Abstract
Application of differential display to cultured rat astrocytes subjected to hypoxia allowed cloning of a novel cDNA, termed stress-associated endoplasmic reticulum protein 1 (SERP1). Expression of SERP1 was enhanced in vitro by hypoxia and/or reoxygenation or other forms of stress, causing accumulation of unfolded proteins in endoplasmic reticulum (ER) stress, and in vivo by middle cerebral artery occlusion in rats. The SERP1 cDNA encodes a 66–amino acid polypeptide which was found to be identical to ribosome-associated membrane protein 4 (RAMP4) and bearing 29% identity to yeast suppressor of SecY 6 protein (YSY6p), suggesting participation in pathways controlling membrane protein biogenesis at ER. In cultured 293 cells subjected to ER stress, overexpression of SERP1/RAMP4 suppressed aggregation and/or degradation of newly synthesized integral membrane proteins, and subsequently, facilitated their glycosylation when the stress was removed. SERP1/RAMP4 interacted with Sec61α and Sec61β, which are subunits of translocon, and a molecular chaperon calnexin. Furthermore, Sec61α and Sec61β, but not SERP1/RAMP4, were found to associate with newly synthesized integral membrane proteins under stress. These results suggest that stabilization of membrane proteins in response to stress involves the concerted action of a rescue unit in the ER membrane comprised of SERP1/RAMP4, other components of translocon, and molecular chaperons in ER.
Keywords: hypoxia, endoplasmic reticulum stress, translocon, aggregation/degradation, refolding
Ischemic stress, due to hypoxemia alone or hypoxemia followed by reperfusion, is one of the most relevant environmental challenges, as it accompanies a spectrum of disorders associated with vascular dysfunction and aging. The central nervous system typifies complexities of the cellular response to ischemia. During ischemia itself, neuronal function is compromised, often resulting in loss of cell viability, due at least in part to the extracellular accumulation of excitatory amino acids and disordered calcium homeostasis (Choi 1995; Rothman and Olney 1995). Subsequent reperfusion exacerbates injury through the formation of reactive oxygen species, which attack membranous and proteinaceous cellular constituents, further compromising their functions (McCord 1987; Solenski et al. 1997). The cellular response to such ischemia occurs on many levels, including modulation of gene expression. In this context, >80 different mRNAs have been identified in ischemic brain (for review see Koistinaho and Hokfelt 1997), and these encode proximal and distal regulators of the cellular stress response, immediate-early gene products, apoptosis-associated proteins, stress proteins, cytokines, and growth factors. Although some of these undoubtedly contribute to pathways underlying neuronal death, others most probably represent adaptive responses promoting cellular survival in response to stress.
In contrast to the extraordinary vulnerability of neurons to ischemic stress, astrocytes have a unique ability to tolerate and even proliferate under such conditions, assigning them a central role in the immediate cellular response and later events associated with tissue remodelling (Hertz 1982). The potential of astrocytes to orchestrate events in the central nervous system triggered by ischemic stress has led us to analyze their response to hypoxia (H) and hypoxia followed by reoxygenation (R), using brain-derived rat astrocytes in order to simulate ischemia and ischemia/reperfusion, respectively (Hori et al. 1994). We have found that astrocytes subjected to these environmental perturbations display upregulation of stress proteins (glucose-regulated protein [GRP] 78, GRP94, heat shock protein [HSP] 72, and heme oxygenase, type 1), and release the neurotrophic cytokine IL-6 (Maeda et al. 1994; Hori et al. 1996). Further dissection of differential gene expression in astrocytes exposed to H/R allowed cloning of three novel cDNAs encoding: (a) a novel stress protein, oxygen-regulated protein (ORP) 150; (b) the rat homologue of the Drosophila splicing factor Tra2 (RA301); and (c) a novel vesicle transporter (RA410) (Matsuo et al. 1995, Matsuo et al. 1997; Kuwabara et al. 1996).
To further probe mechanisms through which astrocytes participate in the response to ischemic stress, we have cloned a novel stress-associated ER protein, termed SERP1, by differential display applied to primary cultures of astrocytes exposed to H. Compared with homeostatic conditions, SERP1 expression is upregulated both in vivo and in vitro in response to H and R (including induction of brain ischemia), as well as under conditions associated with accumulation of unfolded proteins in the endoplasmic reticulum (ER stress). SERP1 was found to be identical to ribosome-associated membrane protein 4 (RAMP4) and bearing ∼30% homology to yeast suppressor of SecY 6 protein (YSY6p), suggesting participation in pathways controlling biogenesis of secretory and membrane proteins at the ER. In cultured 293 cells subjected to ER stress, overexpression of SERP1/RAMP4 suppressed aggregation and/or degradation of integral membrane proteins under stress and facilitates glycosylation after the stress. SERP1/RAMP4 interacted directly with Sec61α and Sec61β, which are subunits of the translocon (Sec61 complex; Görlich et al. 1992; Görlich and Rapoport 1993), and calnexin, a membrane protein and a molecular chaperon in ER that associates with folding intermediates of glycoprotein (Ou et al. 1993). Immunoprecipitation did demonstrate a binding of newly synthesized integral membrane proteins to Sec61α and Sec61β but not to SERP1/RAMP4. These results suggest that the stabilization of membrane proteins in response to stress involves the concerted action of a rescue unit in the ER membrane that appears to be comprised of SERP1/RAMP4, as well as other components of translocon, and molecular chaperons in ER.
Materials and Methods
Cell Culture and Conditions for H/R and Other Stresses
Astrocytes were isolated from the cerebral cortex of E18 rat embryos using a minor modification of previously described methods (McCarthy and de Vellis 1980). In brief, cerebral hemispheres were obtained from E18 brains and the meninges were carefully removed. Brain tissue was digested with papain (Worthington Biochemical Corp.) at 37°C for 15 min and plated in 175-cm2 culture flasks (two brains/flask). Cells were grown in MEM with 10% FCS for 10 d and agitated strongly on a shaking platform to separate astrocytes from microglia and oligodendroglia. Cells were then replated into 150-mm diam dishes and grown for an additional 7 d. Cultures used for experiments were comprised of >95% astrocytes, based on the morphological (fibroblast-like appearance with the formation of a cobblestone cell layer) and immunohistochemical (detection of glial fibrillary acidic protein [GFAP] with anti-GFAP antibody; Sigma Chemical Co.) criteria. When cells achieved confluence, the medium was replaced with serum-free MEM and cultures were subjected to H for the indicated times (up to 22 h) using an incubator equipped with an H chamber (Coy Laboratory Products) as described (Ogawa et al. 1990). Using this chamber, the ambient oxygen tension in culture medium bathing the cells was ∼8–10 Torr (Ogawa et al. 1990). In some experiments, cells were returned to the ambient atmosphere after H and incubated for 4 h (R). In other experiments, cells were maintained in normoxia and exposed to either calcium ionophore A23187 (1 μM for 8 h) (Sigma Chemical Co.), tunicamycin (1 μg/ml for 8 h) (Sigma Chemical Co.), or hydrogen peroxide (80 μM for 4 h) (Wako Chemicals). Alternatively, cultures were subjected to heat shock at 42°C for 2 h and then returned to 37°C and incubated for an additional 2 h.
