Abstract
Merozoite surface protein 1 of Plasmodium vivax (PvMSP-1), a major target for malaria vaccine development, contains six highly polymorphic domains interspersed with conserved sequences. Although there is evidence that the sequence divergence in PvMSP-1 has been maintained over 5 million years by balanced selection exerted by the host's acquired immunity, the variant specificity of naturally acquired antibodies to PvMSP-1 remains poorly investigated. Here, we show that 15 recombinant proteins corresponding to PvMSP-1 variants commonly found in local parasites were poorly recognized by 376 noninfected subjects aged 5 to 90 years exposed to malaria in rural Amazonia; less than one-third of them had detectable immunoglobulin G (IgG) antibodies to at least one variant of blocks 2, 6, and 10 that were expressed, although 54.3% recognized the invariant 19-kDa C-terminal domain PvMSP-119. Although the proportion of responders to PvMSP-1 variants increased substantially during subsequent acute P. vivax infections, the specificity of IgG antibodies did not necessarily match the PvMSP-1 variant(s) found in infecting parasites. We discuss the relative contribution of antigenic polymorphism, poor immunogenicity, and original antigenic sin (the skew in the specificity of antibodies elicited by exposure to new antigenic variants due to preexisting variant-specific responses) to the observed patterns of antibody recognition of PvMSP-1. We suggest that antibody responses to the repertoire of variable domains of PvMSP-1 to which subjects are continuously exposed are elicited only after several repeated infections and may require frequent boosting, with clear implications for the development of PvMSP-1-based subunit vaccines.
Plasmodium vivax is a major public health challenge in Central and South America; the Middle East; Central, South, and Southeast Asia; Oceania; and East Africa, where 2.6 billion people are currently at risk of infection (11), and 70 to 80 million clinical cases are reported each year (22). This human pathogen reappeared in Asian countries where eradication efforts had been successful in the 1960s, such as Uzbekistan (30), Azerbaijan (16), and the Republic of Korea (18). In Brazil, P. vivax surpassed Plasmodium falciparum two decades ago as the main cause of malaria morbidity across the Amazon Basin (19) and caused 80% of the 600,000 malaria cases reported in that country in 2005 (Ministry of Health of Brazil, unpublished data). The emergence of resistance to the first-line antimalarial drug chloroquine is a major concern for the current strategies of P. vivax malaria control (1).
One of the main obstacles to the acquisition of antimalarial immunity is the polymorphism in potential target antigens, which enables parasites to evade immune responses elicited by past exposure to variant forms of the same antigen (6). The 200-kDa merozoite surface protein 1 (MSP-1) of Plasmodium vivax (PvMSP-1), a target of naturally acquired (25) and vaccine-induced (12, 40) immunity, contains six highly polymorphic domains (four of them repetitive) flanked by fairly conserved sequences (27) (Fig. 1A). The extensive sequence divergence in variable domains of PvMSP-1 (amino acid similarity, 21 to 34%) has been maintained by balanced selection over 5 million years, most likely as a result of variant-specific immune pressure (28).
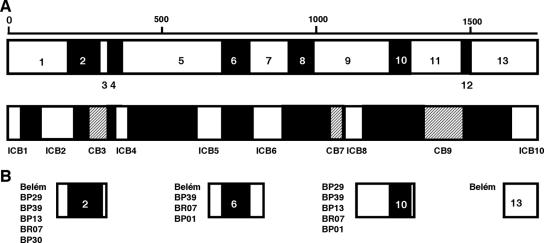
FIG. 1.
Schematic representation of PvMSP-1 and the recombinant antigens used in this study. (A) The PvMSP-1 protein sequence comprises seven conserved blocks (amino acid similarity among PvMSP-1 allele products of 71 to 85% [represented as open boxes]) and six variable blocks (amino acid similarity of 21 to 34% [represented as black boxes]) described previously by Putaporntip et al. (27). For comparison, we also show the original division of PvMSP-1 into interspecies conserved blocks (ICB) (amino acid similarity of >48% in pairwise comparisons of MSP-1 orthologs of P. falciparum, P. vivax, and Plasmodium yoelii [represented as open boxes]), conserved blocks (CB) (amino acid similarity of >50% between P. falciparum and P. vivax but lower in other pairwise comparisons [represented as hatched boxes]), and polymorphic blocks (amino acid similarity of <45% [represented as black boxes]) (5). These two ways of portioning PvMSP-1 differ in that Putaporntip et al. (27) made intraspecific-sequence comparisons, while del Portillo et al. (5) made sequence comparisons among species. Recombinant antigens used in previous studies are often named after interspecies conserved blocks (17, 24, 25, 32); for example, ICB2-5 comprises blocks 1, 2, 3, 4, and 5 defined previously by Putaporntip and others (27). (B) Location of the 16 recombinant antigens used in this study. Note that antigens corresponding to variable domains of PvMSP-1 comprise stretches of conserved flanking sequence. Block 13 antigen corresponds to ICB10, the C-terminal 19-kDa fragment of PvMSP-1.
Although domains under balancing selection have been suggested to represent potential candidates for malaria vaccine development (2, 3), the immune recognition of variable domains of PvMSP-1 and its role in antimalarial immunity remain poorly investigated. A decade ago, Colombian malaria patients were shown to contain antibodies that discriminated between recombinant antigens representing two highly divergent versions of block 6 (20), but no association was made between antibody response and clinical immunity. More recently, naturally acquired antibodies that recognize a recombinant protein (ICB2-5) encompassing variable blocks 2 and 4 of PvMSP-1 were associated with protection against clinical disease in Brazil (25), but it remains unknown whether protective antibodies are variant specific, since a single version of these highly polymorphic domains was used in serology, and PvMSP-1 variants have not been characterized in parasites from incident infections.
Here, we analyze the extent to which PvMSP-1 sequence diversity affects the development of antibody responses to this major malaria vaccine candidate antigen. We compare naturally acquired antibody responses to three polymorphic domains and the highly conserved C-terminal domain of PvMSP-1 in a cohort of malaria-exposed rural Amazonians. We measured specific antibodies at the study baseline, in the absence of P. vivax infection, and during subsequent infections. Sequences of the variable domains of PvMSP-1 used in serology were characterized in local parasites circulating before and during the follow-up to confirm that the variants expressed as recombinant antigens match those to which subjects have been exposed. We discuss the relative contribution of antigenic polymorphism, poor immunogenicity, and original antigenic sin (the skew in the specificity of antibodies elicited by exposure to new antigenic variants due to preexisting variant-specific responses) to the observed patterns of antibody recognition of PvMSP-1 variants.
MATERIALS AND METHODS
Study area and population.
The study protocol was approved by the Ethical Review Board of the Institute of Biomedical Sciences of the University of São Paulo, São Paulo, Brazil (protocol 318, 2002). Blood samples for serum separation and DNA extraction were collected during the first 15 months of follow-up (March 2004 to May 2005) of an ongoing, population-based cohort study in an agricultural settlement (Ramal do Granada [9°41′S-9°49′S, 67°05′W-67°07′W]) in the eastern part of the state of Acre, northwestern Brazil (see Fig. S1 in the supplemental material). Both P. falciparum and P. vivax are transmitted year-round. The study area, cohort recruitment strategies, and baseline data were described in detail elsewhere previously (31). Briefly, all households in the area were visited, and 509 residents (98.5% of the entire population of Ramal do Granada), aged between 1 day and 90 years, were enrolled from March to April 2004 (466 subjects [91.6%]) or September to October 2004 (43 subjects [8.4%]). A questionnaire was given to all study participants to obtain demographic and clinical information and assess their cumulative exposure to malaria. Since most (62.2%) subjects were migrants from malaria-free areas, their ages do not necessarily correlate with exposure to malaria. Cumulative exposure to malaria was therefore estimated as the length of residence in areas where malaria is endemic (either in Acre or elsewhere in Amazonia), while recent exposure to P. vivax was estimated as the number of slide-confirmed P. vivax malaria episodes recorded in the three local malaria diagnosis outposts between January 2001 and March 2004. The 399 study participants aged 5 years or older who had no evidence of P. vivax infection detected by thick-smear microscopy or PCR (15, 39) were considered to be eligible for participation in the serological study. Subjects with P. vivax infection at the baseline (22 infections detected by PCR only and 7 infections detected by both PCR and thick-smear microscopy [total, 29 infections in subjects aged ≥5 years]) were excluded from the serological study because current exposure to the parasite would be a major confounder in the analysis of other determinants (age and cumulative and recent exposure to malaria) of antibody reactivity; 376 (95.7%) eligible subjects had their baseline serum samples tested for antibodies to PvMSP-1.
