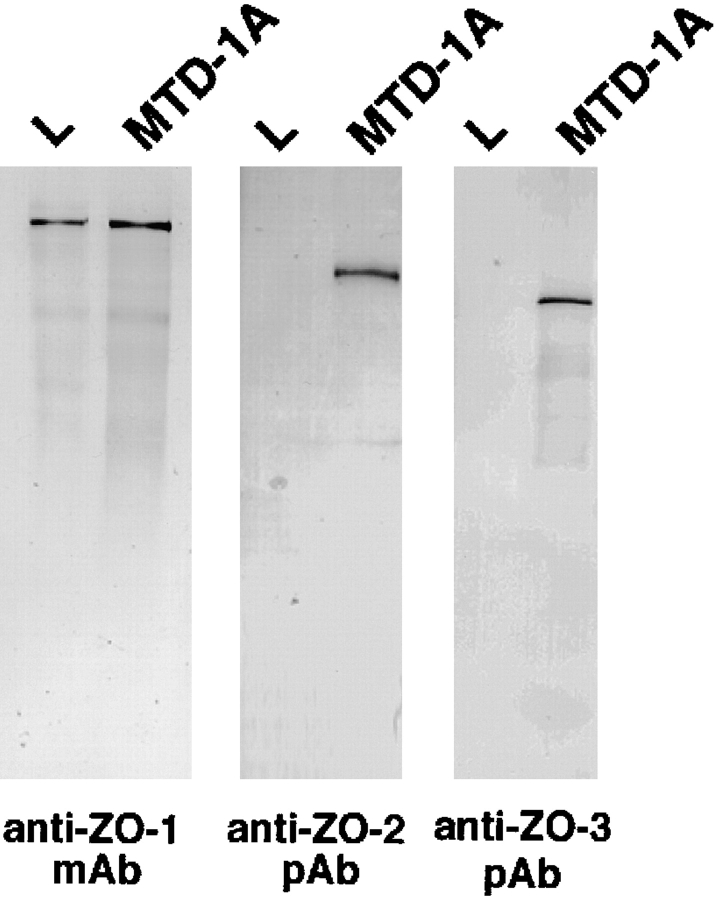
Expression of ZO-1, ZO-2, and ZO-3 in mouse L fibroblasts. The total cell lysate of cultured mouse L fibroblasts and MTD-1A epithelial cells were subjected to immunoblotting with anti–ZO-1 mAb (anti–ZO-1 mAb), anti–ZO-2 pAb (anti–ZO-2 pAb), or anti–ZO-3 pAb (anti–ZO-3 pAb). ZO-1, ZO-2, and ZO-3 were expressed in MTD-1A cells, whereas only ZO-1 was detected in L fibroblasts. Bars indicate molecular masses of 200, 116, 97, 66, 45, and 31 kD, respectively, from the top.
Figure 7.
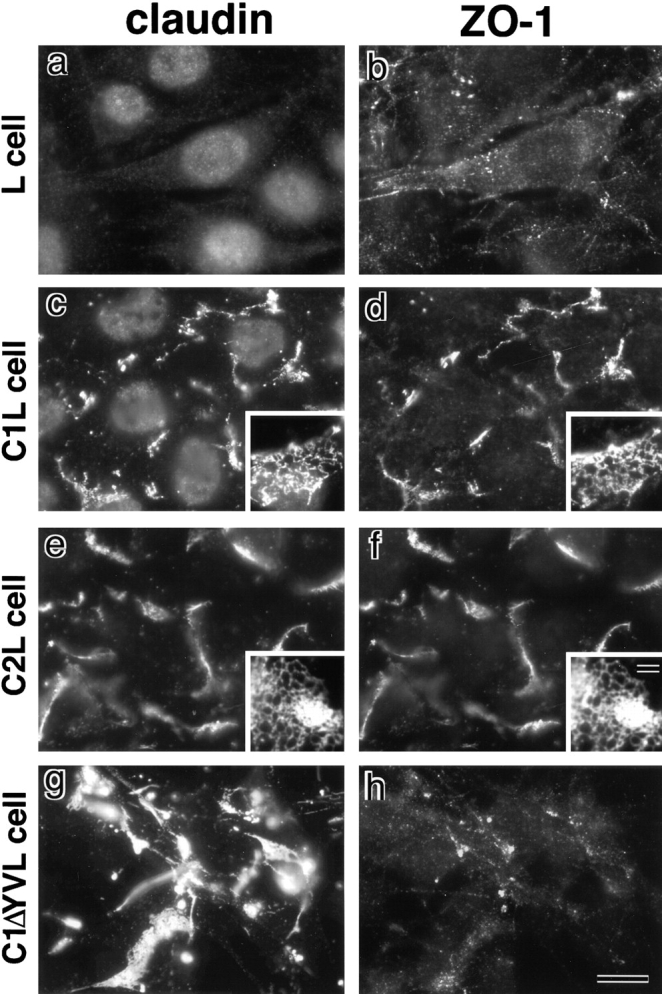
Recruitment of endogenous ZO-1 to claudin-based networks in L transfectants expressing claudin-1 or -2. Parental L cells (L cell; a and b) and L transfectants expressing claudin-1 (C1L cell; c and d) were double stained with rat anti–claudin-1 mAb (claudin; a and c) and mouse anti–ZO-1 mAb (ZO-1; b and d). L transfectants expressing claudin-2 (C2L cell; e and f) were double stained with anti–claudin-2 mAb (e) and anti–ZO-1 mAb (f). L transfectants expressing claudin-1 mutant lacking the COOH-terminal YV sequence (C1ΔYVL cell; g and h) were double stained with anti–claudin-1 pAb (g) and anti–ZO-1 mAb (h). ZO-1 showed no characteristic concentration in L cells (b), whereas in C1L and C2L cells ZO-1 was coconcentrated with claudin-1 (c and d) and claudin-2 (e and f) as planes at cell–cell borders. Close inspection revealed that in these cells both claudins and ZO-1 were concentrated in elaborate network patterns, and that their network patterns were mostly overlapped (insets in c–f). In C1ΔYVL cells, mutant claudin-1 was concentrated at cell–cell borders (g), whereas ZO-1 showed no concentration (h). Bar: (a–h) 10 μm; (insets) 15 μm.
