Abstract
Shiga toxin-producing Escherichia coli (STEC) strains are food-borne pathogens whose ability to produce Shiga toxin (Stx) is due to integration of Stx-encoding lambdoid bacteriophages. These Stx phages are both genetically and morphologically heterogeneous, and here we report the design and validation of a PCR-based multilocus typing scheme. PCR primer sets were designed for database variants of a range of key lambdoid bacteriophage genes and applied to control phages and 70 stx+ phage preparations induced from a collection of STEC isolates. The genetic diversity residing within these populations could be described, and observations were made on the heterogeneity of individual gene targets, including the unexpected predominance of short-tailed phages with a highly conserved tail spike protein gene. Purified Stx phages can be profiled using this scheme, and the lambdoid phage-borne genes in induced STEC preparations can be identified as well as those residing in the noninducible prophage complement. The ultimate goal is to enable robust and realistically applicable epidemiological studies of Stx phages and their traits. The impact of Stx phage on STEC epidemiology is currently unknown.
The emergence of Shiga toxin-producing Escherichia coli (STEC) strains as food-borne pathogens has become a worldwide public health concern. STEC infections can result in diarrheal symptoms that may develop into hemorrhagic colitis and, in severe cases, progress to hemolytic uremic syndrome or thrombotic thrombocytopenic purpura, both of which are potentially fatal complications (15) caused by Shiga toxin (Stx). The genes encoding Stx (stx genes) are located within prophage or remnant prophage sequences in all STEC strains, and the horizontal transfer of stx genes is facilitated by bacteriophages (Stx phages) (35). Over the last 5 years, bacterial genome sequencing has revealed that many bacterial pathogens possess a significant amount of prophage and remnant prophage DNA (2). STEC strains are no exception, and the two sequenced E. coli O157:H7 strains, one isolated from an outbreak in Sakai, Japan (19), and another from an outbreak in Michigan (36) carry an additional ∼1 Mbp of DNA compared to E. coli K-12 (6). Approximately 40% of the variation between the K-12 and O157:H7 strains is due to remnant and inducible prophages (14, 36).
The Stx phages are lambdoid (2), having the ability to infect a host cell and then either replicate or integrate into the bacterial genome, and they share a distinct genetic organization (37) (Fig. 1) with bacteriophage λ (8). Lambdoid bacteriophage genomes have been shown to possess high levels of mosaicism, although their genome organization and orientation remain similar (8, 11). Stx phages are intimately involved in the pathogenic profile of their bacterial lysogens, the survival and dissemination of the stx genes in the environment, and the emergence of new Stx-producing pathogens (2). A classification method for monitoring Stx phage dispersal would enable the generation of epidemiological information to address the relationship between STEC disease outbreaks and the distribution of zoonotic STEC in livestock and the farm environment. There is a wealth of information on STEC strains in this context but little on the occurrence, distribution, and identity of the bacteriophages that disseminate the stx genes and potentially provide a vehicle for their survival in the environment (2, 10, 23, 30). Traditionally, phages have been characterized by morphological characteristics according to a universal system for virus taxonomy (32, 38, 39); but morphologically similar phages may be completely unrelated at the nucleotide sequence level, and morphologically distinct phages can possess large regions of sequence identity or similarity (2). Characterization of bacteriophages can be further complicated by the high levels of recombination that occur between inducible and remnant bacteriophage genomes within a bacterial lysogen (2, 7, 24). Not only do recombination events occur frequently, they are also up-regulated by bacteriophage-encoded proteins; of these, the λ Red recombinase system is used commercially (13, 52) and has also been identified in Stx phages (2). A multilocus typing system designed to identify genetic similarity and disparity would provide the ability to compare and contrast genetic variation in inducible phages without the need to sequence entire phage genomes. The ability to type Stx phages induced from STEC isolates will enable definition of the level of heterogeneity among Stx phages and identification of specific phage genes that are disseminated across a bacterial population or even enable the discovery of phage-borne genes that are associated with enhanced pathogenicity/fitness of their bacterial hosts.
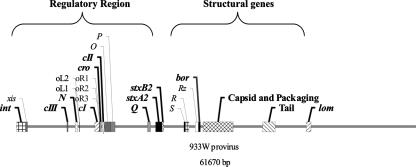
FIG. 1.
A schematic of the well-studied Stx bacteriophage 933W genome, detailing the functional genes that were identified as targets for the multilocus PCR characterization scheme. The target genes used as part of this study, from left to right, include int, cIII, N, cI, cro, cII, Q, stx, capsid and packaging genes, and tail spike/host recognition proteins.