Malaria surveillance, serum samples, and P. vivax isolates.
Malaria episodes were diagnosed during 15 months of follow-up of our cohort population (between March 2004 and May 2005) through both active and passive case detection. For active case detection, all households in the study area were visited 5 days/week by our field team, and blood samples were collected from all subjects who had fever or other symptoms suggestive of malaria since the last visit. Additional malaria episodes were found by passive case detection when symptomatic study participants had a malaria diagnosis confirmed at one of the three government-run malaria outposts in the study area. The combined active and passive case detection strategy identified 157 episodes of symptomatic P. vivax infection confirmed by standardized thick-smear microscopy (38) during the cohort follow-up (489 person-years of follow-up), with an average monthly incidence of 2.67/100 person-months at risk (95% confidence interval [CI], 2.27 to 3.12/100 person-months at risk). P. vivax isolates from 55 (35.0%) infections were obtained for PvMSP-1 gene amplification and sequencing to characterize the PvMSP-1 variants circulating in the study area during the follow-up period. DNA templates for PCR amplification, isolated from 200 μl of whole blood using Wizard DNA purification kits according to the manufacturer's instructions (Promega, Madison, WI), were dissolved in 100 μl of sterile distilled water and stored at −20°C. Serum samples were obtained, at the time when acute, incident P. vivax infection was diagnosed, from 40 cohort subjects who were free of infection at the baseline; levels of antibodies to PvMSP-1 were compared with those measured at the study baseline. To characterize the PvMSP-1 variants to which our study population had recently been exposed, prior to the cohort follow-up, we analyzed 27 P. vivax isolates collected in July 1999 from patients attending malaria clinics in the nearby towns of Acrelândia and Plácido de Castro (see Fig. S1 in the supplemental material). DNA templates for PCR amplification, isolated from 200 μl of whole blood using GFX genomic blood DNA purification kits (Amersham Pharmacia Biotech, Piscataway, NJ), were dissolved in 100 μl of sterile distilled water and stored at −20°C (7) until used for PvMSP-1 gene amplification and sequencing.
PvMSP-1 gene amplification and sequencing.
The analysis of sequence diversity and antibody recognition of PvMSP-1 focused on three polymorphic domains (blocks 2, 6, and 10) (Fig. 1) located in different parts of the molecule. Block 2 is the longest stretch of variable sequence contained in ICB2-5, a recombinant antigen recently shown to be targeted by naturally acquired protective antibodies (25). The centrally located block 6 is the most polymorphic block of PvMSP-1 (27), but little is known about the variant specificity of naturally acquired antibodies to this domain (20). Although block 10 is the longest stretch of variable sequence in the C-terminal part of PvMSP-1 (Fig. 1), no data are currently available about its immune recognition. These polymorphic domains of the PvMSP-1 gene were amplified with the following primers that target conserved flanking sequences (27): PVF1 (5′-CTCTGACAAAGAGCTGGAA-3′ [nucleotides 465 to 483 of the PvMsp-1 sequence of isolate Salvador-I]) (10) and PVR9 (5′-GCTCCTTCAGCACTTTCACGCG-3′ [nucleotides 979 to 958]) for block 2, PVF4 (5′-TACTACTTGATGGTCCTCAAAAG-3′ [nucleotides 2035 to 2057]) and PVR6 (5′-GTGCTTGTGACATGCGTA-3′ [nucleotides 2256 to 2539]) for block 6, and PVF7 (5′-CCTTAAGAATACCGAGATTTTGCTGAAG-3′ [nucleotides 3429 to 3456]) and PVR3 (5′-GCGATTACTTTGTCGTAG-3′ [nucleotides 4007 to 3990]) for block 10. The 25-μl PCR mixture contained 0.2 μM of each primer, 0.5 to 1 μl of template DNA, 2.5 μl of 10× buffer, 100 μM of each deoxynucleoside triphosphate, and 1 unit of Taq DNA polymerase (Amersham Pharmacia Biotech); thermal cycling started with a 5-min incubation at 95°C, followed by 35 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min, with a final 10-min extension step at 72°C. PCR products were purified with GFX PCR purification kits (Amersham Pharmacia Biotech) and sequenced on both strands with BigDye v3.1 terminator chemistry (Applied Biosystems, Foster City, CA) using the forward and reverse primers described above (27). When PCR yielded two or more bands with different sizes, these bands were excised from agarose gels after electrophoretic separation, purified as described above, and sequenced separately. Sequencing reaction products were analyzed on an ABI 3100 automated DNA sequencer (Applied Biosystems). A total of 53 block 2 sequences, 69 block 6 sequences, and 59 block 10 sequences were obtained and analyzed.
PvMSP-1 alleles for antigen expression.
Six versions of block 2 (amplified from isolates Belém, BP29, BP39, BP13, BR07, and BP30), four versions of block 6 (from isolates Belém, BP39, BR07, and BP01), five versions of block 10 (from isolates BP29, BP39, BP13, BR07, and BP01), and one version of block 13 (from isolate Belém) of PvMSP-1 were selected for the expression of recombinant antigens (Fig. 1B). Block 2 antigens comprise the whole sequence of block 2 and short stretches of the conserved blocks 1 and 3. Block 6 antigens comprise the whole sequence of block 6 and short stretches of the conserved blocks 5 and 7. Block 10 antigens comprise the whole sequence of block 10, the 3′ half of the conserved block 9, and a short stretch of the conserved block 11. Finally, the single block 13 antigen comprises the 3′ half of the conserved block 13, which corresponds to the 19-kDa product of the secondary enzymatic processing of PvMSP-1, also known as ICB10 or PvMSP-119 (25, 32), which remains attached to the merozoite surface at the time of red blood cell invasion (9). This region has been shown to be invariant in Brazilian isolates of P. vivax (33). Table S1 in the supplemental material gives the precise locations of the fragments expressed as recombinant antigens in the PvMSP-1 nucleotide sequence of isolates Belém, BP29, BP39, BP13, BR07, BP30, and BP01 available in the GenBank database, while Fig. S2 in the supplemental material shows the translated amino acid sequences of the variable domain of each antigen. Isolate Belém was collected in Belém, the capital of Pará State (Eastern Brazilian Amazonia), in 1980; BR07 was collected in Porto Velho, the capital of Rondônia State (Western Brazilian Amazonia, bordering with Acre), in 1995; and BP29, BP39, BP13, BP30, and BP01 were collected in Porto Velho in 1997.
Recombinant antigen expression.
For expression as recombinant proteins fused to the C terminus of Schistosoma japonicum glutathione S-transferase (GST), DNA coding for blocks 2, 6, 10, and 13 of PvMSP-1 was reamplified by PCR (as described above) from purified whole-gene amplicons kindly supplied by Somchai Jongwutiwes and Chaturong Putaporntip (Chulalongkorn University, Bangkok, Thailand). PCR products were cloned into the pCR 2.1-TOPO vector (Invitrogen, Carlsbad, CA) and subcloned into the expression vector pGEX-3X from Amersham Pharmacia Biotech (blocks 2, 10, and 13) or pGEX-3Y, created from pGEX-3X by BamHI cleavage and subsequent blunting with Klenow polymerase (block 6). Recombinant proteins were expressed by transformed Escherichia coli (strain DH10B) and purified by affinity chromatography as described previously (36). GST was purified from E. coli transformed with the pGEX-3X vector lacking the PvMSP-1 insert for use as a control antigen in serology. The purity of recombinant proteins was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (see Fig. S3 in the supplemental material); their recognition by control sera from malaria-exposed subjects was confirmed by Western blotting.