Recruitment of Exogenous ZO-2 to Claudin-based Networks in L Transfectants
Since ZO-2 was not expressed endogenously in L cells (see Fig. 6), the full-length cDNA encoding c-myc–tagged ZO-2 was introduced into C1L cells. As shown in Fig. 8, a and b, this introduced ZO-2 was recruited to the claudin-based networks. However, since ZO-2 can directly bind to ZO-1 through its PDZ2 domain (Itoh et al. 1999), it was not clear whether this recruitment of full-length ZO-2 was based on the direct interaction of ZO-2 with claudin-1 or endogenous ZO-1 that was recruited to the claudin-based networks as shown in Fig. 7. Therefore, we next transfected cDNAs encoding c-myc–tagged ZO-2 mutant lacking both PDZ1 and -2 (ΔPDZ1,2-ZO-2), PDZ1 alone (ΔPDZ1-ZO-2) or PDZ2 alone (ΔPDZ2-ZO-2) into L transfectants (see Fig. 2), confirmed their expression by immunoblotting, and their subcellular localization was followed by anti–c-myc mAb staining. ΔPDZ1,2-ZO-2 was not recruited to the claudin-based networks (Fig. 8c and Fig. d), whereas ΔPDZ1-ZO-2 and ΔPDZ2-ZO-2 were recruited (Fig. 8, e–h). These findings can be interpreted as indicating that ΔPDZ1-ZO-2 and ΔPDZ2-ZO-2 are recruited by the direct association with endogenous ZO-1 (through PDZ2) and claudin-1 (through PDZ1), respectively. Since ΔPDZ1,2-ZO-2 cannot bind to either endogenous ZO-1 or claudin-1, it would not be recruited. In good agreement with this, in C1ΔYVL cells, exogenous full-length ZO-2 was not concentrated at cell–cell borders, probably because in these cells endogenous ZO-1 as well as exogenous ZO-2 cannot bind to mutant claudin-1 (Fig. 8i and Fig. j). The same results were obtained when C2L cells were used (data not shown).
Figure 8.
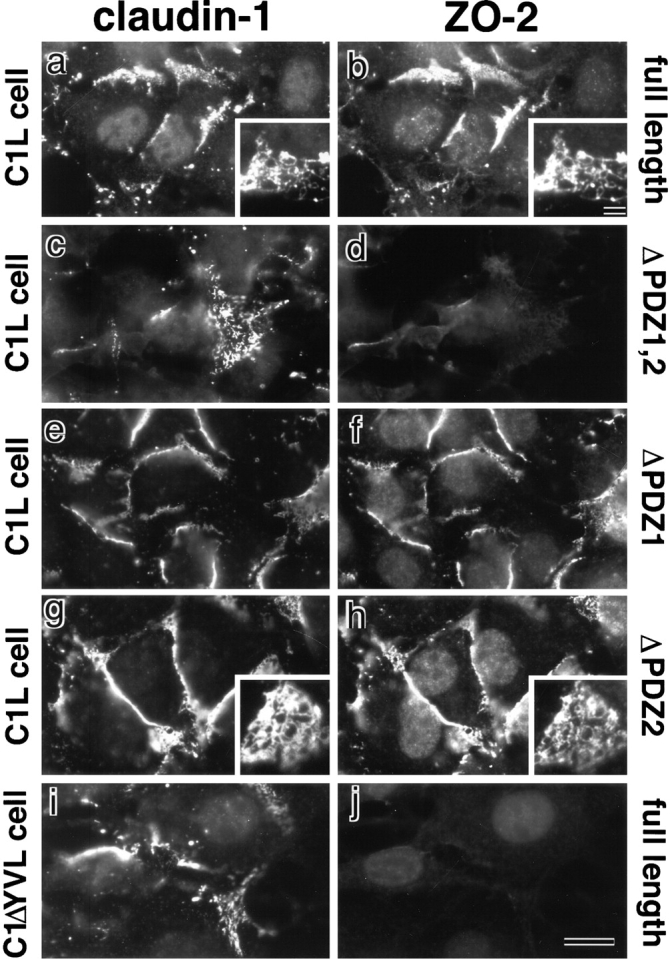
Recruitment of exogenous ZO-2 to claudin-based networks in L transfectants expressing claudin-1. Full-length ZO-2 (full-length; a and b), deletion mutant of ZO-2 lacking both PDZ1 and -2 domains (ΔPDZ1,2; c and d), deletion mutant of ZO-2 lacking PDZ1 domain (ΔPDZ1; e and f), and deletion mutant of ZO-2 lacking PDZ2 domain (ΔPDZ2; g and h) were transfected into L transfectants expressing claudin-1 (C1L cells), and stable transfectants were obtained. Furthermore, full-length ZO-2 (full-length; i and j) was transfected into L transfectants expressing claudin-1 mutant lacking its COOH-terminal YV sequence (C1ΔYVL cell), and stable transfectants were obtained. These introduced proteins were tagged with c-myc epitope. These stable transfectants were double stained with anti–claudin-1 mAb (a, c, e, and g) or pAb (i) and anti–c-myc mAb (b, d, f, h, and j). In C1L cells where claudin-1 was concentrated at cell–cell borders in an elaborate network pattern, full-length ZO-2 (b), ΔPDZ1-ZO-2 (f), and ΔPDZ2-ZO-2 (h), but not ΔPDZ1,2-ZO-2 (d), were recruited to the claudin-1–based networks. Insets represent the network patterns of concentrated claudin-1 (a and g), full-length ZO-2 (b), and ΔPDZ2-ZO-2 (h). No concentration of full-length ZO-2 was observed in C1ΔYVL cells (j). Bar: (a–j) 10 μm; (insets) 15 μm.