293 cells and BHK cells were cultured in DMEM and transfected with the indicated expression vectors as described below.
Differential Display
Total RNA was extracted from cultured astrocytes subjected to normoxia or H for 20 h using Qiagen RNeasy kit, and differential display was performed as described (Liang and Pardee 1992). In brief, RT-PCR with avian myeloblastosis virus reverse transcriptase (Amersham Pharmacia Biotech) and ∼300 random primers (12–21 oligonucleotides) were used to prepare cDNAs. PCR products were separated by acrylamide gel (6%) electrophoresis, and cDNA bands of interests were cut out from the gel. After cloning the cDNAs into pGEM-T vector (Promega), DNA sequencing was performed using a DNA sequencer (model 377; Applied Biosystems). Differential expression of candidate genes in H versus normoxia was confirmed by Northern blotting using 32P-radiolabeled cDNAs as probes.
Cloning of Rat and Human SERP1/RAMP4 cDNAs
One of the cDNA fragments obtained by differential display, termed T41, was used to screen an adult cDNA library (lambda ZapII cDNA library; Stratagene). A cDNA of 2.3 kb was obtained spanning the entire open reading frame (ORF) and polyadenylation signal, and both strands were sequenced. Sequence searches and comparisons were carried out using several databases, including the National Center for Biotechnology Information, FASTA and BLAST molecular analysis systems, and EMBL/ GenBank/DDBJ. The human SERP1/RAMP4 cDNA, which included the complete ORF of rat SERP1/RAMP4, was obtained from a human expression sequence tag (EST) clone through the IMAGE consortium. DNA sequencing was performed on both strands for each clone.
Northern Blot Analysis
Total RNA (10–15 μg), isolated from cultured astrocytes or rat tissues, was separated on agarose/formaldehyde (1%) gels and transferred onto Immobilon N membranes (Millipore). A cDNA fragment of SERP1/RAMP4 was labeled with 32P by the random hexamer procedure (specific activity 0.5–3 × 109 cpm/μg DNA) and was used to probe membranes with immobilized RNA. After washing in 2× SSC and 0.5× SDS for 1 h, membranes were subjected to autoradiography. Northern blots were also performed with rat Sec61α, human Sec61β (see below), rat GRP78 (cloned by PCR with specific primers), rat HSP72 (Imuta et al. 1998), human ORP150 (Kuwabara et al. 1996), and human oligosaccaryltransferase (OST; cloned by PCR with specific primers) cDNA fragments as probes.
Expression of SERP1/RAMP4 in Ischemic Brain
Unilateral middle cerebral artery (MCA) occlusion was performed in male Sprague-Dawley rats (250 g) as described (Garcia et al. 1993). After 12 h of ischemia, rats were killed and brains were frozen at −80°C. Serial coronal sections were cut and SERP1/RAMP4 mRNA was detected by in situ hybridization using previously described techniques (Matsuo et al. 1995). In brief, sense and antisense riboprobes for SERP1/RAMP4 were in vitro transcribed from the rat SERP1/RAMP4 cDNA inserted into the pGEM T vector. After linearizing the vector with NcoI (for the sense probe) or SpeI (for the antisense probe), reaction mixtures were incubated with [35S]UTP (NEG-039H; Dupont-NEN) and SP6 or T7 RNA polymerase (Promega). Brain sections were then hybridized with either sense or antisense probes and washed, dried, and subjected to autoradiography. 2 d later, films were developed and brain images were examined. For some sections, slides were covered with photographic emulsion (Eastman Kodak Co.) for 2 wk, and were then developed and analyzed by dark-field microscopy. SERP1/RAMP4 antigen was also detected by immunoprecipitation followed by the Western blotting with anti–SERP1/RAMP4 antibody as described below.
Plasmid Construction and Generation of Antibodies
SERP1/RAMP4 cDNA encoding the complete ORF was amplified by PCR using primers tagged with FLAG epitope at the NH2 terminus and cloned into pcDNA3 (Invitrogen). The rat Sec61α and human Sec61β cDNAs were cloned as described (Görlich et al. 1992; Hartmann et al. 1994) and hemagglutinin (HA)-tagged, amplified by PCR, and ligated into pcDNA3. All constructs were sequenced before transfection studies (see below). Anti–human Sec61β antibody was generously provided by Dr. Tom A. Rapoport (Harvard University, Boston, MA). Reagents to detect receptor for advanced glycation endproducts (RAGE) at the protein level were prepared as described (Neeper et al. 1992; Schmidt et al. 1995). Antibodies and cDNA for CD8 were kindly provided by Dr. Kazunori Imaizumi (Osaka University Medical School, Osaka, Japan).
Production of Anti-SERP1/RAMP4 Antibodies and Detection of SERP1/RAMP4 Protein
To obtain antibody reactive with SERP1/RAMP4, a peptide with the sequence CTQRGNVAKTSRNAPEEK (containing an extra cysteine residue at the NH2 terminus), was synthesized and conjugated to keyhole limpet hemocyanin. Rabbits were immunized by conventional methods. Once high titer antibody was obtained, the antiserum was affinity-purified using a column with immobilized synthetic peptide (Proton Kit 1; Multiple Peptide Systems). For detecting SERP1/RAMP4 protein, cultured astrocytes or 293 cells transfected with pcDNA3/FLAG-tagged SERP1/RAMP4 were lysed in 1% NP-40/10 mM Tris, pH 7.6, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, and 10 μg/ml aprotinin, and immunoprecipitation or Western blotting was performed using rabbit anti–SERP1/RAMP4 antibody or mouse anti-FLAG antibody. Sites of primary antibody binding were determined by enhanced chemiluminescence method (Amersham Pharmacia Biotech), or using alkaline phosphatase–conjugated secondary antibodies. Immunoblotting for other molecules used specific antibodies for each epitope, and the same general procedure as that for SERP1/RAMP4 was used.
Immunocytochemical Analysis of BHK Cells
To assess the subcellular localization of SERP1/RAMP4, BHK cells transiently transfected to overexpress FLAG-tagged SERP1/RAMP4 or YSY6p, were subjected to double-immunostaining with mouse anti-FLAG (Sigma Chemical Co.) and rabbit anti–protein disulfide isomerase (PDI) antibodies (the latter kindly provided from Dr. Ryuichi Masaki, Kansai Medical University, Osaka, Japan) (Akagi et al. 1988). Sites of primary antibody binding were visualized using TRITC-conjugated anti–mouse antibody (Sigma Chemical Co.) and FITC-conjugated anti–rabbit antibody (Sigma Chemical Co.). In other studies, RAGE was detected using rabbit anti-RAGE IgG followed by FITC-conjugated anti–rabbit antibody.