Enzyme-linked immunosorbent assay (ELISA) for IgG and IgM antibodies.
Assays with all recombinant antigens (including the GST control) were performed essentially as described previously (37). High-binding 96-well microplates (Costar, Cambridge, MA) were coated with 5 ng/well of solid-phase antigens, dissolved in 50 μl of 0.1 M carbonate-bicarbonate buffer (pH 9.6), for 18 h at 4°C. Serum samples (50 μl/well) were tested at a 1:200 dilution. After a 1-h incubation at 37°C, antibody binding to solid-phase antigen was detected with peroxidase-conjugated goat anti-human immunoglobulin G (IgG) (Kirkegaard & Perry, Gaithersburg, MD) at a 1:4,000 dilution and anti-human IgM (BioSys, Compiègne, France) at a 1:1,200 dilution. After the use of ο-phenylenediamine and hydrogen peroxide at acidic pH as a substrate, absorbance values were measured at 492 nm. Corrected absorbance values were obtained by subtracting absorbance readings with GST run on the same microplate. Absorbance data used in statistical analyses correspond to corrected values. Reactivity indices (RIs) were calculated as the ratio between the corrected absorbance values of each test sample and a cutoff value for each antigen, corresponding to the average corrected absorbance for samples from 21 malaria-naïve blood donors plus 3 standard deviations. The cutoff values for IgG antibodies ranged between 0.216 and 0.460 for block 2 antigens, 0.122 and 0.311 for block 6 antigens, and 0.022 and 0.207 for block 10 antigens; the cutoff value for the block 13 antigen was 0.079. Cutoff values for IgM antibodies were 0.177 to 0.667 for block 2 antigens, 0.032 to 0.516 for block 6 antigens, 0.138 to 0.338 for block 10 antigens, and 0.103 for block 13 antigen. Positive samples had RIs greater than 1.
Statistical analyses.
A database was created with SPSS 13.0 (SPSS Inc., Chicago, IL). Correlations were assessed with ρ coefficients obtained with Spearman nonparametric correlation models. Statistical significance was defined at the 5% level. Multiple logistic regression models with stepwise backward deletion were built to describe independent associations between covariates and the presence of antibodies to PvMSP-1 at the study baseline. Age, gender, and other variables associated with P values of <0.20 in unadjusted analysis were included in logistic regression models. Because the data have a nested structure, where individuals are nested within households, the assumption of independence of observations underlying standard logistic regression analysis was violated. We therefore used two-level logistic models with individual-level covariates (age, gender, time of residence in Amazonia, and recent slide-confirmed episode of P. vivax malaria) and household level covariates (socioeconomic status estimated by use of an asset index [8] and sector of residence within the study area). Malaria transmission is heterogeneously distributed across the Ramal do Granada because of different patterns of land use and deforestation rates. To adjust for these differences in logistic models, we divided the study area into four relatively homogeneous sectors with increasing malaria incidence: sector A (0.46 P. vivax episodes/100 person-months at risk between March 2004 and May 2005), sector B (0.79 P. vivax episodes/100 person-months at risk), sector C (3.44 P. vivax episodes/100 person-months at risk), and sector D (9.71 P. vivax episodes/100 person-months at risk). The HML software package (version 6.03; Scientific Software International, Lincolnwood, IL) was used for multilevel analysis. Only variables associated with statistical significance at the 5% level were maintained in the final models.
Nucleotide sequence accession numbers.
DNA sequences obtained during this study were deposited into the GenBank database under accession numbers EF651844 to EF651876.
RESULTS
Antibody recognition of PvMSP-1 variants at the cohort baseline.
We have investigated baseline antibody responses to PvMSP-1 in 376 subjects aged 5 to 90 years (median, 24 years), with a male-to-female ratio of 1.10:1. These subjects had between 1 month and 72 years of residence in Amazonia (median, 14 years), where they are continuously exposed to P. vivax infections; 133 (35.4%) subjects had at least one symptomatic P. vivax malaria episode diagnosed by passive case detection between January 2001 and March 2004. Table 1 shows the proportions of subjects with IgG and IgM antibodies to recombinant antigens representing different PvMSP-1 domains and variants. Variable domains of blood-stage antigens of P. falciparum, such as PfMSP-1 (13) and PfMSP-2 (35), are usually immunodominant, but here, we show that most malaria-exposed subjects had IgG antibodies to the C-terminal region of PvMSP-1, which is conserved, while less than one-third of them had IgG antibodies to at least one of the variants representing variable blocks of the same antigen.
TABLE 1.
Malaria-exposed subjects from rural Amazonia with IgG and IgM antibodies to PvMSP-1 recombinant antigens as detected by ELISA at the cohort baseline
| PvMSP1 domain and antigena | No. (% [95% CI]) of subjects with antibodies to:
|
|
|---|---|---|
| IgG | IgM | |
| Block 2 | ||
| Belém | 43 (11.4 [8.4-15.1]) | 34 (9.0) [6.3-12.4] |
| BP29 | 63 (16.8 [13.1-20.9]) | 22 (5.9) [3.7-8.7] |
| BP39 | 59 (15.7 [12.2-19.8]) | 10 (2.7) [1.3-4.8] |
| BP13 | 53 (14.1 [10.7-18.0]) | 11 (2.9) [1.5-5.2] |
| BR07 | 45 (12.0 [8.9-15.7]) | 19 (5.1) [3.1-7.8] |
| BP30 | 35 (9.3 [6.6-12.7]) | 0 |
| At least one variant | 125 (33.2 [28.5-38.3]) | 70 (18.6) [14.8-22.9] |
| Block 6 | ||
| Belém | 33 (8.8 [6.1-12.1]) | 28 (7.4 [5.0-10.6]) |
| BP39 | 29 (7.7 [5.2-10.9]) | 5 (1.3 [0.4-3.1]) |
| BR07 | 36 (9.6 [6.8-13.0]) | 28 (7.4 [5.0-10.6]) |
| BP01 | 91 (24.2 [20.0-28.9]) | 15 (4.0 [2.3-6.5]) |
| At least one variant | 104 (27.7 [23.2-32.5]) | 62 (16.5 [12.9-20.6]) |
| Block 10 | ||
| BP29 | 48 (12.8 [9.6-16.6]) | 20 (5.3 [3.3-8.1]) |
| BP39 | 52 (13.8 [10.5-17.7]) | 11 (2.9 [1.5-5.2]) |
| BP13 | 15 (4.0) [2.3-6.5]) | 18 (4.8 [2.9-7.5]) |
| BR07 | 37 (9.8 [7.0-13.3]) | 11 (2.9 [1.5-5.2]) |
| BP01 | 49 (13.0 [9.8-16.9]) | 14 (3.7 [2.1-6.2]) |
| At least one variant | 106 (28.2 [23.7-33.0]) | 42 (11.2 [8.2-14.8]) |
| Block 13 | ||
| Belém | 204 (54.3 [49.1-59.4]) | 26 (6.9 [4.6-10.0]) |
Antigens are described in the legend of Fig. 1B.
We next assessed pairwise Spearman's correlations between levels of IgG antibodies (measured as RIs) and different variants belonging to the same PvMSP-1 block. Correlation coefficients varied widely, with the highest estimates usually obtained for pairs of antigens with relatively similar amino acid sequences (Tables 2 to 4). For example, BP13-BP29 and Belém-BP39 are the pairs of block 2 antigens with the highest proportion of amino acid identity (96.4% and 93.0%, respectively) and with the highest pairwise correlation coefficients for IgG levels (0.675 and 0.589, respectively) (Table 2). These findings suggest that a substantial proportion of naturally acquired IgG antibodies recognize polymorphic sequences within the recombinant antigens representing blocks 2, 6, and 10 of PvMSP-1 instead of the flanking conserved sequences that are shared by all variants of the same block.
TABLE 2.