Recruitment of Exogenous ZO-3 to Claudin-based Networks in L Transfectants
ZO-3 was reported to directly bind to ZO-1 (Haskins et al. 1998), but the domain responsible for this binding has not yet been determined. We then transfected cDNAs encoding c-myc–tagged full-length ZO-3 and mutants lacking both PDZ1 and -2 (ΔPDZ1,2-ZO-3), PDZ1 alone (ΔPDZ1-ZO-3) or PDZ2 alone (ΔPDZ2-ZO-3) into C1L cells. When these introduced ZO-3 and ZO-3 mutants were immunoprecipitated with anti–c-myc mAb from the cell lysate of stable transfectants, endogenous ZO-1 was coimmunoprecipitated with full-length ZO-3 and ΔPDZ1-ZO-3, but not with ΔPDZ1,2-ZO-3 or ΔPDZ2-ZO-3 (Fig. 9 A). Therefore, we concluded that ZO-3 binds to ZO-1 through its PDZ2 domain inside cells.
Figure 9.
Recruitment of exogenous ZO-3 to claudin-based networks in L transfectants expressing claudin-1. (A) Domain of ZO-3 responsible for ZO-1/ZO-3 interaction. Full-length ZO-3 (full-length ZO-3), deletion mutant of ZO-3 lacking both PDZ1 and -2 domains (ΔPDZ1,2), deletion mutant of ZO-3 lacking PDZ1 domain (ΔPDZ1), and deletion mutant of ZO-3 lacking PDZ2 domain (ΔPDZ2) were introduced into C1L cells. These introduced proteins were tagged with c-myc epitope. Stable transfectants were metabolically labeled with [35S]methionine and solubilized, then each ZO-3 construct was immunoprecipitated with anti–c-myc mAb. Immunoprecipitates were separated by SDS-PAGE followed by autoradiography (autoradiography) or by immunoblotting with anti–ZO-1 mAb (immunoblots with anti–ZO-1 mAb). Each ZO-3 construct with the expected molecular mass (asterisks) was detected, and endogenous ZO-1 (arrowheads) was coimmunoprecipitated only with full-length ZO-3 and ΔPDZ1-ZO-3 containing the PDZ2 domain. Bars indicate molecular masses of 200, 116, 97, and 66 kD, respectively, from the top. (B) Full-length ZO-3 (full-length; a and b), deletion mutant of ZO-3 lacking PDZ1 and -2 domains (ΔPDZ1,2; c and d), deletion mutant of ZO-3 lacking PDZ1 domain (ΔPDZ1; e and f), and deletion mutant of ZO-3 lacking PDZ2 domain (ΔPDZ2; g and h) were transfected into L transfectants expressing claudin-1, and stable transfectants were obtained. All introduced ZO-3 constructs were tagged with c-myc at their COOH termini. These stable transfectants were double stained with rat anti–claudin-1 mAb (a, c, e, and g) and mouse anti–c-myc mAb (b, d, f, and h). In C1L cells where claudin-1 was concentrated at cell–cell borders in an elaborate network pattern, full-length ZO-3 (b), ΔPDZ1-ZO-3 (f), and ΔPDZ2-ZO-3 (h), but not ΔPDZ1,2-ZO-3 (d), were recruited to the claudin-1–based networks. Insets represent the network patterns of concentrated claudin-1 (a and g), full-length ZO-3 (b), and ΔPDZ2-ZO-3 (h). Bar: (B, a–h) 10 μm; (B, insets) 15 μm.
Then, to examine the direct interaction of ZO-3 with claudins in vivo, we transfected cDNAs encoding c-myc–tagged full-length ZO-3, ΔPDZ1,2-ZO-3, ΔPDZ1-ZO-3, or ΔPDZ2-ZO-3 into C1L cells, confirmed their expression by immunoblotting, and their distribution was examined by immunofluorescence microscopy with anti–c-myc mAb (Fig. 9 B). Similarly to the transfection experiments with ZO-2 (see Fig. 8), full-length ZO-3, ΔPDZ1-ZO-3, and ΔPDZ2-ZO-3, but not ΔPDZ1,2-ZO-3, were recruited to claudin-based networks. Furthermore, full-length ZO-3 was not recruited in C1ΔYVL cells (data not shown), and similar results were obtained in C2L cells (data not shown). Based on these findings, we concluded that ZO-3 also directly binds to the COOH-terminal YV of claudins in vivo, and that ZO-3 can be recruited to the claudin-based TJ strands through interactions either with endogenous ZO-1 or claudins themselves.
