Abstract
Propidium monoazide (PMA) and ethidium monoazide were used for enumeration of viable Listeria monocytogenes cells in the presence of dead cells. PMA had no antimicrobial effect on L. monocytogenes. Viable cell counts were linearly related to real-time PCR threshold cycle values for PMA-treated cells from planktonic and biofilm sources over a 4-log range.
Propidium monoazide (PMA) and ethidium monoazide (EMA) have been reported to be useful for quantifying viable bacteria by real-time PCR (7, 8, 9, 14, 15). Both EMA and PMA can intercalate into double-stranded DNA or RNA and then irreversibly cross-link to the nucleic acids following photoactivation (1, 8). These dyes are reported to be excluded from viable bacterial cells because of their inability to cross intact biological membranes. The viable microorganisms can therefore be enumerated by real-time PCR detection methods once free DNA or DNA from dead cells is inactivated (8, 9, 14, 15).
Real-time PCR methods have been developed to detect and quantify Listeria monocytogenes in food products such as milk, cabbage, and cheese, as well as in biofilms (3, 4, 5, 8, 10, 12, 13). A combination of EMA and real-time PCR (EMA-PCR) has been developed for selective analysis of DNA from live cells of L. monocytogenes (9, 14, 15). However, Nocker et al. (8) observed that EMA, but not PMA, was able to penetrate into viable cells of L. monocytogenes using fluorescence microscopy. Our results showed that EMA, but not PMA, was toxic to viable cells, and we found that temperature and time of exposure can influence the toxicity of EMA but not the toxicity of PMA. In this study we compared the efficiencies of PMA and EMA for enumeration of viable cells by real-time PCR in the presence of various amounts of free DNA or dead cells. From our data, we determined the optimum conditions for PMA treatment of live cell-dead cell populations for enumeration of live cells. We also demonstrated the application of this technique for enumerating viable cells in L. monocytogenes removed from biofilms.
Lethal effects of EMA and PMA on L. monocytogenes.
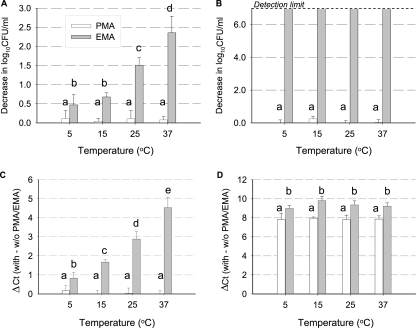
Four strains of L. monocytogenes obtained from different sources and having different serotypes were used, SK1386 (serotype 1/2b; isolated from a clinic in Canada in 2002), SK1389 (serotype 1/2c; isolated from a clinic in England in 2002), SK1403 (serotype 4b; isolated from food in the United States in 2002), and SK1420 (serotype 1/2a; isolated from food in the United States in 2002). These strains were obtained from the culture collection of Sophia Kathariou of the Department of Food Science, North Carolina State University. The bacterial cultures were handled essentially as described by Pan et al. (11), using tryptic soy agar or tryptic soy broth supplemented with yeast extract (TSB-YE) as the growth medium (Difco Laboratories, Detroit, MI). EMA (C21H18BrN5; phenanthridium, 3-amino-8-azide-5-ethl-6-phenyl bromide; Molecular Probes, Inc., Oregon) and PMA (C27H33Cl2N6; Biotium, Inc., California) were prepared, stored, and used as described in previous studies (7, 8, 9). Briefly, EMA and PMA were dissolved in 20% dimethyl sulfoxide to obtain stock solutions having concentrations of 12 and 2.5 mM, respectively. Since temperature plays a role in microbial cell membrane permeability (6, 16), the effect of temperature on the lethality of EMA (or PMA) for L. monocytogenes cells was investigated. In order to obtain a postexponential-stage culture for this study, the bacteria were incubated in TSB-YE for 11 h at 25°C. Equal volumes of the four cell suspensions (ca. 108 CFU/ml) were pooled. The four-strain mixture was preconditioned at four temperatures (5, 15, 25, and 37°C) for 2 h before it was treated with either EMA or PMA (490 μl of cell suspension with 10 μl of a dye stock solution) at room temperature in the dark. The lethal effects of 240 μM (100 μg/ml) EMA increased with the incubation temperature from 5 to 37°C (Fig. 1A). However, 50 μM PMA did not show any lethal effect (Fig. 1A). Following incubation with the dyes for 5 min in the dark, similarly prepared cells were exposed for 5 min to a 600-W halogen light source placed 20 cm directly above 500-μl samples in open microcentrifuge tubes on chipped ice (to prevent heating). This resulted in a >6-log10 reduction in the number of EMA-treated cells (Fig. 1B), but no reduction was seen in the number of PMA-treated cells (Fig. 1B). The MICs for EMA and PMA were determined using twofold serial dilutions of the dyes, and cells were kept at 37°C in TSB-YE in the dark for 16 h. The MICs of EMA and PMA were 240 and 1,600 μM, respectively.