Effect of SERP1/RAMP4 on Stabilization and Glycosylation of Membrane Proteins after Induction of Cell Stress
293 or BHK cells were transiently transfected with expression constructs for RAGE or CD8 (2 μg DNA/ml) alone or with pcDNA/SERP1/RAMP4 (2 μg DNA/ml) using lipofectamine (Life Technologies, Inc.). Where indicated, ER stress was induced by addition of the calcium ionophore A23187 (1 μg/ml) or tunicamycin (1 μM). After overnight incubation, stabilization of membrane proteins (RAGE or CD8) was analyzed by immunoblotting or by immunoprecipitation, the latter after pulse–chase labeling. Metabolic labeling was accomplished by addition of [35S]methionine (American Radiolabeled Chemicals) to the culture medium (0.25 mCi/ml), followed by incubation for 30 min in methionine-free DMEM, and a 1-h chase period in complete medium (i.e., without [35S]methionine). As a control experiment, stabilization of IκBα under stress conditions was also studied using anti-IκBα antibody (Santa Cruz Biotechnology, Inc.). Antiubiquitin antibody (StressGen Biotechnologies Corp.) was used to characterize the smear RAGE antigen under stress condition. In some experiments, cells were released from the stress after overnight incubation with tunicamycin (1 μM) (i.e., the medium was replaced with tunicamycin-free medium) and glycosylation of RAGE was analyzed by metabolic labeling with [35S]methionine (0.25 mCi/ml) for 3 h followed by immunoprecipitation with anti-RAGE antibody.
Interaction of SERP1/RAMP4 with Sec61 Complex, Molecular Chaperons, and the Membrane Protein RAGE
293 cells (5 × 106 cells) were transfected with expression constructs for FLAG-tagged SERP1/RAMP4, or the membrane protein RAGE (see above). After overnight incubation, cultures were lysed in 1% NP-40 or 2% deoxycholate (the latter followed by DNase I treatment)/10 mM Tris, pH 7.6, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, and 10 μg/ml aprotinin, and immunoprecipitation was performed with various antibodies, such as anti-FLAG (Sigma Chemical Co.), anti-Sec61α, anti-Sec61β, anti-RAGE (Neeper et al. 1992; Schmidt et al. 1995), anti-GRP78 (StressGen Biotechnologies Corp.), anti-ORP150, or anticalnexin (StressGen Biotechnologies Corp.) antibodies. Immunoprecipitates were then subjected to SDS-PAGE and immunoblotted with indicated antibodies. Cross-linking with disuccinimidyl suberate (DSS) (Pierce) was performed on membrane fractions isolated from 293 cells cotransfected with expression constructs for SERP1/RAMP4 and HA-tagged Sec61β as described (Schmidt et al. 1994; Kalies et al. 1998; Bebok et al. 1998). In brief, 107 cells were homogenized with a Dounce homogenizer in 0.25 M sucrose, 10 mM acetic acid, 10 mM triethanolamine, 1 mM EDTA, pH 7.4, and 1 mM PMSF, and then centrifuged at 1,000 g for 10 min. Supernatant was spun again at 10,000 g for 30 min and supernatant from the second spinning was subjected to final centrifugation at 100,000 g for 60 min to clarify a cytosolic fraction. Cross-linking was carried out by incubating membrane fractions with 1 mM DSS in PBS for 1 h. Coimmunoprecipitation (co-IP) was then performed as described above.
Laser Densitometric Analysis
Laser densitometric analysis was performed to standardize the results of Western and Northern blotting with Quality One software (Pdi, Inc.) as described previously (Tsukamoto et al. 1996).
Results
Identification of SERP1/RAMP4
Cultured rat astrocytes were exposed to H, and after RT-PCR, a differentially expressed amplicon of 400 bp termed T41 was identified. Northern analysis using this cDNA as a probe and total RNA harvested from hypoxic astrocytes confirmed selective upregulation compared with normoxia (see below), and led us to clone the full-length cDNA. A rat brain cDNA library was screened, and a 2.3-kb cDNA clone was obtained that contained only a single ORF and polyadenylation signal. The cDNA encoded a protein of 66 amino acids, termed SERP1, including a putative transmembrane-spanning domain at the COOH terminus (Fig. 1). Based on sequence homology database searches, SERP1 displayed 29% identity at the amino acid level with the yeast protein YSY6p, which was identified as a high-copy suppressor of SecY/Sec61 mutant (Sakaguchi et al. 1991). Human and Caenorhabditis elegans homologues of SERP1 were encoded in EST clones, and showed 100 and 53% identities, respectively. We further found that SERP1 is identical to a ribosome-associated membrane protein termed RAMP4 which was copurified with Sec61 complex (Görlich and Rapoport 1993). Rat, human, and C. elegans SERP1 cDNA sequences can be obtained from EMBL/GenBank/DDBJ under accession nos. AB018546, AB022427, and Z81095, respectively.
Figure 1.
Amino acid sequence of rat and human SERP1. The putative transmembrane domain is underlined and the sequence used for raising antibodies is indicated by the box.
Expression of SERP1/RAMP4 under Homeostatic Conditions and in Response to Cell Stress
Northern blotting of total RNA harvested from rat tissues demonstrated similar levels of SERP1/RAMP4 transcripts in a range of normal organs, whereas heart, lung, and muscle displayed lower levels of mRNA (Fig. 2 a). Cultured astrocytes exposed to H showed a time-dependent increase in SERP1/RAMP4 transcripts, although this occurred after relatively long-term oxygen deprivation (Fig. 2 b, top, H12 and H22). When cultures were returned to the ambient environment after hypoxia (R), an increase in SERP1/RAMP4 mRNA was kept for at least 4 h (Fig. 2 b, top, H/R). Levels of SERP1/RAMP4 mRNA under H and H/R were then compared with those of Sec61 complex (Sec61α and Sec61β) and the molecular chaperons in ER (GRP78, ORP150), the latter known to be induced in H or other types of ER stress (Fig. 2 b, top). Transcripts of Sec61α and Sec61β increased in a manner paralleling SERP1/RAMP4, whereas GRP78 and ORP150 displayed enhanced expression at earlier times and began to decline soon after replacement into normoxia. Quantification of Northern analysis demonstrated that SERP1/RAMP4 and GRP78 mRNAs increased by ∼5.1- and 6.0-fold in H, compared with normoxia, respectively (Fig. 2 b, bottom).
Figure 2.