Recruitment of GFP-Fusion Proteins with PDZ1 and PDZ2 Domains of ZO-1/ZO-2/ZO-3 to TJs in Cultured Epithelial Cells
A question naturally arose as to whether PDZ1 domains of ZO-1/ZO-2/ZO-3 interact with the cytoplasmic domain of claudins in epithelial cells. To evaluate this point, we constructed expression vectors for green fluorescent protein (GFP)-fusion proteins with PDZ1, PDZ2, or PDZ3 domains of ZO-1 and introduced them into cultured MDCK cells (Fig. 10, a–f). For an unknown reason, all these fusion proteins were concentrated in the nucleus. In addition to the nuclear staining, PDZ1-GFP and PDZ2-GFP were clearly recruited to and concentrated at claudin-1–positive TJs (Fig. 10, a–d). Similarly to L transfectants, these findings can be interpreted as indicating that PDZ1-GFP and PDZ2-GFP are recruited by the direct association with endogenous claudins and ZO-2/ZO-3, respectively. In epithelial cells, PDZ3-GFP also appeared to be concentrated at TJs, although very faintly as compared with PDZ1-GFP and PDZ2-GFP (Fig. 10e and Fig. f), probably due to unidentified binding partners for PDZ3 domain of ZO-1 localized at TJs in epithelial cells. GFP-fusion proteins with PDZ1 (Fig. 10, g–j) or PDZ2 (data not shown) domains of ZO-2/ZO-3 also showed significant concentration at claudin-1–positive TJs in MDCK cells.
Figure 10.
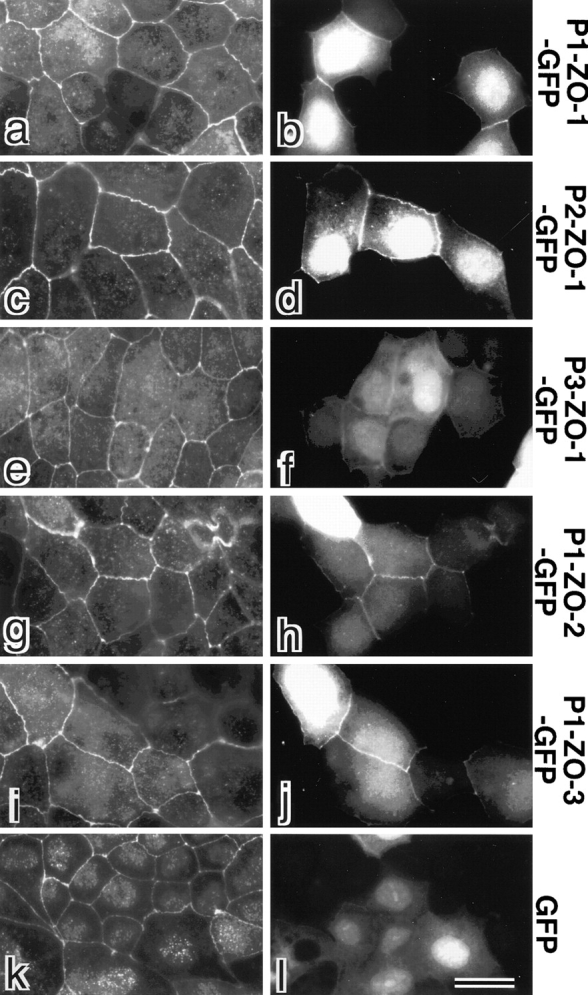
Behavior of GFP-fusion proteins with PDZ1, -2, and -3 domains of ZO-1/ZO-2/ZO-3 in cultured epithelial cells (MDCK cells). GFP (GFP) or GFP-fusion proteins with PDZ1 domain of ZO-1 (P1-ZO-1-GFP), PDZ2 domain of ZO-1 (P2-ZO-1-GFP), PDZ3 domain of ZO-1 (P3-ZO-1-GFP), PDZ1 domain of ZO-2 (P1-ZO-2-GFP) or PDZ1 domain of ZO-3 (P1-ZO-3-GFP) were exogenously and transiently expressed in MDCK cells. These cells were fixed and stained with anti–claudin-1 pAb (a, c, e, g, i, and k) in red. Expressed GFP or GFP-fusion proteins were visualized by green fluorescence (b, d, f, h, j, and l). PDZ1 domains of ZO-1 (b, P1-ZO-1-GFP), ZO-2 (h, P1-ZO-2-GFP), ZO-3 (j, P1-ZO-3-GFP), and PDZ2 domain of ZO-1 (d, P2-ZO-1-GFP) were recruited to claudin-1–positive TJs. PDZ3 domain of ZO-1 (f, P3-ZO-1-GFP) also appeared to be concentrated at TJs, although very faintly as compared with b, d, h, and j. No concentration of GFP at cell–cell borders was observed (l). Bar, 10 μm.
Discussion
TJs are essential intercellular junctions, because their barrier and fence functions are indispensable for the establishment of compositionally distinct compartments in multicellular organisms (Schneeberger and Lynch 1992). Furthermore, evidence has accumulated that TJs recruit various important intracellular signaling molecules to their cytoplasmic surface, which may be involved in the polarization of epithelial cells (Tsukita et al. 1999; Hazuka et al. 1999). Thus, elucidation of the molecular architecture of the TJ plaque is very important to understand the molecular mechanism of the regulation of barrier and fence functions of TJs as well as the relationship between TJs and epithelial polarization. To date, three MAGUKs, ZO-1, ZO-2, and ZO-3, have been identified as components of the TJ plaque (Mitic and Anderson 1998), and occludin has been thought to be a binding partner for these MAGUKs to recruit them to TJs (Furuse et al. 1994; Itoh et al. 1999; Haskins et al. 1998). However, even in occludin-deficient visceral endoderms (Saitou et al. 1998) and human Sertoli cells (Moroi et al. 1998), ZO-1 was shown to be concentrated at TJs. Furthermore, in this study, we found that not only ZO-1 but also ZO-2 and ZO-3 were localized at TJs of highly polarized intestinal epithelial cells in occludin-deficient mice. These findings prompted us to examine the interaction between these MAGUKs and claudins, major constituents of TJ strands in vitro as well as in vivo. We then found that the PDZ1 domains of ZO-1, ZO-2, and ZO-3 directly bind to the COOH-terminal YV sequence of claudins. Interestingly, we detected no differences in affinity for claudin-1 among ZO-1, ZO-2, and ZO-3 (K d = ∼13, 11 and 18 nM, respectively). ZO-1, ZO-2, and ZO-3 shared the same portion of the cytoplasmic domain of claudins (i.e., their COOH-terminal YV sequence) as their binding sites. Furthermore, ZO-1/ZO-2/ZO-3 bound to claudin-1 to -8, although it is possible that the binding affinity varies depending on the claudin species.