FIG. 1.
Effects of PMA and EMA on DNA amplification from viable and dead cells of L. monocytogenes. (A and B) Viable cell density was determined following 5 min of incubation in the dark (A) and after 5 min of exposure to light (B). (C and D) Effects of PMA and EMA on DNA amplification from viable cells (C) and heat-killed cells (D). The bars indicate the mean values, and the error bars indicate the standard deviations (n = 3). Different letters indicate a significant difference (P < 0.05).
Rudi et al. (15) investigated the exclusion of EMA from viable Campylobacter jejuni cells by using inhibitors of efflux systems. They found no evidence that efflux pump systems were responsible for excluding EMA from viable cells (15). The effects of temperature that we observed for EMA toxicity may have reflected changes in cell membrane fluidity or permeability. PMA apparently cannot penetrate through the viable cell membrane of L. monocytogenes. This may be due to the higher positive charge of the PMA molecule than of the EMA molecule (8).
Effects of PMA and EMA on DNA amplification in PCR.
The four-strain mixture of exponential-stage cultures was subjected to heat treatment for 10 min at 80°C to generate a dead cell population. Aliquots of viable or dead cell suspensions were treated with EMA or PMA as described above. After light exposure, the cell suspensions were harvested by centrifugation at 6,000 × g for 5 min. Genomic DNA from the cell pellet was isolated using a DNeasy blood and tissue kit (Qiagen) by following the supplier's protocol for gram-positive bacteria. Forward primer 5′-GCGGATGTGATTGATTTAC-3′ and reverse primer 5′-AAACTGCACTAACTCTTGAAT-3′ were used to target a 78-bp fragment of the prs gene which is present in all Listeria species (2). Real-time PCR amplification was performed with a 96-well thermocycler (iCycler; Bio-Rad Laboratories). The reaction mixtures (25 μl) consisted of the primers (300 nM), 4.0 μl of template, and 12.5 μl of 2× iQ SYBR green Supermix (catalog no. 170-8882; Bio-Rad Laboratories, California). The cycling conditions were as follows: 94°C for 3 min, followed by 35 cycles of 94°C for 20 s, 58°C for 30 s, and 72°C for 25 s and then a final step of 72°C for 10 min after the cycling was completed. Threshold cycle (CT) values were automatically generated by the MyiQ (Bio-Rad) software.
The CT values for EMA (240 μM)-treated viable cells increased by 0.8 cycle to 4.5 cycles as the preconditioning temperature was raised from 5 to 37°C (Fig. 1C). However, there was no difference in CT values for PMA (50 μM)-treated cells (Fig. 1C). The CT values for killed cell aliquots treated with EMA or PMA were not different with different preincubation temperatures (Fig. 1D). The concentrations of the two dyes used in this study have been reported to be the optimal concentrations for enumeration of viable cells (7, 9, 14, and 15). A lower concentration of EMA (50 μM) was tested along with a higher concentration (240 μM), as well as with 50 μM PMA, to examine the efficiency of viable cell enumeration. For L. monocytogenes viable cells, there was no statistically significant difference between the higher concentration of EMA (240 μM) and the lower concentration of EMA (50 μM) (data not shown).