Expression of SERP1/RAMP4 in vivo and in vitro. (a) Distribution of SERP1 mRNA in normal rat organs. Total RNA (12 μg/lane) from the indicated organ of an adult male rat was applied to each lane. B, brain; H, heart; Lu, lung; L, liver; K, kidney; S, spleen; M, muscle; and T, testes. The lower panel shows ethidium bromide staining of the gel. (b) Expression of SERP1/RAMP4 mRNA under H/R and comparison with those of translocon (Sec61α and Sec61β) and molecular chaperons (ORP150 and GRP78). Cultured rat astrocytes were subjected to H for 12 h (H12), 22 h (H22), H for 22 h followed by R for 4 h (H/R), and the same amount of total RNA (15 μg/lane) was used for Northern analysis. The sixth panel shows ethidium bromide staining of the gel. (c) Expression of SERP1/RAMP4, GRP78, or HSP72 mRNA in astrocytes subjected to other forms of stress. Total RNA (10 μg/lane) from cultured rat astrocytes treated with A23187 (1 μM; A), tunicamycin (1 μg/ml; Tu), hydrogen peroxide (80 μM; H2O2), and heat shock (42°C; HS) or without any treatment (C) were used for Northern blotting to detect SERP1/RAMP4 transcripts. The middle panel shows ethidium bromide staining of the gel. (d) Expression of SERP1/RAMP4 protein in cultured rat astrocytes exposed to H, H/R, and ER stress. Protein extracts from cultured rat astrocytes (107 cells/condition) exposed to H (H22), H followed by R (H/R) (left panel), or treated with A23187 (1 μM; A) or tunicamycin (1 μg/ml; Tu) were immunoprecipitated with anti-SERP1/RAMP4 antibody and subjected to Western blotting with the same antibody. The binding of primary antibody was detected by enhanced chemiluminescence method. Migration of simultaneously run molecular mass standards is indicated on the far left in kD. Quantification of SERP1/RAMP4 mRNA, GRP78 mRNA, and SERP1/RAMP4 antigen was performed by laser densitometry and is expressed as fold increase in intensity under each condition compared with normoxic control (results of quantification are shown in the graphs at the bottom of b, c, and d). Mean ± SD of three experiments is shown.
Treatment of normoxic cultures with tunicamycin or A23187, both of which result in accumulation of unfolded polypeptides within the ER (Bush et al. 1994), caused a ∼4.2- and 5.8-fold increase in SERP1/RAMP4 transcripts, respectively, whereas hydrogen peroxide and HSP were without effect (Fig. 2 c). Accompanying H/R- and ER stress–induced elevation of SERP1/RAMP4 mRNA, there was a five- to sixfold increase in SERP1/RAMP4 antigen (∼10 kD) as shown using immunoprecipitation followed by immunoblotting with anti-SERP1 antibody (Fig. 2 d).
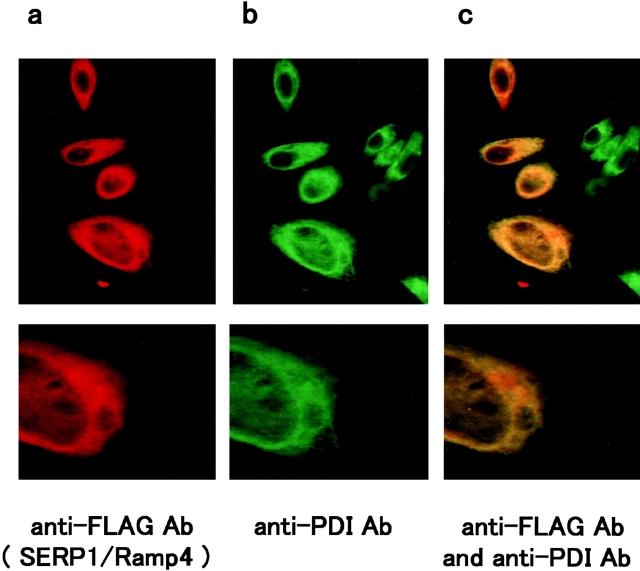
Immunostaining of BHK cells transiently transfected with FLAG-tagged SERP1/RAMP4 displayed an overlapping distribution of SERP1/RAMP4 antigen and the ER marker PDI (Fig. 3, a–c). Although SERP1/RAMP4 has a putative n-glycosylation site at its NH2 terminus, treatment of SERP1/RAMP4-transfected cells with tunicamycin or incubation of SERP1/RAMP4 protein itself with endoglycosidase H revealed no change in SERP1/RAMP4 (with respect to either its subcellular distribution or migration on SDS-PAGE), suggesting that glycosylation had not occurred (data not shown).
Figure 3.
Immunostaining of BHK cells transfected with the FLAG-tagged pcDNA/SERP1/RAMP4 using anti-FLAG antibody (a), anti-PDI antibody (b), and both (c). SERP1/RAMP4-transfected BHK cells were fixed in 0.1% NP-40/4% paraformaldehyde solution and subjected to the immunostaining protocol (see text). Sites of primary antibody binding were visualized with TRITC-conjugated anti–mouse antibody for FLAG epitope (a), FITC conjugated anti–rabbit antibody for PDI (b), or using both detection systems (c). Upper panels show lower magnification and lower panels higher magnification. Note that the two epitopes colocalize throughout the interior, and even in peripheral regions of the cells.
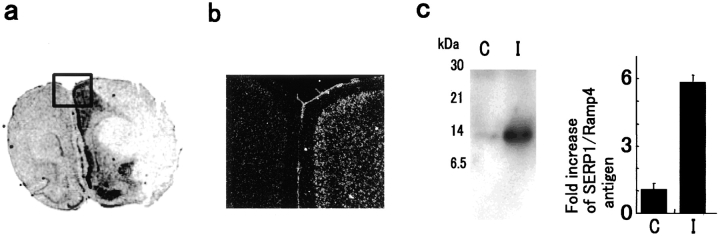
To extend our studies of SERP1/RAMP4 expression in response to oxygen deprivation in vivo, distribution of SERP1/RAMP4 transcripts and the expression of SERP1/RAMP4 antigen were assessed in rat brain after MCA occlusion. In situ hybridization showed increased SERP1/RAMP4 transcripts, especially in the periischemic penumbral region, compared with lower levels of expression in nonischemic areas (Fig. 4 a). Microautoradiography suggested that induction of SERP1/RAMP4 transcripts occurred in both neurons and astrocytes in the ischemic hemisphere (Fig. 4 b). Immunoprecipitation followed by Western blotting with anti-SERP1/RAMP4 antibody confirmed ∼5.7-fold induction of SERP1/RAMP4 antigen in rat ischemic brain (Fig. 4 c).
Figure 4.
Expression of SERP1/RAMP4 in ischemic rat brain. Brain ischemia was induced in rats by unilateral (left) MCA occlusion. After 12 h of ischemia, rats were killed and brain slices were studied by in situ hybridization (a and b) with riboprobes derived from SERP1 cDNA to detect SERP1/RAMP4 transcripts. SERP1/RAMP4 antigen was also studied with the homogenates of the same brain samples using immunoprecipitation followed by Western blotting with anti–SERP1/RAMP4 antibody (c). b shows a microautoradiogram of boxed area in a, displaying enhanced SERP1/RAMP4 mRNA in the periischemic penumbral region. c shows the induction of SERP1/RAMP4 at the protein level in ischemic brain. C, control hemisphere; I, ischemic hemisphere. Migration of simultaneously run molecular mass standards is indicated on the left side in kD. Quantification of SERP1/RAMP4 antigen in ischemic brain (c, right) was showed as fold increase (mean ± SD of three experiments).