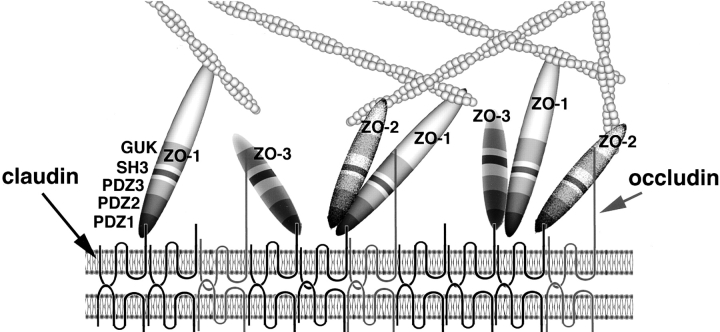
These findings led to the molecular architecture model for the TJ plaque shown schematically in Fig. 11. In this scheme, TJ strands are represented as linear co-polymers of occludin and various species of claudin (Furuse et al. 1999; Tsukita and Furuse 1999). It is likely that these kinds of co-polymers present large linear clusters of YV sequences toward the cytoplasm, which may strongly attract cytoplasmic proteins containing PDZ domains with high affinity to the COOH-terminal sequence of claudins. Since the PDZ1 domains, but not PDZ2 or PDZ3 domains, of ZO-1, ZO-2, and ZO-3 has an affinity to the COOH-terminal sequence of claudins, they are recruited to TJ strands (see Fig. 11). In good agreement, GFP-fusion proteins with PDZ1 domains of ZO-1/ZO-2/ZO-3 were recruited to TJs in MDCK cells (see Fig. 10). These three MAGUKs were also reported to bind to the COOH-terminal 150–amino acid sequence of the cytoplasmic domain of occludin (Furuse et al. 1994; Haskins et al. 1998; Itoh et al. 1999). Since occludin does not end in valine, the molecular basis for the occludin-MAGUKs interaction may be different from that of the claudin-MAGUKs interaction. In ZO-1, the GUK domain was reported to be responsible for occludin-ZO-1 binding (Fanning et al. 1998). If this is the case also for ZO-2 and ZO-3, each MAGUK molecule can bind to occludin and claudins simultaneously through GUK and PDZ1 domains, respectively. Furthermore, to complicate the molecular architecture model for the TJ plaque, ZO-1/ZO-2 and ZO-1/ZO-3 interactions were also reported, suggesting the existence of heterodimers of MAGUKs (Fanning et al. 1998; Haskins et al. 1998; Itoh et al. 1999). The results of this and previous studies revealed that these interactions were dependent on the PDZ2/PDZ2 interaction (Itoh et al. 1999). GFP-fusion proteins with PDZ2 domains of ZO-1/ZO-2/ZO-3 were recruited to TJs in MDCK cells (see Fig. 10), probably through this PDZ2/PDZ2 interaction. ZO-1 does not form homodimers reportedly (Fanning et al. 1998), but no information is available regarding the homodimers of ZO-2 and ZO-3. Furthermore, it remains unclear whether ZO-1, ZO-2, and ZO-3 exist in the TJ plaque as monomers. On the other hand, the COOH-terminal region of ZO-1 and ZO-2 directly binds to actin filaments (Itoh et al. 1997, Itoh et al. 1999; Fanning et al. 1998). These findings indicated that ZO-1, ZO-2 and ZO-3 function as scaffolds of the TJ plaque to cross-link TJ strands to the actin-based cytoskeleton.
Figure 11.
Schematic drawing of the molecular architecture of the TJ plaque. TJ strands are represented as linear co-polymers of claudins and occludin with short and long COOH-terminal tails, respectively. ZO-1, ZO-2, and ZO-3 are directly associated with the COOH termini of claudins at their PDZ1 domains. These molecules are depicted here to interact with occludin at their GUK domains. ZO-1/ZO-2 and ZO-1/ZO-3 heterodimers are depicted to be formed through direct PDZ2/PDZ2 interaction, but the possibility cannot be completely excluded that these interaction requires some linker protein. It remains unclear whether ZO-1, ZO-2, and ZO-3 exist as monomers and/or homodimers (not depicted here), except that ZO-1/ZO-1 homodimers were reported to be undetectable. The COOH-terminal region of ZO-1 and ZO-2 has a binding affinity to actin filaments to function as cross-linkers between TJ strands and actin filaments. The interaction between ZO-3 and actin filaments has not yet been examined.
The molecular linkage described in Fig. 11 would be dynamically regulated within cells. For example, phosphorylation is expected to play an important role in the protein-protein interactions in the TJ plaque. The ZO-1/ZO-2 interaction has been detected by immunoprecipitation from cell lysates, but not by in vitro binding assay with recombinant proteins, probably because some modifications such as phosphorylation on ZO-1 and/or ZO-2 are required for the interaction (Itoh et al. 1999). Furthermore, occludin was shown to be heavily serine/threonine phosphorylated when it was incorporated into TJ strands (Sakakibara et al. 1997). Therefore, it should be examined whether these phosphorylated occludin still showed binding affinity for ZO-1/ZO-2/ZO-3.