Optimization of PMA treatments for L. monocytogenes.
To optimize the PMA treatments, viable cell aliquots and killed cell aliquots were treated with different concentrations of PMA or subjected to repeated PMA treatments. The CT values for the killed cell aliquots (approximately 2.4 × 107 cells/ml) treated with PMA (50 μM) two or three times were similar, 10.7 ± 0.35, and the average value was 2.8 cycles more than the value for the aliquots treated only once. Varying the PMA concentration (50, 100, and 200 μM) or the incubation temperature (23 and 40°C) did not result in a significant difference in CT values for either viable cell samples or killed cell samples. Based on these data, repeating the PMA (50 μM) treatment twice at room temperature was considered the optimal conditions for quantifying viable cells of L. monocytogenes with the PMA-PCR assay.
Determination of the dynamic range of the killed cell/viable cell ratio with which viable L. monocytogenes cells could be quantified by real-time PCR.
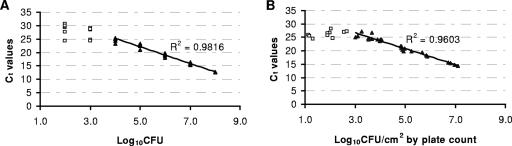
Mixtures of viable cells and killed cells (Table 1) were subjected to real-time PCR as described above. Each mixture was subject to PMA treatment twice at room temperature to ensure maximum binding of PMA to free DNA or DNA from dead cells. Three replicates of each mixture were used to determine correlation coefficients (R2) for CT values and viable cell counts (CFU/ml) with various concentrations of dead cells (Table 1 and Fig. 2A). A linear relationship between CT and the number of viable cells (R2, 0.9816) was observed as long as the ratio of dead cells to viable cells was no greater than 104 and the minimum number of viable cells was not less than 103 CFU/ml (Fig. 2A and Table 1). This is similar to the results of the EMA-PCR analysis of C. jejuni reported by Rudi et al. (15). We found that the minimum number of DNA copies that are available for PCR analysis and the fraction of dead cells in a viable cell-dead cell mixture are two critical factors that could limit the range of the PMA-PCR assay.
TABLE 1.
Dynamic range test for the dead cell/viable cell ratio of L. monocytogenes with the PMA-PCR assay
| Dead cell density (log10 CFU/ml) |
CT value with a viable cell density ofa:
|
R2 | ||||||
|---|---|---|---|---|---|---|---|---|
| 8 log10 CFU/ml | 7 log10 CFU/ml | 6 log10 CFU/ml | 5 log10 CFU/ml | 4 log10 CFU/ml | 3 log10 CFU/ml | 2 log10 CFU/ml | ||
| 0 | 12.85 ± 0.07 | 16.03 ± 0.14 | 19.78 ± 0.12 | 23.19 ± 0.35 | 25.38 ± 0.81 | 28.57 ± 0.14 | 30.69 ± 1.98 | 0.9952 |
| 3 | 12.83 ± 0.17 | 16.02 ± 0.26 | 19.17 ± 0.52 | 22.95 ± 0.17 | 25.12 ± 0.17 | 28.22 ± 0.39 | 29.65 ± 1.62 | 0.9964 |
| 4 | 12.77 ± 0.33 | 16.42 ± 0.42 | 19.33 ± 0.21 | 23.19 ± 0.43 | 25.42 ± 0.53 | 29.01 ± 0.85 | 29.24 ± 2.30 | 0.9967 |
| 5 | 12.74 ± 0.38 | 15.68 ± 0.33 | 18.94 ± 0.28 | 22.64 ± 0.32 | 24.99 ± 0.70 | 28.32 ± 0.84 | 29.68 ± 2.89 | 0.9978 |
| 6 | 12.56 ± 0.27 | 15.68 ± 0.45 | 18.91 ± 0.31 | 23.06 ± 0.60 | 25.28 ± 0.56 | 28.47 ± 0.98 | 30.02 ± 1.74 | 0.9953 |
| 7 | 12.57 ± 0.36 | 15.56 ± 0.28 | 19.09 ± 0.34 | 22.32 ± 0.47 | 24.26 ± 0.82 | 25.79 ± 1.34 | 27.75 ± 1.82 | 0.9918 |
| 8 | 12.58 ± 0.32 | 15.26 ± 0.52 | 18.08 ± 0.35 | 21.05 ± 0.76 | 23.34 ± 0.46 | 24.28 ± 1.24 | 24.38 ± 1.91 | 0.9986 |
The boldface values were outliers and were not included in the linear regression analysis.