Multilocus sequence typing is an established molecular tool for typing bacteria through the generation of an allelic profile. Here, we take a similar approach to produce a scheme based on lambdoid phage genome organization using loci that represent key modules involved in phage infection and propagation. This system makes it possible to identify inducible phage genes that are present in an STEC background and the genes that impinge upon the fluidity of the mosaic Stx phage genome. The core functional genes controlling the biology of Stx phages (int, cIII, N, cI, cro, cII, Q, stx, capsid structural genes, packaging genes, and tail spike/host recognition protein genes) (Fig. 1 and Table 1) were identified from published genomic Stx phage sequences (NC_004813, NC_000924, AF034975, NC_003525, NC_004914, NC_000902, and NC_003356), as well as lambda phage. The sequences of these target genes were subjected to BLASTN analysis (4) against sequenced STEC strains. Matches from these analyses, from both inducible and remnant Stx prophages, were then subjected to further rounds of BLASTN (4) analyses to identify all complete, phage-related genes within the genetic databases. Sequences for each gene were aligned (21, 48) and grouped into clades (27), and oligonucleotide primers capable of differentiating between variants of each target gene were designed. These primer pairs (Table 1), upon which the multilocus typing scheme is based, were validated by PCR amplification of target genes from bacteriophages possessing the specific sequence for which the primer pair was designed, as well as against phages that did not harbor the particular gene variant, to ensure specificity of the primer pair (data not shown). All amplification products from these controls were sequenced by Macrogen, Inc., Seoul, Korea, to confirm their identity. DNA amplification (30 cycles) was performed using Phusion (New England BioLabs) according to the manufacturer's instructions (1× HF buffer containing a 200 μM concentration of each deoxynucleoside triphosphate, a 0.5 μM concentration of each oligonucleotide primer, and 0.02 U of Phusion DNA polymerase). Amplification reactions involved denaturation at 98°C, annealing for 30 s (temperatures listed in Table 1), and an extension period of 1 min at 72°C. Amplification products were produced from PCR using 2 or 4 μl of each phage preparation as the template source. Failure to amplify the E. coli housekeeping gene gapA (encoding glyceraldehyde-3-phosphate dehydrogenase) from the phage preparations indicated that host genomic DNA did not contaminate the phage preparations to any significant level.
TABLE 1.
Oligonucleotide primers comprising the multilocus identification scheme
| Gene target | Phage namea | Primer name | Sequence (5′ → 3′) | Gene type | Tmb | Annealing temp | Amplicon size (bp) | Reference or source |
|---|---|---|---|---|---|---|---|---|
| int | Φ24B | Φ24Bint 5′ | GCCAGGCTTTCTGAGCTACG | 24B int | 55.0 | 16 | ||
| Φ24Bint 3′ | GCCTAAAATCATGCGTTCTCC | 55.0 | 16 | |||||