Percent amino acid similarity in pairwise comparisons of PvMSP-1 block 2 variants expressed as recombinant antigens and Spearman's ρ correlation coefficients of levels of IgG antibodies to these variantsa
| Antigen | Pairwise correlation or % amino acid similarity withb:
|
|||||
|---|---|---|---|---|---|---|
| Belém | BR07 | BP13 | BP29 | BP30 | BP39 | |
| Belém | 66.9 | 67.5 | 67.5 | 63.7 | 93.0 | |
| BR07 | 0.494 | 68.9 | 69.6 | 71.4 | 67.5 | |
| BP13 | 0.327 | 0.497 | 96.4 | 69.8 | 70.7 | |
| BP29 | 0.470 | 0.569 | 0.675 | 71.0 | 70.7 | |
| BP30 | 0.318 | 0.333 | 0.443 | 0.522 | 65.0 | |
| BP39 | 0.589 | 0.560 | 0.427 | 0.564 | 0.455 | |
All Spearman's ρ correlation coefficients are significant(P < 0.001).
Pairwise correlation values are presented in lightface; percent amino acid similarities are presented in boldface.
TABLE 3.
Percent amino acid similarity in pairwise comparisons of PvMSP-1 block 6 variants expressed as recombinant antigens and Spearman's ρ correlation coefficients of levels of IgG antibodies to these variantsa
| Antigen | Pairwise correlation or % amino acid similarity withb:
|
|||
|---|---|---|---|---|
| Belém | BR07 | BP01 | BP39 | |
| Belém | 97.9 | 59.5 | 80.4 | |
| BR07 | 0.707 | 61.4 | 81.4 | |
| BP01 | 0.468 | 0.551 | 73.9 | |
| BP39 | 0.687 | 0.736 | 0.576 | |
All Spearman's ρ correlation coefficients are significant (P < 0.001).
Pairwise correlation values are presented in lightface; percent amino acid similarities are presented in boldface.
TABLE 4.
Percent amino acid similarity in pairwise comparisons of PvMSP-1 block 10 variants expressed as recombinant antigens and Spearman's ρ correlation coefficients of levels of IgG antibodies to these variantsa
| Antigen | Pairwise correlation or % amino acid similarity withb:
|
||||
|---|---|---|---|---|---|
| BR07 | BP01 | BP13 | BP29 | BP39 | |
| BR07 | 80.1 | 93.4 | 87.3 | 86.7 | |
| BP01 | 0.245 | 74.2 | 71.7 | 77.1 | |
| BP13 | 0.545 | 0.270 | 88.7 | 82.3 | |
| BP29 | 0.319 | 0.023 | 0.450 | 71.4 | |
| BP39 | 0.468 | 0.213 | 0.563 | 0.505 | |
All Spearman's ρ correlation coefficients are significant (P < 0.001) except for that corresponding to antibodies to the block 10 antigens BP29 and BP01 (ρ = 0.023; P > 0.05).
Pairwise correlation values are presented in lightface; percent amino acid similarities are presented in boldface.
We also investigated how cumulative or recent exposure to malaria affected antibody recognition of PvMSP-1. First, we calculated Spearman's regression coefficients between levels of IgG antibodies (measured as RIs) and time of residence in Amazonia (range, 30 days to 72 years). We found significant correlations (P < 0.05) with levels of IgG antibodies to the BP39 version of block 2 (ρ = 0.107), the BR07 and BP01 versions of block 6 (ρ = 0.108 and ρ = 0.137, respectively), and the BR07 and BP29 versions of block 10 (ρ = 0.138 and ρ = 0.125, respectively), showing that levels of IgG antibodies to at least some variable domains correlate with cumulative exposure to malaria. Next, we found that the number of slide-confirmed episodes of P. vivax infections recorded between January 2001 and March 2004 correlated positively with the levels of IgG antibodies to the BP1 version of block 6 (ρ = 0.108), the BP29 version of block 10 (ρ = 0.229), and the Belém version of block 13 (ρ = 0.420) and negatively with the levels of antibodies to the BP1 version of block 10 (ρ = −0.230). No correlation was found between age and levels of IgG antibodies to any antigen. The total number of variants of blocks 6 and 10 recognized by IgG antibodies correlated with their time of residence in Amazonia (ρ = 0.162 and ρ = 0.214, respectively), while the number of block 2 variants recognized correlated with the number of recent slide-confirmed P. vivax episodes (ρ = 0.116).
We used multiple logistic regression models to determine whether cumulative or recent exposure to malaria predicted the presence of IgG antibodies to PvMSP-1 after controlling for several covariates putatively associated with malaria risk in our population, such as age, gender, place of residence, and socioeconomic status. The number of years of residence in Amazonia, a surrogate measure of cumulative exposure to malaria, was a strong predictor of the presence of IgG antibodies to at least one version of blocks 6 and 10 (adjusted odds ratio, 1.04; 95% CI, 1.02 to 1.07; P = 0.001 [identical results for blocks 6 and 10]). In other words, each additional year of exposure to malaria increased the probability of having IgG antibodies to at least one block 6 or block 10 antigen by 4% (95% CI, 2 to 7%). Recent exposure to P. vivax, as inferred by the occurrence of at least one slide-confirmed P. vivax infection between January 2001 and March 2004, was a strong predictor of the presence of IgG antibodies to block 13 at the study baseline (adjusted odds ratio, 4.16; 95% CI, 2.41 to 7.18; P < 0.0001) but not to other antigens. As expected, the sector of residence was also significantly associated with the presence of IgG antibodies in all models. We conclude that cumulative exposure to malaria predicted the presence and levels of IgG antibodies to many PvMSP-1 variants, while recent exposure to malaria was a strong predictor of the presence and levels of IgG antibodies to the C-terminal region of PvMSP-1.
Local repertoire of PvMSP-1 variants.
To determine whether the poor recognition of variable domains of PvMSP-1 at the study baseline resulted from a mismatch between the sequences of recombinant antigens and those actually found in local parasites, we sequenced blocks 2, 6, and 10 of the PvMSP-1 gene from local P. vivax isolates circulating before (1999) and during (2004 to 2005) our cohort study. The translated amino acid sequences and their absolute frequencies in 1999 and 2004 to 2005 are shown in Fig. 2, 3, and 4. We found 8 unique block 2 sequences, 14 unique block 6 sequences, and 11 unique block 10 sequences. Note that none of these blocks can be defined as strictly dimorphic (i.e., not all sequences can be grouped as either Belém type [5] or Salvador-I type [10]), since some sequences clearly diverge from both putative prototypes, and many of them are generated by recombination between the prototypes (27). Sequences of four block 2 antigens (BP29, BP39, BP13, and BR07), two block 6 antigens (BP39 and BR07), and three block 10 antigens (BP39, BP13, and BR07) used in serological analyses were found in local parasites; the remaining antigens were very similar (amino acid similarity, 91.2 to 98.2%), although not identical, to at least one of the variants found in local isolates (Fig. 2 to 4). These results suggest that the panel of recombinant antigens used in serology properly represents the repertoire of PvMSP-1 variants to which subjects have been exposed before and during the study. As a consequence, the poor recognition of the block 2, block 6, and block 10 antigens at the study baseline cannot be ascribed to the fact that subjects are rarely exposed to these or quite similar PvMSP-1 variants.
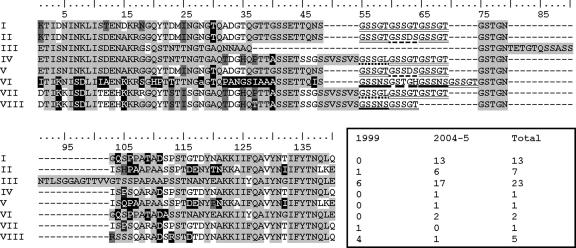
FIG. 2.
The eight unique PvMSP-1 block 2 sequences found in P. vivax isolates circulating in the study area in 1999 and 2004 to 2005 and their absolute frequencies. The lowercase letter indicates a codon with a synonymous nucleotide replacement. Repetitive motifs are underlined. Four sequences (sequences II, III, VI, and VIII, corresponding to 69.8% of all block 2 sequences) match sequences found in recombinant antigens used in serology (BR07, BP39, BP13, and BP30, respectively). The remaining block 2 antigens, Belém and BP29, are quite similar to sequences III (91.5% amino acid identity) and VI (96.3% amino acid identity), respectively.