Effect of SERP1/RAMP4 on the Stabilization of Membrane Proteins in Response to ER Stress
The effect of SERP1/RAMP4 on processing of two integral membrane glycoproteins, RAGE (Neeper et al. 1992) and CD8, was studied by cotransfecting 293 cells with expression vectors for each along with pcDNA/SERP1/RAMP4. Transient transfection of cultures with pcDNA3/RAGE followed by treatment of cells with tunicamycin (Fig. 5 a) or A23187 (data not shown) resulted in trapping of RAGE in the ER followed by degradation. Similar results were observed when CD8 was overexpressed and cultures were treated with these drugs. Intracellular levels of RAGE were analyzed by immunoblotting (Fig. 5 b) and metabolic labeling with [35S]methionine followed by immunoprecipitation (Fig. 5 c). 293 cells overexpressing RAGE and exposed to tunicamycin or A23187 showed a decrease in the amount of RAGE antigen (∼58 and 36%, respectively, compared with quiescent cultures; Mr ∼55 kD in glycosylated form and 52 kD in unglycosylated form), as well as a poorly defined smear of RAGE immunoreactivity in the upper portion of the membrane, the latter likely due to aggregation of RAGE (Fig. 5 b, lanes 2 and 3), and/or several more rapidly migrating immunoreactive bands, consistent with the occurrence of degradation (Fig. 5 b, lane 3). Immunoprecipitation of cell lysates with anti-RAGE antibody followed by Western blotting with antiubiquitin antibody showed that the smear of RAGE immunoreactivity includes polyubiquitinated RAGE (data not shown). Although cotransfection of quiescent 293 cells with pcDNA3/RAGE and pcDNA3/SERP1/RAMP4 had no apparent effect on RAGE antigen (Fig. 5 b, lane 4), overexpression of SERP1/RAMP4 in tunicamycin- or A23187-treated cultures increased the intensity of the major RAGE band to ∼80 and 86% of the intensity in untreated controls, respectively. Furthermore, both higher and lower molecular weight immunoreactive RAGE bands disappeared in cotransfectants overexpressing RAGE and SERP1/RAMP4 (Fig. 5 b, lanes 5 and 6). In contrast, overexpression of Sec61β instead of SERP1/RAMP4 did not have a similar effect on RAGE and overexpression of SERP1/RAMP4 did not stabilize cytosolic proteins, such as IκBα, which also undergoes degradation in response to stress conditions under study (data not shown).
Figure 5.
Effect of SERP1/RAMP4 on the stabilization and refolding of membrane proteins during and after ER stress. (a) Subcellular localization of RAGE in control and in tunicamycin-treated cells. RAGE-transfected BHK cells were fixed in 0.1% NP-40/4% paraformaldehyde and subjected to the immunostaining with rabbit anti-RAGE antibody (primary antibody) and FITC-conjugated anti–rabbit IgG (secondary antibody). Addition of tunicamycin caused RAGE to be distributed mainly in the perinuclear region. (b) Western blotting with anti-RAGE IgG using cell extracts from RAGE-transfected 293 cells with or without cotransfection with pcDNA/SERP1/RAMP4 and/ or treatment with tunicamycin and A23187 (top), and quantification of the intensity of RAGE-immunoreactive bands as a percentage of the intensity of the bands in control RAGE-transfected cultures (bottom). Mean ± SD of three experiments is shown. Lane 1, RAGE transfection alone; lane 2, RAGE transfection plus tunicamycin; lane 3, RAGE transfection plus A23187; lane 4, RAGE plus SERP1/RAMP4 cotransfection; lane 5, RAGE plus SERP1/RAMP4 cotransfection plus tunicamycin; and lane 6, RAGE plus SERP1/RAMP4 cotransfection plus A23187. (c) Immunoprecipitation with either anti-RAGE or anti-CD8 IgG after metabolic labeling for 30 min alone or with additional chasing for 60 min (top), and quantification of the intensity of the corresponding RAGE-CD8 immunoreactive bands as a percentage of the same band in control RAGE-CD8–transfected cultures (bottom). Mean ± SD of three experiments is shown. Lane 1, transfection with pcDNA3/RAGE or pcDNA3/CD8 alone; lane 2, transfection as in lane 1 followed by treatment with tunicamycin; lane 3, transfection as in lane 1 followed by treatment with A23187; lane 4, transfection as in lane 1 followed by cotransfection with pcDNA/SERP1/RAMP4 and treatment with tunicamycin; and lane 5, transfection as in lane 1 followed by cotransfection with pcDNA/SERP1/RAMP4 and treatment with A23187. (d) Metabolic labeling of RAGE-transfected 293 cells after quenching tunicamycin treatment. Control cells (lane 4) were not treated with tunicamycin or cotransfected with pcDNA3/SERP1/RAMP4. Cultures were transfected or cotransfected with constructs encoding RAGE and/or SERP1. Glycosylated and unglycosylated forms of RAGE are indicated by CHO+ and CHO−.
293 cells transfected with pcDNA3/RAGE were labeled with [35S]methionine for 30 min after treatment of cell cultures with tunicamycin or A23187. Immunoprecipitation of RAGE resulted in the appearance of the expected immunoreactive species (Fig. 5 c, top). However, RAGE present after 60 min into the chase period was substantially reduced in tunicamycin or A23187-treated cells (32 and 5% of untreated control, respectively), presumably due to degradation (Fig. 5 c, lanes 2 and 3). Cotransfection with pcDNA/SERP1/RAMP4, along with pcDNA3/RAGE, maintained, at least in part, levels of RAGE in cells exposed to tunicamycin (Fig. 5 c, lane 4) (∼73% of untreated control) or A23187 (Fig. 5 c, lane 5; ∼60% of untreated control). Similar results were obtained when pcDNA3/CD8 was transfected instead of pcDNA3/RAGE (Fig. 5 c).
Effect of SERP1/RAMP4 on the Glycosylation of Membrane Proteins after the ER Stress
In view of the enhanced expression of SERP1/RAMP4 in both H and R, and its homology with the yeast protein YSY6p (Sakaguchi et al. 1991), further studies were performed to assess the contribution of SERP1/RAMP4 to protein refolding (glycosylation) after stress. SERP1/RAMP4 overexpression had no detectable effect on the glycosylation of RAGE in the presence of tunicamycin (Fig. 5b and Fig. c). However, metabolic labeling with [35S]methionine for 3 h using pcDNA3/RAGE-transfected 293 cells after quenching tunicamycin treatment demonstrated that SERP1/RAMP4 overexpression not only stabilized unglycosylated RAGE but also facilitated, at least in part, the glycosylation of RAGE (Fig. 5 d, lanes 2 and 3; ∼24% of total RAGE was glycosylated). These effects of SERP1/RAMP4 overexpression during and after the stress were not due to induction of the OST complex, the enzymes that are required for (N-linked) glycosylation (data not shown).