Elucidation of the molecular architecture of the TJ plaque will provide insight into the functions of ZO-1, ZO-2, and ZO-3. As discussed above, the model shown in Fig. 11 favors the notion that these TJ-specific MAGUKs function as cross-linkers between TJ strands and actin filaments, although the actin binding ability has not been examined for ZO-3. Accumulating evidence has suggested that the barrier function of TJs is regulated through the association of actin filaments with TJs (Madara 1998). Therefore, it appears reasonable to speculate that ZO-1, ZO-2, and ZO-3 are directly involved in the mechanism of regulation of the barrier function of TJs by modulating the actin filament/TJ strand association.
In general, MAGUKs are believed to cluster integral membrane proteins to establish and maintain specialized plasma membrane domains such as pre- and post-synaptic regions (Woods and Bryant 1993; Anderson 1996; Sheng 1996). Therefore, it is interesting to speculate that ZO-1, ZO-2, and ZO-3 are required for the clustering of claudins and occludin within the plasma membranes, i.e., the formation of TJ strands. In our previous study (Furuse et al. 1998b), we obtained L transfectants expressing claudin-1 and -2 tagged with a FLAG sequence at their COOH termini (C1FL and C2FL cells, respectively), and found that the tagged claudin-1 and -2 were concentrated at cell–cell borders to reconstitute TJ strands. Interestingly, however, in contrast to C1L and C2L cells used in this study, in these C1FL and C2FL cells endogenous ZO-1 was not recruited to the claudin-based TJ strands because the COOH-terminal end of claudin-1 and -2 was masked by the FLAG sequence causing loss of its binding activity to the PDZ1 domain of ZO-1 (data not shown). Furthermore, recently we found that claudin-1 deletion mutant lacking almost all portion of its COOH-terminal cytoplasmic domain still reconstituted a well-developed network of TJ strands in L transfectants (Furuse et al. 1999). Therefore, we concluded that claudins can be polymerized to form TJ strands without interacting with ZO-1 (and probably ZO-2 and ZO-3).
This study improved our understanding of the molecular architecture of the major scaffolds of the TJ plaque. Further studies are required to identify various proteins that are recruited to these ZO-1/ZO-2/ZO-3–based scaffolds directly or indirectly. To date, three other TJ-specific peripheral membrane proteins, cingulin, 7H6 antigen and symplekin have been identified (Citi et al. 1988; Zhong et al. 1993; Keon et al. 1996). An integral membrane protein called JAM, which belongs to the immunoglobulin superfamily, has also been shown to be concentrated at TJs (Martin-Padura et al. 1998). Furthermore, TJs were reported to recruit various types of molecules involved in intracellular signaling (heterotrimeric G proteins, atypical protein kinase C λ/ζ and their specific binding protein called ASIP) (Stuart and Nigam 1995; Denker et al. 1996; Saha et al. 1998; Izumi et al. 1998) and vesicle targeting/fusion (Rab3B/Rab13 and sec6/8) (Weber et al. 1994; Zahraoui et al. 1994; Grindstaff et al. 1998). On the other hand, as shown in Fig. 11, the binding partners for the PDZ3 and SH3 domains of ZO-1, ZO-2, and ZO-3 have not yet been identified, although a putative kinase, ZAK (ZO-1–associated kinase), was reported to be associated with the SH3 domain of ZO-1 (Balda et al. 1996). Therefore, it is possible that these PDZ3 and SH3 domains have some important roles in recruiting the molecules mentioned above to TJs. Further studies to clarify the molecular linkage within the TJ plaque will lead to a better understanding of the functions of TJs as well as the molecular mechanism of their regulation.
Acknowledgments
We thank all the members of our laboratory (Department of Cell Biology, Faculty of Medicine, Kyoto University) for helpful discussions. Our thanks are also due to Ms. K. Kamikubo-Tsuchihashi for her excellent technical assistance.
This study was supported in part by a Grant-in-Aid for Cancer Research and a Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Science, and Culture of Japan.
Footnotes
Abbreviations used in this paper: aa, amino acids; TJ, tight junction; MAGUKs, membrane-associated guanylate kinase-like homologues; pAb, polyclonal antibody.