FIG. 3.
The 14 unique PvMSP-1 block 6 sequences found in P. vivax isolates circulating in the study area in 1999 and 2004 to 2005 and their absolute frequencies. Two of the sequences (sequences I and VII, corresponding to 36.2% of all block 6 sequences) match sequences found in recombinant antigens used in serology (BR07 and BP39, respectively). The remaining block 6 antigens, Belém and BP1, are quite similar to sequences I (97.5% amino acid identity) and XII (98.1% amino acid identity), respectively.
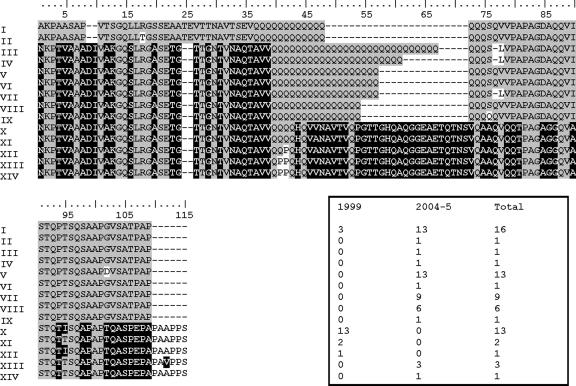
FIG. 4.
The 11 unique PvMSP-1 block 10 sequences found in P. vivax isolates circulating in the study area in 1999 and 2004 to 2005 and their absolute frequencies. The lowercase letter indicates a codon with a synonymous nucleotide replacement. Three sequences (sequences IV, V, and IX, corresponding to 37.3% of all block 10 sequences) match sequences found in recombinant antigens used in serology (BP13, BR07, and BP39, respectively). The remaining block 10 antigens, BP1 and BP29, are quite similar to sequences XI (91.2% amino acid identity) and VI (98.2% amino acid identity), respectively.
Although all PvMSP-1 sequence types expressed as recombinant antigens were identical or quite similar to those circulating in local parasites, the reverse is not necessarily true: some relatively common local PvMSP-1 variants differed substantially from the sequence types represented in the panel of recombinant antigens. For example, the amino acid identity between block 2 sequence type I, which was particularly abundant (35.5%) among parasites collected from 2004 to 2005 (Fig. 2), and the most similar block 2 recombinant antigen, BP30 (see Fig. S2 in the supplemental material), is only 83.0%. Similarly, block 10 sequence type I (found in 7 of 42 parasites collected from 2004 to 2005) (Fig. 4) and the most similar block 10 recombinant antigen, BP29 (see Fig. S2 in the supplemental material), are only 78% identical at the amino acid level. Therefore, the recombinant antigens used in serology cover only part of the repertoire of sequences of blocks 2, 6, and 10 found in local parasites.
Interestingly, however, we found no clear association between the abundance of a given PvMSP-1 sequence type in 1999 or 2004 to 2005 and the proportion of responders to the corresponding recombinant antigen at the study baseline. For example, the BP1 version was the most frequently recognized block 6 variant at baseline (24.2% subjects had detectable IgG antibodies) (Table 1), but its sequence was not found in parasites circulating in 1999 or 2004 to 2005; the closest sequence type (sequence type XII, with 98.1% amino acid identity) was detected in only 1 of 19 parasites analyzed in 1999 and none of the 50 parasites analyzed from 2004 to 2005 (Fig. 3). In addition, block 2 sequence type III was the most abundant variant in both 1999 and 2004 to 2005 (Fig. 2) but was recognized by IgG antibodies from only 15.7% of study subjects at baseline (the corresponding recombinant antigen is BP39) (Table 1). In fact, all block 2 antigens were recognized by similar proportions of subjects (Table 1) irrespective of the relative abundance of the corresponding sequences in local parasites.
The proportion of responders to a given PvMSP-1 variant at the study baseline did not predict the risk of subsequent infections with parasites carrying that variant. For example, although block 10 antigens BR07 and BP39 were recognized by 13.8% and 9.8% of subjects at baseline (Table 1), their sequences (types V and IX) accounted for 29 of 50 (58.0%) block 10 variants observed in subsequent infections (Fig. 3). Conversely, the poor recognition of block 10 antigen BP13 at baseline (only 4.0% of subjects had specific IgG antibodies) (Table 1) did not translate into an increased risk of subsequent infection with parasites carrying the corresponding sequence (type IV), which was found in only 2 of 50 (4.0%) isolates analyzed from 2004 to 2005 (Fig. 4). However, the number of infections with known PvMSP-1 variants is too small for meaningful statistical analyses.
Antibody recognition of PvMSP-1 variants during P. vivax malaria episodes.
Most or all serum samples from subjects experiencing acute P. vivax infection had IgG antibodies recognizing at least one variant of each block analyzed; the proportions of responders were 91.3% (21 of 23) for block 2 antigens, 78.6% (22 of 28) for block 6, and 100% (27 of 27) for block 10 (Fig. 5). These results suggest that the low proportion of responders detected at the study baseline, in the absence of P. vivax infection, cannot be ascribed to the poor immunogenicity of these variable domains. However, IgG antibodies present during acute infections often failed to match the PvMSP-1 variant characterized in infecting parasites while recognizing other, structurally nonrelated variants.
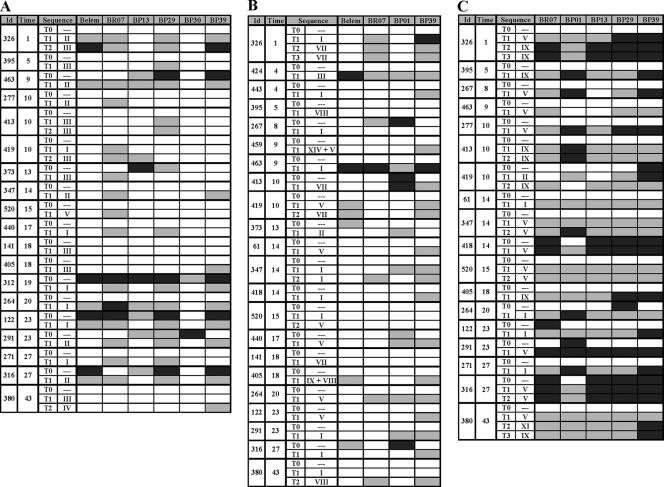
FIG. 5.
IgG antibodies to PvMSP-1 variants in sequential samples from malaria-exposed rural Amazonians. “Id” refers to the identification number of each subject, and “Time” refers to the time of residence (in years) in Amazonia, a proxy of the cumulative exposure to malaria. “Sequence” indicates the PvMSP-1 sequence type(s) detected during the acute malaria infection (numbered as in Fig. 2 [block 2], Fig. 3 [block 6], and Fig. 4 [block 10]); dashes indicate the absence of malaria parasites at the time of sample collection. T0 denotes samples collected at the study baseline in the absence of P. vivax infection; other serum samples (T1, T2, and T3) were collected during incident P. vivax episodes that had the PvMSP-1 variant present in infecting parasites characterized by DNA sequencing. Serological results for incident P. vivax infections with parasites of undetermined PvMSP-1 type are not shown. Each box represents levels of antibodies, in an individual sample, to a given antigen. Box shading is proportional to the concentration of IgG antibodies measured by ELISA; RIs between 1.1 and 1.3 are represented with light gray shading, and RIs above 1.3 are represented with dark gray shading; negative results are represented as open boxes. (A) Data for six block 2 antigens (19 subjects; 42 samples analyzed). (B) Data for four block 6 antigens (22 subjects; 49 samples analyzed). (C) Data for five block 10 antigens (18 subjects; 45 samples analyzed). Corresponding data for IgM antibodies are shown in Fig. S4 in the supplemental material.