| lambda, H19J, | 1F | GTTACMGGGCARMGAGTHGG | 1 int | 50.0 | 5 | |||
| HKO22, P434 (group 1) | 1R | ATGCCCGAGAAGAYGTTGAGC | 5 | |||||
| P21, e14 (group 2) | 2F | GTTACTGGWCARCGKTTAGG | 2 int | 50.0 | 5 | |||
| 2R | GATCATCATKRTAWCGRTCGGT | 5 | ||||||
| ST64B (group 3) | 3F | AATGGARATWKCYTATYTVTGTGC | 3 int | 50.0 | 5 | |||
| 3R | TCRTARTCTGARATYCCYTTBGC | 5 | ||||||
| P27, 933W, Fels1, BP4795, Gifsy2, EH297 (group 4) | 4F | CTBGCMTGGGARGATATHGA | 50.0 | 5 | ||||
| 4R | GMCCAGCABGCATARGTRTG | 5 | ||||||
| P4, HK620, Sf6 (group 5a) | 5AF | TGGRAKRAMKTCGAYTTYGA | 5A int | 50.0 | 5 | |||
| 5AR | CAGTTGCMYYTCWATMGCGTCA | 5 | ||||||
| P4, epsilon 15 | 5BF | TWGTKCGTWMMAGTGAATT | 5B int | 50.0 | 5 | |||
| (group 5b) | 5BR | TKGWTRTATRCCGCWCGYAC | 5 | |||||
| Phi80, Gifsy1 | 5CF | GGRMARTYATAAAACKSG | 5C int | 50.0 | 5 | |||
| (group 5c) | 5CR | TGCCCGAGCAKCWTYTCA | 5 | |||||
| P2, L413C, Wphi | 6AF | CTGAGYACWGGAGSAMGWTGG | 6A int | 50.0 | 5 | |||
| (Group 6a) | 6AR | CCBCCRTTMATCATRAARTG | 5 | |||||
| Fels2, PSP3, P186 | 6BF | TVGCWACYGGCGCMMGRTGG | 6B int | 50.0 | 5 | |||
| (Group 6b) | 6BR | CCBCCRTTMATCATRAARTG | 5 | |||||
| P22, SfV, SfII, | 7F | AACATYATMAAYCTKGARTGGCA | 7 int | 50.0 | 5 | |||
| ST64T | 7R | CGAACCATTTCKATRGACTCCCA | 5 | |||||
| P1 | 8F | TGCTTATAACACCCTGTTACGTAT | 8 int | 50.0 | 5 | |||
| 8R | CAGCCACCAGCTTGCATGATC | 5 | ||||||
| cIII | All listed | cIII 5′ | ATGCAATATGCCATTGCA | cIII | 43.0 | 53.0 | 168 | This study |
| cIII 3′ | TTAGTCTGGATAGCCATA | 43.0 | This study | |||||
| N | N (1) 5′ | ATGACACGCAGAACTCAG | N1 | 51.4 | 50.0 | 384 | This study | |
| N (2) 5′ | ATGCAATGCCGAAGCAAC | N2 | 56.5 | 57.5 | 295 | This study | ||
| N 3′ | TYACCTYGCYGTCAGTTG | 54.4 | This study | |||||
| cI | BP4795, H19B | cI (1a) 5′ | ATGGAAAACAAAGATATTCGC | cI 1a | 59.4 | 54.0 | 705 | This study |
| 933W, Stx2(I) | cI (1b) 5′ | ATGGTTCAGAATGAAAAAGTG | cI 1b | 58.1 | 61.0 | 708 | This study | |
| BP4795, H19B, 933W, Stx2(I) | cI (1) 3′ | TCACGAACTTTTCAGCCACTC | 64.2 | This study | ||||
| HK97, Lambda | cI (2a) 5′ | ATGAGCRCAAAAAAGAAACCA | cI 2a | 60.2 | 60.0 | 714 | This study | |
| Nil 2 | cI (2b) 5′ | ATGAAATGGTATGAACTGGCT | cI 2b | 59.9 | 60.0 | 654 | This study | |
| Stx2(II) | cI (2c) 5′ | ATGGATGGTTCCAGTACAGAG | cI 2c | 60.4 | 59.0 | 598 | This study | |
| VT2-Sa | cI (2d) 5′ | GTGGTGTTTAAATACCTTGGT | cI 2d | 57.0 | 59.0 | 510 | This study | |
| HK97, Lambda, Nil 2, Stx2(II), VT2-Sa | cI (2) 3′ | TYAACCAAACGTCTCTTCAGG | cI 2d | 59.3 | 510 | This study | ||
| HK620 | cI (3) 5′ | ATGGAAAATAAAAAATCACTG | cI 3 | 54.9 | 53.0 | 714 | This study | |
| cI (3) 3′ | TCAAACCAGCCTTAGTTTTGT | 61.3 | This study | |||||
| D3112 | cI (4) 5′ | GTGAAATCAGACACTTACGGA | cI 4 | 59.2 | 62.2 | 665 | This study | |
| cI (4) 3′ | CTAAACCATCCAGCGGCTAGC | 66.7 | This study | |||||
| P27 | cI (5) 5′ | ATGAAATCTTTAGGTGAACGC | cI 5 | 59.6 | 62.6 | 723 | This study | |
| cI (5) 3′ | TCAGAAAATATCCCACCTGGC | 64.3 | This study | |||||
| phi 105 | cI (6) 5′ | ATGACTGTAGGGCAAAGAATC | cI 6 | 59.6 | 53.0 | 437 | This study | |