Interactions of SERP1/RAMP4, RAGE, Sec61 Complex, and Molecular Chaperons
To analyze the mechanism underlying the chaperon-like functions of SERP1/RAMP4 (by stabilizing and facilitating glycosylation of membrane proteins) under and after ER stress, we first studied the interaction of SERP1/RAMP4 with integral membrane protein RAGE. However, co-IP of 293 cells transfected with pcDNA3/RAGE and pcDNA3/SERP1/RAMP4 did not show a complex comprised of both molecules under either normal and stress conditions. Next, the binding of SERP1/RAMP4 to Sec61 complex was examined, since RAMP4 was originally copurified with Sec61 complex (Görlich and Rapoport 1993). Co-IP study using whole cell lysates in NP-40–containing buffer (Fig. 6 a) or co-IP after cross-linking of membrane fractions (Fig. 6 b) revealed that SERP1/RAMP4 formed a stable complex with Sec61β both in transfected cells and under endogenous conditions. In Fig. 6 b, lane 4, the two major bands at ∼42 and 27 kD may represent the heterotrimer and heterodimer of Sec61β and SERP1/RAMP4 complex based on the molecular masses of both molecules. It should be noted that transfection of cells with pcDNA3/SERP1/RAMP4 resulted in at least 10 times higher expression of proteins, based on immunoprecipitation and Western blotting (data not shown). To rule out the possibility that Sec61β formed a complex with SERP1/RAMP4 after its dissociation from Sec61 complex in NP-40–containing buffer, similar co-IP studies were performed with cell lysates made in the presence of deoxycholate (Fig. 6 c). Under these conditions, not only Sec61β but also Sec61α was coimmunoprecipitated with SERP1/RAMP4.
Figure 6.
Interaction of SERP1/RAMP4, RAGE, Sec61 complex and molecular chaperon calnexin. (a and c–f) 293 cells were transfected or cotransfected with constructs encoding FLAG-SERP1/RAMP4 and/or RAGE, and co-IP was performed with the indicated antibodies after lysis of cultures in buffer containing NP-40 (a, d, and e) or deoxycholate (c and f). (b) Co-IP study after cross-linking membrane fractions from 293 cells cotransfected with pcDNA3/FLAG-SERP1/ RAMP4 and pcDNA3/HA-Sec61β. Cross-linking was performed with DSS as described in the text, and co-IP was carried out using anti-FLAG and anti-HA antibodies. Migration of simultaneously run molecular mass standards is indicated on the left in kD.
Formation of a complex between RAGE and Sec61 complex was also studied by co-IP; Sec61β–RAGE complex was detected in response to stress in both NP-40– and deoxycholate-containing buffer, although Sec61α–RAGE binding was detected only in the presence of deoxycholate (Fig. 6d and Fig. e). SERP1/RAMP4 also became associated with calnexin, which is a membrane protein and a molecular chaperon in ER (Fig. 6 f). Such an interaction occurred under control (endogenous) conditions and after overexpression of SERP1/RAMP4. In these co-IP studies, ∼10 and 15% of Sec61β was coimmunoprecipitated with SERP1 and unglycosylated RAGE, respectively (data not shown).
Discussion
A key facet of the cellular response to the environmental stress imposed by oxygen deprivation appears to be induction of molecular chaperons. In this context, previous studies from our and other laboratories have demonstrated induction of molecular chaperons in ER, GRP78, GRP94, and a novel 150-kD polypeptide termed ORP150 (Lowenstein et al. 1994; Massa et al. 1995; Kuwabara et al. 1996) in hypoxic and ischemic conditions. Suppression of ORP150 transcripts in oxygen-deprived cultures of human embryonic kidney cells increased their vulnerability to H-induced apoptosis (Ozawa et al. 1999), suggesting that a stress response in ER might have a important role in adaptation to ischemic challenge. The current studies describe another polypeptide, SERP1, whose expression is increased by H/R although the induction is preceded by those of molecular chaperons in ER, and which participates in the ER stress response due to the accumulation of unfolded proteins in the ER.
SERP1 encodes a polypeptide localized to the ER and comprised of only 66 amino acids, which was identical to RAMP4. RAMP4 was originally copurified with the core component of the protein-translocation machinery of the ER, the Sec61 complex, and it was recently reported that RAMP4 controls the glycosylation of major histocompatbility complex class II–associated invariant chain (Schroder et al. 1999). Sec61 is a complex composed of α, β, and γ subunits, which assemble into a channel through which nascent proteins enter the ER (Görlich et al. 1992; Wilkinson et al. 1997; Kalies et al. 1998). A putative transmembrane-spanning domain at the COOH terminus probably anchors SERP1/RAMP4 in the ER membrane. The orientation of SERP1/RAMP4 in the ER is likely to present the NH2 terminus to the cytosol, as predicted by previous studies of other similar proteins (Hartmann et al. 1989; Kutay et al. 1993), and consistent with the apparent lack of glycosylation of the putative N-glycosylation site at the SERP1/RAMP4 NH2 terminus. SERP1/RAMP4 displays homology with yeast YSY6p, identified based on its capacity to suppress the defect in protein export of a SecY (Sec61 homologue in bacteria) mutant (Sakaguchi et al. 1991). The identity between SERP1 and RAMP4, and the homology between SERP1 and YSY6, as well as induction of SERP1 in response to ER stress at mRNA and protein levels, suggested the hypothesis that SERP1 might participate in the biosynthesis or degradation of secretory and membrane proteins.
To evaluate the effect of SERP1/RAMP4 on protein processing, experiments were performed to address whether SERP1/RAMP4 has any effect on stabilization and refolding of membrane proteins during stress conditions (i.e., ER stress). Induction of ER stress with tunicamycin or A23187 in 293 cells promoted aggregation and degradation of two integral membrane proteins, RAGE and CD8, members of the Ig superfamily of cell surface molecules with a single transmembrane-spanning domain. Cotransfection of 293 cells to overexpress SERP1/RAMP4 prevented accelerated aggregation and degradation of RAGE and CD8 in the setting of ER stress (Fig. 5b and Fig. c). Although it is not yet clear whether these effects of SERP1/RAMP4 occurred by directly preventing the degradation of membrane proteins or by promoting their refolding in the ER, our results support the latter concept, because SERP1/RAMP4 did facilitate, at least in part, glycosylation of integral membrane proteins in the ER after ER stress was relieved (Fig. 5 d) without induction of the OST.
Co-IP and cross-linking studies revealed that SERP1/RAMP4 interacts with the Sec61α and Sec61β subunits, which bind to RAGE (and by analogy, other membrane proteins) in response to cellular perturbation (Fig. 6, a–e). It is important to note that SERP1/RAMP4 is likely to interact with other ER proteins in addition to Sec61α and Sec61β, as indicated by the presence of several bands after immunoblotting of SERP1/RAMP4 after cross-linking (Fig. 6 b). However, in a previous cross-linking study, RAMP4 was not detected in a complex with Sec61β (Kalies et al. 1998). We speculate that the reason for this difference with our results is likely to derive from the use of different cross-linking agents. Kalies et al. 1998 used bis-maleimidohexane, which depends on optimal juxtaposition of cysteines, whereas our study used DSS, which does not interact with sulfhydryl groups. SERP1/RAMP4 is coprecipitated with calnexin (Fig. 6 f), a membrane protein and a molecular chaperon in ER known to associate with folding intermediates of glycoproteins (monomeric glycoproteins) and believed to play a major role for the quality control apparatus in ER (Ou et al. 1993; Wada et al. 1997). Together these results suggest that the stabilization of membrane proteins during and after ER stress involves the concerted action of a rescue unit in the ER membrane comprised of SERP1/RAMP4, components of the translocon such as Sec61α and Sec61β, and ER chaperons, such as calnexin.