The most frequent block 2 sequences found in 23 acute malaria episodes with serum samples available for serology were types II and III (Fig. 5A), which are identical to those of antigens BR07 and BP39, respectively (Fig. 2). All seven subjects exposed to type II block 2 recognized the homologous antigen BR07, but only two of eight subjects exposed to type III block 2 sequences had detectable IgG antibodies to the homologous antigen BP39. These subjects who failed to mount IgG responses to BP39, despite the documented exposure to type III sequences, also failed to generate IgM antibodies to the same antigen (see Fig. S4 in the supplemental material). Comparable differences were found in the homologous recognition of block 6 antigens. Block 6 sequence types I and VII are identical to block 6 antigens BR07 and BP39 (Fig. 3), but only 3 of 10 subjects exposed to sequence type I had detectable IgG antibodies to the homologous antigen BR07, while four of five subjects exposed to the homologous antigen BP39 had specific antibodies (Fig. 5B). None of them had IgM antibodies to BR07 or BP39 (see Fig. S4 in the supplemental material). Block 10 antigens BR07 and BP39 match block 10 sequence types V and IX (Fig. 4); all subjects exposed to these sequence types had IgG antibodies to the homologous antigen, but they usually recognized other, unrelated variants as well (Fig. 5C). These findings are not unexpected, since block 10-derived recombinant proteins include the 3′ half of the conserved block 9 (Fig. 1B), which may contain common B-cell epitopes. Data shown in Fig. 5 do not reveal a clear association between patterns of IgG antibody responses to PvMSP-1 variants during acute P. vivax infection and cumulative exposure to malaria (measured as the time of residence in Amazonia), although the sample size is too small for formal statistical testing. The high immunogenicity of conserved block 13 (PvMSP-119) (32) was confirmed in our population: 37 of 40 (92.5%) serum samples collected during acute P. vivax infections had detectable IgG antibodies. The proportions of acute-phase samples with detectable IgM antibodies recognizing at least one variant of each block were low: 17.4% for block 2, none for block 6, and 44.4% for block 10 (see Fig. S4 in the supplemental material). Only 4 of 40 subjects tested (5.0%) had IgM antibodies to block 13.
Data shown in Fig. 5 do not support the original antigenic sin hypothesis, since there is no evidence that successive infections with different variants might boost preexisting antibody responses instead of generating antibodies with new specificities (34). For example, subjects 463 and 316 had IgG antibodies to block 2 variants other than BR07 at baseline but were able to seroconvert to BR07 when exposed to parasites carrying the type II sequence, while subject 373, who failed to seroconvert to BP39 when exposed to the homologous type III sequence, did not present a boost of preexisting heterologous antibodies (Fig. 5A). Although the small number of subjects with detectable IgG antibodies at baseline limits the power of this analysis, our data suggest that the original antigenic sin hypothesis (34) is unlikely to account for the observed mismatches in the variant specificities of IgG antibodies to PvMSP-1 elicited during acute P. vivax infections.
DISCUSSION
One of the critical issues when designing PvMSP-1-based subunit vaccines is to determine whether sequence polymorphism translates into variant-specific immunity. The notion that protective immunity targets predominantly highly polymorphic domains of MSP-1 received recently is further supported by elegant experiments with rodent malaria parasites showing that these variable domains are selected, in a variant-specific manner, in mice previously immunized with parasites carrying heterologous MSP-1 variants (21). However, the recognition of variable domains of PvMSP-1 by naturally acquired antibodies remains poorly investigated. Here, we show that a panel of recombinant proteins corresponding to PvMSP-1 variants commonly found in local parasites was poorly recognized by noninfected subjects who are exposed to malaria in rural Amazonia (Table 1). Few subjects had detectable IgG antibodies to at least one of the several variants of blocks 2, 6, and 10 expressed as recombinant proteins, although most of them recognized the conserved C-terminal domain of PvMSP-1. The mechanisms underlying the limited immune recognition of antigenic variants that are known to circulate in a given area of endemicity remain unclear. Sequence polymorphism has surely affected the analysis of antibody recognition of PvMSP-1 variants, since the panel of recombinant antigens did not include the whole repertoire of PvMSP-1 variants found in local parasites (Fig. 2 to 4). In addition, variable antigens may be poorly immunogenic or may elicit antibody responses that are short-lived in the absence of frequent boosting (23).
The proportion of responders to PvMSP-1 variants increased substantially during subsequent P. vivax infections, suggesting that the poor immunogenicity of variable domains may not represent a major obstacle to variant-specific immunity (Fig. 5). Acute-phase antibody responses to block 10 antigens were particularly strong compared to those to the other variable domains analyzed despite the fact that several block 10 variants found in local parasites were not included in the panel of recombinant proteins used for serology (variants I, II, III, VII, and VIII) (Fig. 4). Because the block 10 recombinant antigens contain a long stretch of conserved sequence (see Fig. S2 in the supplemental material), these antibodies may be either cross-reactive among different variable domains or directed against conserved epitopes within these recombinant antigens. The poor correlation observed between the baseline levels of IgG antibodies to the block 10 antigens BP29 and BP01 (Table 2) suggests that the conserved sequences included in these antigens may not be a major target of naturally acquired antibodies.
However, the specificity of acute-phase IgG antibodies did not necessarily match the PvMSP-1 variants found in infecting parasites. These mismatches, which have also been observed in antibodies to P. falciparum MSP-1 (4, 29), might be explained (i) by the presence of PvMSP-1 variants in infecting parasites that were missed during the typing procedure, (ii) by the presence of IgG antibodies generated following recent infections with other PvMSP-1 variants, which remained undetected during the follow-up, (iii) by the generation of IgM-restricted antibody responses to some PvMSP-1 domains (24), particularly during the early phase of infection, or (iv) by clonal imprinting or antigenic original sin. We next examine these hypotheses in greater detail because of their possible implications for naturally acquired immunity and malaria vaccine development.
The first hypothesis has been suggested to explain similar mismatches of antibody specificity in P. falciparum infections, since parasites carrying some antigenic variants may be missed by typing procedures (the so-called “hidden genotypes”) because they are sequestered in deep organs or in the placenta at the time of blood collection. We argue that hidden genotypes are very unlikely to be present in our P. vivax isolates. First, since deep organ sequestration does not occur in P. vivax infection, all variants present in a given isolate are expected to circulate in the peripheral blood, being accessible to PCR amplification and subsequent DNA sequencing. Second, additional PvMSP-1 alleles present in our isolates would have affected the quality of DNA sequencing of uncloned PCR products. The comparison of IgG and IgM responses to individual PvMSP-1 variants lends little support to the third hypothesis, since all P. vivax-infected subjects who failed to generate IgG antibodies to homologous antigens (Fig. 5) also failed to produce specific IgM antibodies to the same antigens (see Fig. S4 in the supplemental material). No definitive conclusion can be drawn, from our results, about the last hypothesis, since the numbers of individuals and sequential serum samples examined are limited. However, we found no evidence for antibodies with fixed specificity to some PvMSP-1 variants being detected in sequential serum samples (Fig. 5), as it would be expected under the clonal imprinting hypothesis (34). In fact, most mismatches did not result from the persistence of preexisting heterologous antibodies since the study baseline but rather resulted from seroconversions to heterologous antigens. Therefore, it seems more parsimonious to explain the frequent mismatches between the PvMSP-1 sequence found in infecting parasites and the antigenic variants recognized by IgG antibodies as a consequence of undetected infections with other variants that occurred between the study baseline and the subsequent blood draws.