References
- Anderson J.M. Cell signallingMAGUK magic. Curr. Biol. 1996;6:382–384. doi: 10.1016/s0960-9822(02)00501-8. [DOI] [PubMed] [Google Scholar]
- Ando-Akatsuka Y., Saitou M., Hirase T., Kishi M., Sakakibara A., Itoh M., Yonemura S., Furuse M., Tsukita Sh. Interspecies diversity of the occludin sequencecDNA cloning of human, mouse, dog, and rat-kangaroo homologues. J. Cell Biol. 1996;133:43–47. doi: 10.1083/jcb.133.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda M.S., Anderson J.M., Matter K. The SH3 domain of the tight junction protein ZO-1 binds to a serine protein kinase that phosphorylates a region C-terminal to this domain. FEBS Lett. 1996;399:326–332. doi: 10.1016/s0014-5793(96)01352-x. [DOI] [PubMed] [Google Scholar]
- Balda M.S., González-Mariscal L., Matter K., Cereijido M., Anderson J.M. Assembly of the tight junctionThe role of diacylglycerol. J. Cell Biol. 1993;123:293–302. doi: 10.1083/jcb.123.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citi S., Sabanay H., Jakes R., Geiger B., Kendrick-Jones J. Cingulin, a new peripheral component of tight junctions. Nature. 1988;333:272–276. doi: 10.1038/333272a0. [DOI] [PubMed] [Google Scholar]
- Cereijido M., Valdes J., Shoshani L., Contreras R.G. Role of tight junctions in establishing and maintaining cell polarity. Annu. Rev. Physiol. 1998;60:161–177. doi: 10.1146/annurev.physiol.60.1.161. [DOI] [PubMed] [Google Scholar]
- Denker B.M., Saha C., Khawaja S., Nigam S.K. Involvement of a heterotrimeric G protein α subunit in tight junction biogenesis. J. Biol. Chem. 1996;271:25750–25754. doi: 10.1074/jbc.271.42.25750. [DOI] [PubMed] [Google Scholar]
- Fanning A.S., Jameson B.J., Jesaitis L.A., Anderson J.M. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J. Biol. Chem. 1998;273:29745–29753. doi: 10.1074/jbc.273.45.29745. [DOI] [PubMed] [Google Scholar]
- Furuse M., Fujita T., Hiiragi T., Fujimoto K., Tsukita Sh. Claudin-1 and -2novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin J. Cell Biol. 141 1998. 1539 1550a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Hirase T., Itoh M., Nagafuchi A., Yonemura S., Tsukita Sa., Tsukita Sh. Occludina novel integral membrane protein localizing at tight junctions. J. Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Itoh M., Hirase T., Nagafuchi A., Yonemura S., Tsukita Sa., Tsukita Sh. Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J. Cell Biol. 1994;127:1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Sasaki H., Fujimoto K., Tsukita Sh. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts J. Cell Biol. 143 1998. 391 401b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Sasaki H., Tsukita Sh. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J. Cell Biol. 1999;147:891–903. doi: 10.1083/jcb.147.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindstaff K.K., Yeaman C., Anandasabapathy N., Hsu S.C., Rodriguez-Boulan E., Scheller R.H., Nelson W.J. Sec6/8 complex is recruited to cell–cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell. 1998;93:731–740. doi: 10.1016/s0092-8674(00)81435-x. [DOI] [PubMed] [Google Scholar]
- Gumbiner B., Lowenkopf T., Apatira D. Identification of a 160-kDa polypeptide that binds to the tight junction protein ZO-1. Proc. Natl. Acad. Sci. USA. 1991;88:3460–3464. doi: 10.1073/pnas.88.8.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskins J., Gu L., Wittchen E.S., Hibbard J., Stevenson B.R. ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J. Cell Biol. 1998;141:199–208. doi: 10.1083/jcb.141.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazuka C.D., Foletti D.L., Scheller R.H. Targeting vesicles to specific sites on the plasma membranethe role of the sec6/8 complex. Trends Cell Biol. 1999;9:150–153. doi: 10.1016/s0962-8924(99)01516-0. [DOI] [PubMed] [Google Scholar]
- Itoh M, Morita K., Tsukita Sh. Characterization of ZO-2 as a MAGUK family member associated with tight and adherens junctions with a binding affinity to occludin and α catenin. J. Biol. Chem. 1999;274:5981–5986. doi: 10.1074/jbc.274.9.5981. [DOI] [PubMed] [Google Scholar]
- Itoh M., Nagafuchi A., Moroi S., Tsukita Sh. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to α catenin and actin filaments. J. Cell Biol. 1997;138:181–192. doi: 10.1083/jcb.138.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Nagafuchi A., Yonemura S., Kitani-Yasuda T., Tsukita Sa., Tsukita Sh. The 220-kD protein colocalizing with cadherins in non-epithelial cells is identical to ZO-1, a tight junction-associated protein in epithelial cellscDNA cloning and immunoelectron microscopy. J. Cell Biol. 1993;121:491–502. doi: 10.1083/jcb.121.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Yonemura S., Nagafuchi A., Tsukita Sa., Tsukita Sh. A 220-kD undercoat-constitutive proteinits specific localization at cadherin-based cell–cell adhesion sites. J. Cell Biol. 1991;115:1449–1462. doi: 10.1083/jcb.115.5.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y., Hirose T., Tamai Y., Hirai S., Nagashima Y., Fujimoto T., Tabuse Y., Kemphues K.J., Ohno S. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J. Cell Biol. 1998;143:95–106. doi: 10.1083/jcb.143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesaitis L.A., Goodenough D.A. Molecular characterization and tissue distribution of ZO-2, a tight junction protein homologous to ZO-1 and the Drosophila discs-large tumor suppresser protein. J. Cell Biol. 1994;124:949–961. doi: 10.1083/jcb.124.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keon B.H., Schäfer S., Kuhn C., Grund C., Franke W.W. Symplekin, a novel type of tight junction plaque protein. J. Cell Biol. 1996;134:1003–1018. doi: 10.1083/jcb.134.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Niethammer M., Rothschild A., Jan Y.N., Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- Kornau H.C., Schenker L.T., Kennedy M.B., Seeburg P.H. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Madara J.L. Intestinal absorptive cell tight junctions are linked to cytoskeleton. Am. J. Physiol. 1987;253:C171–C175. doi: 10.1152/ajpcell.1987.253.1.C171. [DOI] [PubMed] [Google Scholar]