FIG. 2.
Standard curves for quantifying viable L. monocytogenes cells in a viable cell-dead cell mixture (A) and in stressed biofilms (B). The solid standard line and the linear regression coefficient factor (R2) were generated using the solid triangles. The open squares are data points not included in the linear regression analysis.
Quantification of viable cells of L. monocytogenes in sanitizer-treated biofilms by real-time PCR.
Stainless steel coupons (T-316; no. 7 finish; 82.5 by 25 by 1.6 mm; M. G. Newell Corp., Greensboro, NC) were used to form biofilms of the four-strain mixture of L. monocytogenes as described previously (11). Briefly, following 3 h of cell attachment in a prepared suspension of the four-strain mixture (ca. 108 CFU/ml), the stainless steel coupons (a total of 40 coupons) were submerged in 10-fold-diluted rich medium (TSB-YE) and incubated at 37°C for 48 h, allowing biofilm formation. After biofilm formation, three sets of 10 coupons were then treated with a peroxide-based sanitizing agent (Matrixx; 100 ppm; pH 3.8; Ecolab, St. Paul, MN) (one set each for 1, 2, and 3 min). A control group (10 coupons) was treated with saline (0.85% NaCl) in place of the sanitizer. Following neutralization with a 0.1% sodium thiosulfate-phosphate solution (pH 7.0), biofilms were then detached from the surfaces of the coupons using sterile cotton-tipped swabs (11). The detached biofilm cell suspension from each coupon was concentrated to 1.5 ml by centrifugation. Two aliquots of the concentrated cell suspension from each coupon were transferred into 1.5-ml microcentrifuge tubes for PMA treatment (two treatments with 50 μM PMA) in 500 μl (total volume), followed by DNA extraction and real-time PCR. The remaining cell suspension from each sample was used to determine the viable cell count. The viable cell density in non-sanitizer-treated biofilms on stainless steel surfaces was approximately 107 CFU/cm2. The viable cell count decreased with increasing exposure to the sanitizer. There was a linear relationship (R2 = 0.96) between the CT value and the number of viable biofilm cells when the estimated ratio of dead cells to viable cells was no more than 104 (Fig. 2B).
We found that PMA, in combination with real-time PCR, could be used for quantification of viable cells of L. monocytogenes in suspensions in which the ratio of dead cells to viable cells was no more than 104 and the concentration of live cells was no less than 103 CFU/ml. Cell suspensions prepared from broth cultures and from biofilms gave similar results. Compared with EMA, PMA was not found to penetrate live cells, as determined by the toxicity of the two dyes. Use of PMA with real-time PCR may be useful for quantifying viable cells of L. monocytogenes in food, pharmaceutical, and environmental applications. Further studies to refine PMA treatment with real-time PCR are needed to overcome the limitations and maximize the utilization of the method for DNA-based quantitative analysis.
Acknowledgments
This investigation was partially supported by Pickle Packers International, Inc., Washington, DC.
We recognize Trevor Phister of North Carolina State University for his helpful discussions and Sandra Parker for her excellent secretarial assistance.
Mention of a trademark or proprietary product does not constitute a guarantee or warranty of the product by the U.S. Department of Agriculture or North Carolina Agricultural Research Service, nor does it imply approval to the exclusion of other products that may be suitable.
Footnotes
Published ahead of print on 12 October 2007.
Paper no. FSR07-09 of the Journal Series of the Department of Food Science, North Carolina State University, Raleigh.