We suggest that the acquisition of IgG antibody responses to variable (blocks 2, 6, and 10) and conserved (block 13) domains of PvMSP-1 in malaria-exposed subjects may be determined by different factors. The presence of naturally acquired IgG antibodies to ICB2-5, a recombinant antigen comprising three conserved blocks (blocks 1, 3, and 5) and two variable blocks (blocks 2 and 4) of the Belém PvMSP-1 allele, but not the recognition of the conserved PvMSP-119 domain, was recently associated with cumulative exposure to malaria in Brazil (25). Interestingly, previous analyses suggested that antibodies to the recombinant protein ICB2-5 recognize predominantly its variable domains, blocks 2 and 4 (17). A similar association between cumulative exposure to malaria and antibody levels was found for variable PvMSP-1 domains in our population. In contrast, recent, rather than cumulative, exposure to P. vivax was associated with IgG antibody recognition of PvMSP-119 in our noninfected subjects assessed at the study baseline. These differences may be relevant for malaria vaccine development, since natural exposure to the parasite is expected to boost immune responses elicited by immunization to maintain long-term protection. Although there are persuasive data showing that antibodies to MSP-1 may confer protection against malaria, the relative contribution of different targets to antibody-mediated protection remains unclear. The conserved 19-kDa domain of P. falciparum MSP-1 is a major target of naturally acquired antibodies that inhibit parasite growth in vitro (26) and reduce the risk of subsequent infection (14), but antibodies to the highly polymorphic block 2 domain of MSP-1 also seem to provide clinical protection (3). Our data suggest that antibodies to the local repertoire of variable domains of PvMSP-1 are elicited after several repeated infections and require frequent boosting, while those to PvMSP-119 are more frequently detected in noninfected subjects, possibly because of their longer half-life.
This study has several limitations. First, further testing of hypotheses about the development of variant-specific antibody responses to PvMSP-1 is restricted by the relatively small number of consecutive serum samples, which were obtained during most, but not all, P. vivax infections experienced by each subject during the follow-up. Second, these sera were tested against a panel of recombinant antigens representing a limited number of variants of only three of six variable domains of PvMSP-1; antibody responses to variants of blocks 4, 8, and 12 have not been investigated. Third, to characterize the repertoire of PvMSP-1 variants to which the study subjects have been exposed, we were able to partially sequence a relatively small number of alleles. Fourth, the study does not have enough power to test whether the presence of variant-specific antibodies conferred protection against P. vivax isolates carrying homologous PvMSP-1 variants. Despite these limitations, we have provided a detailed description of naturally acquired variant-specific antibody responses to PvMSP-1. The ongoing follow-up of our cohort of malaria-exposed subjects in rural Amazonia may provide further biological material and epidemiological data for additional, in-depth analyses of naturally acquired immunity and clinical protection against P. vivax.
Supplementary Material
Acknowledgments
This study was supported by grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (grant 470067/2004-7) and the Fundação de Amparo à Pesquisa do Estado de São Paulo (grants 03/09719-6 and 05/51988-0). M.D.S.-N. and E.H.E.H. were recipients of doctoral scholarships from the Fundação de Amparo à Pesquisa do Estado de São Paulo, while G.W. and M.U.F. are recipients of research scholarships from the Conselho Nacional de Desenvolvimento Científico e Tecnológico.
We thank the population of our field site, Ramal do Granada, for their enthusiastic participation in the study; Sebastião Bocalom Rodrigues, Damaris de Oliveira, and Nésio M. Carvalho (Municipal Government of Acrelândia); Raimundo A. Costa and the malaria control team in Acrelândia for their logistic support; Adamílson Luís de Souza, Pascoal Torres Muniz, and Natal Santos da Silva for help in fieldwork; Kézia K. G. Scopel, Erika M. Braga, Natália T. Komatsu, and Rosane Rezende D'Arcádia for PCR-based malaria diagnosis; Rogério Láuria da Silva for help with recombinant protein expression; and Francisco das Chagas O. Luz (Ministry of Health, Brasília, Brazil) for reviewing malaria slides.
Footnotes
Published ahead of print on 15 August 2007.
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1.Baird, J. K. 2004. Chloroquine resistance in Plasmodium vivax. Antimicrob. Agents Chemother. 48:4075-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conway, D. J. 1997. Natural selection on polymorphic malaria antigens and the search for a vaccine. Parasitol. Today 13:26-29. [DOI] [PubMed] [Google Scholar]
- 3.Conway, D. J., D. R. Cavanagh, K. Tanabe, C. Roper, Z. S. Mikes, N. Sakihama, K. A. Bojang, A. M. J. Oduola, P. G. Kremsner, D. E. Arnot, B. M. Greenwood, and J. S. McBride. 2000. A principal target of human immunity to malaria identified by molecular population genetic and immunological analyses. Nat. Med. 6:689-692. [DOI] [PubMed] [Google Scholar]
- 4.da Silveira, L. A., M. L. Dorta, E. A. S. Kimura, A. M. Katzin, F. Kawamoto, K. Tanabe, and M. U. Ferreira. 1999. Allelic diversity and antibody recognition of Plasmodium falciparum merozoite surface protein 1 during hypoendemic malaria transmission in the Brazilian Amazon region. Infect. Immun. 67:5906-5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.del Portillo, H. A., S. Longacre, E. Khouri, and P. H. David. 1991. Primary structure of the merozoite surface antigen 1 of Plasmodium vivax reveals sequences conserved between different Plasmodium species. Proc. Natl. Acad. Sci. USA 88:4030-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira, M. U., M. da Silva-Nunes, and G. Wunderlich. 2004. Antigenic diversity and immune evasion by malaria parasites. Clin. Diagn. Lab. Immunol. 11:987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira, M. U., N. D. Karunaweera, M. da Silva-Nunes, N. S. Silva, D. F. Wirth, and D. L. Hartl. 2007. Population structure and transmission dynamics of Plasmodium vivax in rural Amazonia. J. Infect. Dis. 195:1218-1226. [DOI] [PubMed] [Google Scholar]
- 8.Filmer, D., and L. H. Pritchett. 2001. Estimating wealth effects without expenditure data—or tears: an application to educational enrollments in states of India. Demography 38:115-132. [DOI] [PubMed] [Google Scholar]
- 9.Freeman, R. R., and A. A. Holder. 1983. Surface antigens of malaria merozoites. A high molecular weight precursor is processed to an 83,000 mol wt form expressed on the surface of Plasmodium falciparum merozoites. J. Exp. Med. 158:1647-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson, H. L., J. E. Tucker, D. C. Kaslow, A. U. Krettli, W. E. Collins, M. C. Kiefer, I. C. Bathurst, and P. J. Barr. 1992. Structure and expression of the gene for Pv200, a major blood-stage surface antigen of Plasmodium vivax. Mol. Biochem. Parasitol. 50:325-334. [DOI] [PubMed] [Google Scholar]
- 11.Guerra, C. A., R. W. Snow, and S. L. Hay. 2006. Mapping the global extent of malaria in 2005. Trends Parasitol. 22:353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrera, S., G. Corradin, and M. Arévalo-Herrera. 2007. An update on the search for a Plasmodium vivax vaccine. Trends Parasitol. 23:122-128. [DOI] [PubMed] [Google Scholar]
- 13.Holder, A. A., and E. M. Riley. 1996. Human immune response to MSP-1. Parasitol. Today 12:173-174. [DOI] [PubMed] [Google Scholar]