Increased expression of components of the Sec61 complex begins relatively late during H (>12 h) and is sustained during R. This suggests that induction of the Sec61 complex probably has an integral role for increased biosynthesis of luminal and membrane proteins in replacement of those damaged during stress. It is noteworthy that expression of SERP1/RAMP4 parallels that of components of the Sec61 complex and not that of luminal ER chaperons, which declines during R. This may indicate that the functions of the Sec61 complex and SERP1/RAMP4 during de novo protein synthesis are tightly coupled. These considerations underline the importance of further studies to determine the detailed function of SERP1/RAMP4 in the context of cell stress, and the contribution of SERP1/RAMP4 to cellular processing of native polypeptides or disease-related proteins, such as the cystic fibrosis transmembrane conductance regulator (CFTR) (Bebok et al. 1998; Ward et al. 1995; Jensen et al. 1995) or amyloid-beta precursor protein (APP) (Coor et al. 1997; Hartmann et al. 1997).
Acknowledgments
We are grateful to Dr. Tom A. Rapoport for providing anti-Sec61α and anti-Sec61β antibodies. We thank Drs. Kazunori Imaizumi and Ryuichi Masaki for providing cDNAs and/or antibodies for CD8 and PDI. We also thank Drs. Masao Sakaguchi (Kyusyu University, Fukuoka, Japan) and Koreaki Ito (Kyoto University, Kyoto, Japan) for providing the information for yeast YSY6p and Dr. Naoya Sato (Osaka University) for giving valuable suggestions.
Footnotes
A. Yamaguchi and O. Hori contributed equally to this work.
Abbreviations used in this paper: co-IP, coimmunoprecipitation; DSS, disuccinimidyl suberate; GRP, glucose-regulated protein; H, hypoxia; HA, hemagglutinin; HSP, heat shock protein; MCA, middle cerebral artery; ORF, open reading frame; ORP, oxygen-regulated protein; OST, oligosaccaryltransferase; PDI, protein disulfide isomerase; R, reoxygenation; RAGE, receptor for advanced glycation endproducts; RAMP, ribosome-associated membrane protein; SERP, stress-associated endoplasmic reticulum protein; YSY6p, yeast suppressor of SecY 6 protein.
References
- Akagi S., Yamamoto A., Yoshimori T., Masaki R., Ogawa R., Tashiro Y. Distribution of protein disulfide isomerase in rat hepatocytes. J. Histochem. Cytochem. 1988;36:1533–1542. doi: 10.1177/36.12.3192937. [DOI] [PubMed] [Google Scholar]
- Bebok Z., Mazzochi C., King S.A., Hong J.S., Sorscher E.J. The mechanism underlying cystic fibrosis transmembrane conductance regulator transport from the endoplasmic reticulum to the proteasome includes Sec61β and a cytosolic, deglycosylated intermediary. J. Biol. Chem. 1998;273:29873–29878. doi: 10.1074/jbc.273.45.29873. [DOI] [PubMed] [Google Scholar]
- Bush K.T., Hendrickson B.A., Nigam S.K. Induction of FK506-binding protein, FKBP13, under conditions which misfold proteins in endoplasmic reticulum. Biochem. J. 1994;303:705–708. doi: 10.1042/bj3030705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D.W. Calcium still center-stage in hypoxic-ischemic neuronal death. Trends Neurosci. 1995;18:58–60. [PubMed] [Google Scholar]
- Coor D.G., Forman M.S., Sung J.C., Leight S., Kolson D.L., Iwatsubo T., Lee V.M.Y., Doms R.W. Alzheimer's Aβ (1-42) is generated in the endoplasmic reticulum/intermediate compartment of NT2N cells. Nat. Med. 1997;3:1021–1023. doi: 10.1038/nm0997-1021. [DOI] [PubMed] [Google Scholar]
- Garcia J.H., Yoshida Y., Chen H., Li Y., Zhang Z.G., Lian J., Chen S., Chopp M. Progression from ischemic injury to infarct following middle cerebral artery occlusion in the rat. Am. J. Pathol. 1993;142:623–635. [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Rapoport T.A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- Görlich D., Prehn S., Hartmann E., Kalies U.K., Rapoport T.A. A mammalian homologue of sec61p and secYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- Hartmann E., Rapoport T.A., Lodish H.F. Predicting the orientation of eukaryotic membrane spanning proteins. Proc. Natl. Acad. Sci. USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E., Sommer T., Prehn S., Gorlich D., Jentsch S., Rapoport T.A. Evolutionary conservation of components of the protein translocation complex. Nature. 1994;367:654–657. doi: 10.1038/367654a0. [DOI] [PubMed] [Google Scholar]
- Hartmann T., Bieger S.C., Bruhl B., Tienari P.J., Ida N., Allsop D., Roberts G.W., Masters C.L., Dotti C.G., Unsicker K., Beyreuther K. Distinct sites of intracellular production of Alzheimer's disease Aβ40/42 amyloid peptides. Nat. Med. 1997;3:1016–1020. doi: 10.1038/nm0997-1016. [DOI] [PubMed] [Google Scholar]
- Hertz L. Astrocytes. In: Lajtha A., editor. Handbook of Neurochemistry. Vol. 1. 2nd ed. Plenum Press; New York: 1982. pp. 603–661. [Google Scholar]
- Hori O., Matsumoto M., Maeda Y., Ueda H., Kinoshita T., Stern D., Ogawa S., Kamada T. Metabolic and biosynthetic alteration in cultured astrocytes exposed to hypoxia/reoxygenation. J. Neurochem. 1994;62:1489–1495. doi: 10.1046/j.1471-4159.1994.62041489.x. [DOI] [PubMed] [Google Scholar]
- Hori O., Matsumoto M., Kuwabara K., Maeda Y., Ueda H., Ohtsuki T., Kinoshita T., Ogawa S., Stern D., Kamada T. Exposure of astrocytes to hypoxia/reoxygenation enhances expression of glucose-regulated protein 78 facilitating astrocyte release of the neuroprotective cytokine interleukin 6. J. Neurochem. 1996;66:973–979. doi: 10.1046/j.1471-4159.1996.66030973.x. [DOI] [PubMed] [Google Scholar]
- Imuta N., Ogawa S., Maeda Y., Kuwabara K., Hori O., Ueda H., Yanagihara T., Tohyama M. Induction of 72-kDa inducible heat shock protein (HSP72) in cultured rat astrocytes after energy depletion. J. Neurochem. 1998;70:550–557. doi: 10.1046/j.1471-4159.1998.70020550.x. [DOI] [PubMed] [Google Scholar]
- Jensen T.J., Loo M.A., Pind S., Williams D.B., Goldberg A.L., Riordan J.R. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- Kalies U.K., Rapoport T.A., Hartmann E. The β subunit of the sec61 complex facilitates cotranslational protein transport and interacts with the signal peptidase during translocation. J. Cell Biol. 1998;141:887–894. doi: 10.1083/jcb.141.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koistinaho J., Hokfelt T. Altered gene expression in brain ischemia. Neuroreport. 1997;8:1–8. [PubMed] [Google Scholar]