REFERENCES
- 1.Bolton, P. H., and D. R. Kearns. 1978. Spectroscopic properties of ethidium monoazide: a fluorescent photoaffinity label for nucleic acids. Nucleic Acids Res. 5:4891-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doumith, M., C. Buchrieser, P. Glaser, C. Jacquet, and P. Martin. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42:3819-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guilbaud, M., P. de Coppet, F. Bourion, C. Rachman, H. Prevost, and X. Dousset. 2005. Quantitative detection of Listeria monocytogenes in biofilms by real-time PCR. Appl. Environ. Microbiol. 71:2190-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hein, I., D. Klein, A. Lehner, A. Bubert, E. Brandl, and M. Wagner. 2001. Detection and quantification of the iap gene of Listeria monocytogenes and Listeria innocua by a new real-time quantitative PCR assay. Res. Microbiol. 152:37-46. [DOI] [PubMed] [Google Scholar]
- 5.Hough, A. J., S.-A. Harbison, M. G. Savill, L. D. Melton, and G. Fletcher. 2002. Rapid enumeration of Listeria monocytogenes in artificially contaminated cabbage using real-time polymerase chain reaction. J. Food Prot. 65:1329-1332. [DOI] [PubMed] [Google Scholar]
- 6.Konings, W. N., S.-V. Albers, S. Koning, and A. J. Driessen. 2002. The cell membrane plays a crucial role in survival of bacteria and archaea in extreme environments. Antonie Leeuwenhoek 81:61-72. [DOI] [PubMed] [Google Scholar]
- 7.Nocker, A., and A. K. Camper. 2006. Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl. Environ. Microbiol. 72:1997-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nocker, A., C. Cheung, and A. K. Camper. 2006. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67:310-320. [DOI] [PubMed] [Google Scholar]
- 9.Nogva, H. K., S. M. Dromtorp, H. Nissen, and K. Rudi. 2003. Ethidium monoazide for DNA-based differentiation of viable and dead bacteria by 5′-nuclease PCR. BioTechniques 34:804-813. [DOI] [PubMed] [Google Scholar]
- 10.Nogva, H. K., K. Rudi, K. Naterstad, A. Holck, and D. Lillehaug. 2000. Application of 5′-nuclease PCR for quantitative detection of Listeria monocytogenes in pure cultures, water, skim milk, and unpasteurized whole milk. Appl. Environ. Microbiol. 66:4266-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan, Y., F. Breidt, Jr., and S. Kathariou. 2006. Resistance of Listeria monocytogenes biofilms to sanitizing agents in a simulated food processing environment. Appl. Environ. Microbiol. 72:7711-7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodríguez-Lázaro, D., M. Hernández, M. Scortti, T. Esteve, J. A. Vázquez-Boland, and M. Pla. 2004. Quantitative detection of Listeria monocytogenes and Listeria innocua by real-time PCR: assessment of hly, iap, and lin02483 targets and AmpliFluor technology. Appl. Environ. Microbiol. 70:1366-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez-Lázaro, D., M. Pla, M. Scortti, H. J. Monzó, and J. A. Vázquez-Boland. 2005. A novel real-time PCR for Listeria monocytogenes that monitors analytical performance via an internal amplification control. Appl. Environ. Microbiol. 71:9008-9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudi, K., K. Naterstad, S. M. Dromtorp, and H. Holo. 2005. Detection of viable and dead Listeria monocytogenes on gouda-like cheeses by real-time PCR. Lett. Appl. Microbiol. 40:301-306. [DOI] [PubMed] [Google Scholar]
- 15.Rudi, K., B. Moen, S. M. Dromtorp, and A. L. Holck. 2005. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 71:1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van de Vossenberg, J. L., T. Ubbink-Kok, M. G. Elferink, A. J. Driessen, and W. N. Konings. 1995. Ion permeability of the cytoplasmic membrane limits the maximum growth temperature of bacteria and archaea. Mol. Microbiol. 18:925-932. [DOI] [PubMed] [Google Scholar]