- 14.John, C. C., R. A. O'Donnell, P. O. Sumba, A. M. Moormann, T. F. de Koning-Ward, C. L. King, J. W. Kazura, and B. S. Crabb. 2004. Evidence that invasion-inhibitory antibodies specific for the 19-kDa fragment of merozoite surface protein-1 (MSP-1 19) can play a protective role against blood-stage Plasmodium falciparum infection in individuals living in a malaria endemic area. J. Immunol. 173:666-672. [DOI] [PubMed] [Google Scholar]
- 15.Kimura, M., O. Kaneko, Q. Liu, M. Zhou, F. Kawamoto, Y. Wataya, S. Otani, Y. Yamaguchi, and K. Tanabe. 1997. Identification of the four species of human malaria parasites by nested PCR that targets variant sequences in the small subunit rRNA gene. Parasitol. Int. 46:91-95. [Google Scholar]
- 16.Leclerc, M. C., M. Menegon, A. Cligny, J. L. Noyer, S. Mammadov, N. Aliyev, E. Gasimov, G. Majori, and C. Severini. 2004. Genetic diversity of Plasmodium vivax isolates from Azerbaijan. Malar. J. 3:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levitus, G., F. Mertens, M. A. Sperança, L. M. A. Camargo, M. U. Ferreira, and H. A. del Portillo. 1994. Characterization of naturally acquired human IgG responses against the N-terminal region of the merozoite surface protein 1 of Plasmodium vivax. Am. J. Trop. Med. Hyg. 51:68-76. [DOI] [PubMed] [Google Scholar]
- 18.Lim, C. S., S. H. Kim, J. W. Song, K. J. Song, and K. N. Lee. 2000. Analysis of Plasmodium vivax merozoite surface protein-1 gene sequences from resurgent Korean isolates. Am. J. Trop. Med. Hyg. 62:261-265. [DOI] [PubMed] [Google Scholar]
- 19.Loiola, C. C., C. J. da Silva, and P. L. Tauil. 2002. Malaria control in Brazil: 1965 to 2001. Rev. Panam. Salud Publica 11:235-244. [DOI] [PubMed] [Google Scholar]
- 20.Mancilla, L. I., G. Levitus, K. Kirchgatter, F. Mertens, S. Herrera, and H. A. del Portillo. 1994. Plasmodium vivax: dimorphic DNA sequences form the MSP-1 gene code for regions that are immunogenic in natural infections. Exp. Parasitol. 79:148-158. [DOI] [PubMed] [Google Scholar]
- 21.Martinelli, A., S. Cheesman, P. Hunt, R. Culleton, A. Raza, M. Mackinnon, and R. Carter. 2005. A genetic approach to the de novo identification of targets of strain-specific immunity in malaria parasites. Proc. Natl. Acad. Sci. USA 102:814-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendis, K. N., B. J. Sina, P. Marchesini, and R. Carter. 2001. The neglected burden of Plasmodium vivax malaria. Am. J. Trop. Med. Hyg. 64:97-106. [DOI] [PubMed] [Google Scholar]
- 23.Mercereau-Puijalon, O., T. Fandeur, M. Guillotte, and S. Bonnefoy. 1991. Parasite features impeding malaria immunity: antigenic diversity, antigenic variation and poor immunogenicity. Res. Immunol. 142:690-697. [DOI] [PubMed] [Google Scholar]
- 24.Mertens, F., G. Levitus, L. M. A. Camargo, M. U. Ferreira, A. P. Dutra, and H. A. del Portillo. 1993. Longitudinal study of naturally acquired humoral immune responses against the merozoite surface protein 1 of Plasmodium vivax in patients from Rondônia, Brazil. Am. J. Trop. Med. Hyg. 49:383-392. [DOI] [PubMed] [Google Scholar]
- 25.Nogueira, P. A., F. P. Alves, C. Fernandez-Becerra, O. Pein, N. R. Santos, L. H. Pereira da Silva, E. P. Camargo, and H. A. del Portillo. 2006. A reduced risk of infection with Plasmodium vivax and clinical protection against malaria are associated with antibodies against the N terminus but not the C terminus of merozoite surface protein 1. Infect. Immun. 74:2726-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Donnell, R. A., T. E. de Koning-Ward, R. A. Burt, M. Bockarie, J. C. Reeder, A. F. Cowman, and B. S. Crabb. 2001. Antibodies against merozoite surface protein (MSP-)119 are a major component of the invasion-inhibitory response in individuals immune to malaria. J. Exp. Med. 193:1403-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Putaporntip, C., S. Jongwutiwes, N. Sakihama, M. U. Ferreira, W.-G. Kho, A. Kaneko, H. Kanbara, T. Hattori, and K. Tanabe. 2002. Mosaic organization and heterogeneity in frequency of allelic recombination of the Plasmodium vivax merozoite surface protein-1 locus. Proc. Natl. Acad. Sci. USA 99:16348-16353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Putaporntip, C., S. Jongwutiwes, T. Iwasaki, H. Kanbara, and A. L. Hughes. 2006. Ancient common ancestry of the merozoite surface protein 1 of Plasmodium vivax as inferred from its homologue in Plasmodium knowlesi. Mol. Biochem. Parasitol. 146:105-108. [DOI] [PubMed] [Google Scholar]
- 29.Scopel, K. K. G., C. J. F. Fontes, M. U. Ferreira, and E. M. Braga. 2005. Plasmodium falciparum: IgG subclass antibody response to merozoite surface protein-1 among Amazonian gold miners, in relation to infection status and disease expression. Exp. Parasitol. 109:124-134. [DOI] [PubMed] [Google Scholar]
- 30.Severini, C., M. Menegon, M. Di Luca, I. Abdullaev, G. Majori, S. A. Razakov, and L. Gradoni. 2004. Risk of Plasmodium vivax malaria reintroduction in Uzbekistan: genetic characterization of parasites and status of potential malaria vectors in the Surkhandarya region. Trans. R. Soc. Trop. Med. Hyg. 98:585-592. [DOI] [PubMed] [Google Scholar]
- 31.Silva-Nunes, M., R. S. Malafronte, B. A. Luz, E. A. Souza, L. C. Martins, S. G. Rodrigues, J. O. Chiang, P. F. C. Vasconcelos, P. T. Muniz, and M. U. Ferreira. 2006. The Acre Project: the epidemiology of malaria and arthropod-borne virus infections in a rural Amazonian population. Cad. Saude Publ. 22:1325-1334. [DOI] [PubMed] [Google Scholar]
- 32.Soares, I. S., G. Levitus, J. M. Souza, H. A. del Portillo, and M. M. Rodrigues. 1997. Acquired immune responses to the N- and C-terminal regions of Plasmodium vivax merozoite surface protein 1 in individuals exposed to malaria. Infect. Immun. 65:1606-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soares, I. S., J. W. Barnwell, M. U. Ferreira, M. G. Cunha, J. P. Laurino, B. A. Castilho, and M. M. Rodrigues. 1999. A Plasmodium vivax vaccine candidate displays limited allele polymorphism, which does not restrict recognition by antibodies. Mol. Med. 5:459-470. [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor, R. R., A. Egan, D. McGuiness, A. Jepson, R. Adair, C. Drakeley, and E. Riley. 1996. Selective recognition of malaria antigens by human serum antibodies is not genetically determined but demonstrates some features of clonal imprinting. Int. Immunol. 8:905-915. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, R. R., D. B. Smith, V. J. Robinson, J. S. McBride, and E. M. Riley. 1995. Human antibody response to Plasmodium falciparum merozoite surface protein 2 is serogroup specific and predominantly of the immunoglobulin G3 subclass. Infect. Immun. 63:4382-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tonhosolo, R., G. Wunderlich, and M. U. Ferreira. 2001. Differential antibody recognition of four allelic variants of the merozoite surface protein-2 (MSP-2) of Plasmodium falciparum. J. Eukaryot. Microbiol. 48:556-564. [DOI] [PubMed] [Google Scholar]
- 37.Tonon, A. P., E. H. E. Hoffmann, L. A. da Silveira, A. G. Ribeiro, C. R. S. Gonçalves, P. E. M. Ribolla, G. Wunderlich, and M. U. Ferreira. 2004. Plasmodium falciparum: sequence diversity and antibody recognition of the merozoite surface protein-2 (MSP-2) in Brazilian Amazonia. Exp. Parasitol. 108:114-125. [DOI] [PubMed] [Google Scholar]
- 38.Trape, J.-F. 1985. Rapid evaluation of malaria parasite density and standardization of thick smear examination for epidemiological investigations. Trans. R. Soc. Trop. Med. Hyg. 79:181-184. [DOI] [PubMed] [Google Scholar]
- 39.Win, T. T., K. Lin, S. Mizuno, M. Zhou, Q. Liu, M. U. Ferreira, I. S. Tantular, S. Kojima, A. Ishii, and F. Kawamoto. 2002. Wide distribution of Plasmodium ovale in Myanmar. Trop. Med. Int. Health 7:231-239. [DOI] [PubMed] [Google Scholar]
- 40.Yang, C., W. E. Collins, J. S. Sullivan, D. C. Kaslow, L. Xiao, and A. A. Lal. 1999. Partial protection against Plasmodium vivax blood-stage infection in Saimiri monkeys by immunization with a recombinant C-terminal fragment of merozoite surface protein 1 in block copolymer adjuvant. Infect. Immun. 67:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.