- Kutay U., Hartmann E., Rapoport T.A. A class of membrane proteins with a c-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- Kuwabara K., Matsumoto M., Ikeda J., Hori O., Ogawa S., Maeda Y., Kitagawa K., Imuta N., Kinoshita T., Stern D. Purification and characterization of a novel stress protein, the 150-kDa oxygen-regulated protein (ORP150), from cultured rat astrocytes and its expression in ischemic mouse brain. J. Biol. Chem. 1996;27:5025–5032. doi: 10.1074/jbc.271.9.5025. [DOI] [PubMed] [Google Scholar]
- Liang P., Pardee A.B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Lowenstein D.H., Gwinn R.P., Seren M.S. Increased expression of mRNA encoding calbindin-D28K, the glucose-regulated proteins, or 72 kDa heat shock protein in three models of acute CNS injury. Mol. Brain Res. 1994;22:299–308. doi: 10.1016/0169-328x(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Maeda Y., Matsumoto M., Ohtsuki T., Kuwabara K., Ogawa S., Hori O., Yan S.D., Kinoshita T., Kamada T., Stern D. Hypoxia/reoxygenation-mediated induction of interleukin 6a paracrine mechanism potentially enhancing neuron survival. J. Exp. Med. 1994;180:2297–2308. doi: 10.1084/jem.180.6.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa S.M., Longo F.M., Zuo J., Chen W.J., Sharp F.R. Cloning of rat grp75, an hsp 70-family member, and its expression in neuronal and ischemic brain. J. Neurosci. Res. 1995;40:807–819. doi: 10.1002/jnr.490400612. [DOI] [PubMed] [Google Scholar]
- Matsuo N., Ogawa S., Imai Y., Takagi T., Tohyama M., Stern D., Wanaka A. Cloning of a novel RNA binding poly-peptide (RA301) induced by hypoxia/reoxygenation. J. Biol. Chem. 1995;270:28216–28222. doi: 10.1074/jbc.270.47.28216. [DOI] [PubMed] [Google Scholar]
- Matsuo N., Ogawa S., Takagi T., Wanaka A., Mori T., Matsuyama M., Pinsky D., Stern D., Tohyama M. Cloning of a putative vesicle transport-related protein, RA410, from cultured rat astrocytes and its expression in ischemic rat brain. J. Biol. Chem. 1997;272:16438–16444. doi: 10.1074/jbc.272.26.16438. [DOI] [PubMed] [Google Scholar]
- McCarthy K.D., de Vellis J. Preparation of separate and astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord J.M. Oxygen-derived radicalsa link between reperfusion injury and inflammation. Fed. Proc. 1987;46:2402–2406. [PubMed] [Google Scholar]
- Neeper M., Schmidt A.M., Brett J., Yan S.D., Wang F., Pan Y.C., Elliston K., Stern D., Shaw A. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J. Biol. Chem. 1992;267:14998–15004. [PubMed] [Google Scholar]
- Ogawa S., Herwig G., Esposito C., Macaulay A.P., Brett J., Stern D. Hypoxia modulates the barrier and coagulant function of cultures of bovine endothelium. J. Clin. Invest. 1990;69:1090–1098. doi: 10.1172/JCI114540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou W.J., Cameron P.H., Thomas D.Y., Bergeron J.J.M. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature. 1993;364:771–776. doi: 10.1038/364771a0. [DOI] [PubMed] [Google Scholar]
- Ozawa K., Kuwabara K., Tamatani M., Takatsuji K., Tsukamoto Y., Kaneda S., Yanagi H., Stern D., Eguchi Y., Tsujimoto Y. 150 kDa oxygen-regulated protein (ORP150) suppressed hypoxia-induced apoptotic cell death. J. Biol. Chem. 1999;274:6397–6404. doi: 10.1074/jbc.274.10.6397. [DOI] [PubMed] [Google Scholar]
- Rothman S.M., Olney J.W. Excitotoxity and the NMDA receptor—still lethal after eight years. Trends Neurosci. 1995;18:57–58. doi: 10.1016/0166-2236(95)93869-y. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M., Ueguchi C., Ito K., Omura T. Yeast gene which suppresses the defect in protein transport of a secY mutant of E. coli . J. Biochem. 1991;109:799–802. doi: 10.1093/oxfordjournals.jbchem.a123460. [DOI] [PubMed] [Google Scholar]
- Schmidt A.M., Mora R., Cao R., Yan S.D., Brett J., Ramakrishnan R., Tsang T.C., Simionescu M., Stern D. The endothelial cell binding site for advanced glycation end products consists of a complexan integral membrane protein and a lactoferrin-like polypeptide. J. Biol. Chem. 1994;269:9882–9888. [PubMed] [Google Scholar]
- Schmidt A.M., Hori O., Chen J.X., Li J.F., Crandall J., Zhang J., Cao R., Yan S.D., Brett J., Stern D. Advanced glycation endproducts interacting with their endothelial receptor induce expression of vascular cell adhesion molecule-1 (VCAM-1) in cultured human endothelial cells and in micea potential mechanism for the accelerated vasculopathy of diabetes. J. Clin. Invest. 1995;96:1395–1403. doi: 10.1172/JCI118175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K., Martoglio B., Hofmann M., Holscher C., Hartmann E., Prehn S., Rapoport T.A., Dobberstein B. Control of glycosylation of MHC class II-associated invariant chain by translocon-associated RAMP4. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:4804–4815. doi: 10.1093/emboj/18.17.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solenski N.J., Kwan A.L., Yamamoto H., Bennett J.P., Kassell N.F., Lee K.S. Differential hydroxylation of salicylate in core and penumbra regions during focal reversible cerebral ischemia. Stroke. 1997;28:2545–2552. doi: 10.1161/01.str.28.12.2545. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y., Kuwabara K., Hirota S., Ikeda J., Stern D., Yanagi H., Matsumoto M., Ogawa S., Kitamura Y. 150-kD oxygen-regulated protein is expressed in human atherosclerotic plaques and allows mononuclear phagocytes to withstand cellular stress on exposure to hypoxia and modified low density lipoprotein. J. Clin. Invest. 1996;98:1930–1941. doi: 10.1172/JCI118994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada I., Kai M., Imai S., Sakane F., Kanoh H. Promotion of transferrin folding by cyclic interaction with calnexin and calreticulin. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:5420–5432. doi: 10.1093/emboj/16.17.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward C.L., Omura S., Kopito R.R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Wilkinson B.M., Esnault Y., Craven R.A., Skiba F., Fieschi J., Kepes F., Stirling C.J. Molecular architecture of the ER translocase probed by chemical crosslinking of Sss1p to complementary fragments of sec61p. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:4549–4559. doi: 10.1093/emboj/16.15.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]