| cI (6) 3′ | GTATTCTTGATCGTCATTTCT | 54.8 | This study | |||||
| cro | HK97, Sakai | Cro (1) 5′ | ATGGAACAACGCATAACCCTG | cro 1 | 65.6 | 59.4 | 201 | This study |
| Cro (1) 3′ | TTATGCAGTTGTTTTTTTGTT | 56.4 | This study | |||||
| HK620 | Cro (2) 5′ | ATGATTCGAATGACACTTGCC | cro 2 | 63.7 | 57.0 | 195 | This study | |
| Cro (2) 3′ | CTATTTGTTTTTCTTGTTGCT | 55.3 | This study | |||||
| 933W, StxII | Cro (3) 5′ | ATGCAAAATCTTGATGAGCCG | cro 3 | 66.0 | 65.0 | 228 | This study | |
| Cro (3) 3′ | TTATGCAGCCAGAAGGTTCTT | 62.6 | This study | |||||
| Sakai, ST64T, VT2Sa | Cro (4) 5′ | ATGAGCAAYCTWCGRAAAWWY | cro 4 | 56.9 | 59.9 | 216 | This study | |
| Cro (4) 3′ | TTARGCRGCWTTRWGYTCMGG | 61.7 | This study | |||||
| P22 | Cro (5) 5′ | ATGTACAAGAAAGATGTTATCGAC | cro 5 | 57.7 | 55.0 | 186 | This study | |
| Cro (5) 3′ | CTTCATGGTTCTTTTGCG | 59.4 | This study | |||||
| ST104 | Cro (6) 5′ | ATGACTAACAAAGCAATACAA | cro 6 | 54.0 | 54.0 | 216 | This study | |
| Cro (6) 3′ | TTAACTTGCTGCCAGTAAGTC | 58.6 | This study | |||||
| SfV | Cro (7) 5′ | ATGAAAGCGTATTGGGACTCT | cro 7 | 61.4 | 54.8 | 201 | This study | |
| Cro (7) 3′ | TTAATCTTTCGGATAGATATC | 55.0 | This study | |||||
| N15 | Cro (8) 5′ | ATGAAACCCGAAGAACTTGTG | cro 8 | 62.8 | 56.0 | 216 | This study | |
| Cro (8) 3′ | CTATTTAGTTCCACTGTTATG CCC | 61.4 | This study | |||||
| BP4795 | Cro (9) 5′ | ATGAGTAATGAACTACTACGCTGG | cro 9 | 60.1 | 56.0 | 234 | This study | |
| Cro (9) 3′ | TTATGCAGCCGATGCTCT | 62.1 | This study | |||||
| cII | All listed | CII - 5′ | ATGRMACRARCAAGYTACAGC | cII | 56.0 | 53.0 | 297 | This study |
| CII - 3′ | TCAGAATTGCATATCAAT | 50.7 | This study | |||||
| Q | All listed | Q ATG 5′ | ATGTTCTTATGGTTCACCG | Q | 53.0 | 55.5 | 435 | This study |
| Q 3′ | TTACGATCGTAAACTATTTTT | 50.0 | This study | |||||
| stx | All sequenced stx-genes | Degen StxA (Type I&II) | TTTGTYACYGTSAYAGCWGAAG | stx | 47.2 | 47.5 | 675 | This study |
| Degen StxB (Type I&II) | TYMTCATTATAYTTDGWRWACT | 49.0 | This study | |||||
| Capsid | CP933X Capsid | CP933X Capsid 5′ | TGGGRCCGGSAWRACATSCTG | capX | 63.3 | 63.3 | 1,572 | This study |
| CP933X Capsid 3′ | TTACGCAGCTCTGCTGTC | 60.48 | This study | |||||
| CP933I Capsid | CP933I Capsid 5′ | ATGACAATCCCAGAACAG | capI | 55.6 | 54.9 | 753 | This study | |
| CP933I Capsid 3′ | TCATACTGCTTTCTCCTT | 51.9 | This study | |||||
| CP933X (3) Capsid | CP933X (3) Capsid 5′ | ATGTCSRTKTACACMACYGCC | capX3 | 55.0 | 58.0 | 1,026 | This study | |
| CP933X (3) Capsid 3′ | TTAYGCCAGYTKKACGGASAC | 58.1 | This study | |||||
| CP933X (2) Capsid | CP933X (2) Capsid 5′ | RTGRCAGCAGAGCTGCGT | capX2 | 63.8 | 55.0 | 1,204 | This study | |
| CP933X (2) Capsid 3′ | TTAMACTGGKGTGTTYARCAA | 52.1 | This study | |||||
| CP933C Capsid | CP933C Capsid 5′ | ATGCMGAGAATAATCGAATTAC | capC | 56.5 | 56.0 | 1,158 | This study | |
| CP933C Capsid 3′ | TTAATCGTCGTCYTSYGGCAG | 63.8 | This study | |||||
| CP933R (3) Capsid | CP933R (3) Capsid 5′ | ATGAAACGAACGCCTGTC | capR3 | 61.3 | 53.0 | 1,590 | This study | |