- Madara J.L. Regulation of the movement of solutes across tight junctions. Annu. Rev. Physiol. 1998;60:143–159. doi: 10.1146/annurev.physiol.60.1.143. [DOI] [PubMed] [Google Scholar]
- Martin-Padura I., Lostaglio S., Schneemann M., Williams L., Romano M., Fruscella P., Panzeri C., Stoppacciaro A., Ruco L., Villa A., Simmons D., Dejana E. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 1998;142:117–127. doi: 10.1083/jcb.142.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitic L.L., Anderson J.M. Molecular architecture of tight junctions. Annu. Rev. Physiol. 1998;60:121–142. doi: 10.1146/annurev.physiol.60.1.121. [DOI] [PubMed] [Google Scholar]
- Morita K., Furuse M., Fujimoto K., Tsukita Sh. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc. Natl. Acad. Sci. USA. 1999;96:511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroi S., Saitou M., Fujimoto K., Sakakibara A., Furuse M., Yoshida O., Tsukita Sh. Occludin is concentrated at tight junctions of mouse/rat but not human/guinea pig Sertoli cells in testes. Am. J. Physiol. 1998;274:C1708–C1717. doi: 10.1152/ajpcell.1998.274.6.C1708. [DOI] [PubMed] [Google Scholar]
- Niethammer M., Kim E., Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J. Neurosci. 1996;16:2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Saha C., Nigam S.K., Denker B.M. Involvement of Gαi2 in the maintenance and biogenesis of epithelial cell tight junctions. J. Biol. Chem. 1998;273:21629–21633. doi: 10.1074/jbc.273.34.21629. [DOI] [PubMed] [Google Scholar]
- Saitou M., Fujimoto K., Doi Y., Itoh M., Fujimoto T., Furuse M., Takano H., Noda T., Tsukita Sh. Occludin-deficient embryonic stem cells can differentiate into polarized epithelial cells bearing tight junctions. J. Cell Biol. 1998;141:397–408. doi: 10.1083/jcb.141.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara A., Furuse M., Saitou M., Ando-Akatsuka Y., Tsukita Sh. Possible involvement of phosphorylation of occludin in tight junction formation. J. Cell Biol. 1997;137:1393–1401. doi: 10.1083/jcb.137.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger E.E., Lynch R.D. Structure, function, and regulation of cellular tight junctions. Am. J. Physiol. 1992;262:L647–L661. doi: 10.1152/ajplung.1992.262.6.L647. [DOI] [PubMed] [Google Scholar]
- Sheng M. PDZs and receptor/channel clusteringRounding up the latest suspects. Neuron. 1996;17:575–578. doi: 10.1016/s0896-6273(00)80190-7. [DOI] [PubMed] [Google Scholar]
- Simon D.B., Lu Y., Choate K.A., Velazquez H., Al-Sabban E., Praga M., Casari G., Bettinelli A., Colussi G., Rodriguez-Soriano J. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285:103–106. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- Staehelin L.A. Structure and function of intercellular junctions. Int. Rev. Cytol. 1974;39:191–283. doi: 10.1016/s0074-7696(08)60940-7. [DOI] [PubMed] [Google Scholar]
- Stevenson B.R., Siliciano J.D., Mooseker M.S., Goodenough D.A. Identification of ZO-1a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 1986;103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart R.O., Nigam S.K. Regulated assembly of tight junctions by protein kinase C. Proc. Natl. Acad. Sci. USA. 1995;92:6072–6076. doi: 10.1073/pnas.92.13.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita Sh., Furuse M. Occludin and claudins in tight junction strandsLeading or supporting players? Trend. Cell Biol. 1999;9:268–273. doi: 10.1016/s0962-8924(99)01578-0. [DOI] [PubMed] [Google Scholar]
- Tsukita Sh., Furuse M., Itoh M. Structural and signaling molecules interact at tight junctions. Curr. Opin. Cell Biol. 1999;11:628–633. doi: 10.1016/s0955-0674(99)00016-2. [DOI] [PubMed] [Google Scholar]
- Weber E., Berta G., Tousson A., St. John P., Green M.W., Gopalokrishnan U., Jilling T., Sorscher E.J., Elton T.S., Abrahamson D.R., Kirk K.L. Expression and polarized targeting of a rab3 isoform in epithelial cells. J. Cell Biol. 1994;125:583–594. doi: 10.1083/jcb.125.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willott E., Balda M.S., Fanning A.S., Jameson B., Van Itallie C., Anderson J.M. The tight junction protein ZO-1 is homologous to the Drosophila discs-large tumor suppresser protein of septate junctions. Proc. Natl. Acad. Sci. USA. 1993;90:7834–7838. doi: 10.1073/pnas.90.16.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D.A., Bryant P.J. ZO-1, DlgA and PSD95/SAP90homologous proteins in tight, septate and synaptic cell junctions. Mech. Dev. 1993;44:85–89. doi: 10.1016/0925-4773(93)90059-7. [DOI] [PubMed] [Google Scholar]
- Yap A.S., Mullin J.M., Stevenson B.R. Molecular analysis of tight junction physiologyinsights and paradoxes. J. Membr. Biol. 1998;163:159–167. doi: 10.1007/s002329900380. [DOI] [PubMed] [Google Scholar]
- Zahraoui A., Joberty G., Arpin M., Fontaine J.J., Hellio R., Tavitian A., Louvard D. A small rab GTPase is distributed in cytoplasmic vesicles in nonpolarized cells but colocalizes with the tight junction marker ZO-1 in polarized epithelial cells. J. Cell Biol. 1994;124:101–115. doi: 10.1083/jcb.124.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y., Saitoh T., Minase T., Sawada N., Enomoto K., Mori M. Monoclonal antibody 7H6 reacts with a novel tight junction-associated protein distinct from ZO-1, cingulin, and ZO-2. J. Cell Biol. 1993;120:477–483. doi: 10.1083/jcb.120.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]