| CP933R (3) Capsid 3′ | TTACGTCTCACGKGRTGT | 54.6 | This study | |||||
| CP933R Capsid | CP933R Capsid 5′ | ATGGGATTGTTTACGACC | capR | 57.1 | 53.0 | 1,029 | This study | |
| CP933R Capsid 3′ | TTATTTCACCTGTACCAC | 50.0 | This study | |||||
| CP933O/R Capsid 5′ | CP933O/R Capsid 5′ | ATGGTRACGAAAAMATCACTGA | capO/R | 57.5 | 60.6 | 348 | This study | |
| CP933O/R Capsid 3′ | TTACGGCAGCGCYGC | 63.8 | This study | |||||
| P27 Capsid | P27 Capsid 5′ | RYGGYTGATRTTAAAGATGTG | capP27 | 60.0 | 56.9 | 1,224 | This study | |
| P27 Capsid 3′ | AATTTTCAKCAGCTTRATMGC | 57.0 | This study | |||||
| HK620 Major Capsid | HK620 Major Capsid 5′ | ATGGCCTAACAATCTCGA | cap620 | 51.0 | 51.0 | 1,272 | This study | |
| HK620 Major Capsid 3′ | TTACGGATTACCGAAGAA | 49.0 | This study | |||||
| Host recognition proteins | This study | |||||||
| Short-tailed | 933W, VT2-Sa, | VTTF1 5′ | GTTGTTGTTTCGGGGACG | VTTF1 | 64.0 | 55.0 | 1,900 | This study |
| Stx phages | Φ24B | VTUTF 3′ | TCATTCTCCTGTTCTGCC | VTTF2 | 58.8 | This study | ||
| VTTF3 5′ | TGCAGAGGAAAGCTCGAC | VTTF3F | 54.9 | 55.0 | 800 | This study | ||
| VTTF3 3′ | GCAGCCTCTTCTGCCTTT | VTTF3R | 62.3 | This study | ||||
| Long-tailed | P27 TF | P27 (p56) (TF) 5′ | ATGTCTGTAGTGATATCAGGT | TFP27 | 52.4 | 55.3 | 400 | This study |
| Stx phage | P27 (p56) (TF) 3′ | TCATGCCAATCCTCACAA | 61.0 | This study | ||||
| N15 HRP | N15 (TF) 5′ | ATGGCTACATCTACTCCG | TFN15 | 48.0 | 47.0 | 3,200 | This study | |
| N15 (TF) 3′ | TCAAAATACCCCCGTAAT | 43.0 | This study | |||||
| (CP)EcS1650, | (CP)EcS1650 (TF) 5′ | ATGGCAGTAAAGATTTCA | TFcp | 52.3 | 55.8 | 2,916 | This study | |
| CP933M, λ (TF) 5 | (CP)EcS1650 (TF) 3′ | TTATGCAAGCCTCACAAT | 56.6 | This study | ||||
| T7 (TF) | T7 (TF) 5′ | ATGGCTTAACGTAATTAA | TFT7 | 50.0 | 48.0 | 1,662 | This study | |
| T7 (TF) 3′ | TTACACGTCCTCTACGGC | 58.0 | This study | |||||
| HK022, | HK022 (TF) 5′ | TTGCCTGGAGAAAATATG | TF022 | 56.0 | 57.7 | 1,116 | This study | |
| HK022 (TF) 3′ | TTAGTCAACAAGCTCCCT | 54.6 | This study | |||||
| HK97 (31111) (TF) | HK97 (31111)(TF) 5′ | ATGATTTATAGCACCGGA | TF97 | 55.3 | 58.3 | 795 | This study | |
| HK97 (31111)(TF) 3′ | TTATACTGCCCTTACAATGTA | 54.0 | This study | |||||
| Terminase | 933W Major | 933W Mterm 5′ | ATGACATTCCGGAAGAAT | Term1 | 56.7 | 51.0 | 1,272 | This study |
| Terminase | 933W MTerm 3′ | TCAGTGAGCCATGCAGTG | 62.5 | This study | ||||
| 933W Minor | 933W 5′ MinTerm | ATGGCAAAGCTGGACTGG | Term2 | 63.9 | 64.9 | 807 | This study | |
| Terminase | 933W MinTerm 3′ | TCATTCTTCCGGAATGTCAT | 61.9 | This study |
Phage Accession numbers: BP 4795, NC_004813; 933W, NC_000924; partial sequence H19B, AF034975; Stx II bacteriophage I, NC_003525; HK97, NC_002167; Lambda, NC_001416; Nil 2, AJ413274; Stx II bacteriophage II, NC_004914; VT2-Sa, NC_000902; HK620, NC_002730; D3112, NC_005178; P27, NC_003356; phi 105, NC_004167; ST64T, NC_004348; Sakai, NC_002695; P22, NC_002371; ST104, NC_005841; SfV, NC_003444; N15, NC_001901; EcS1650, NC_002695. All CP933 sequences are from EDL 933, NC_002655; T7, NC_001604; HK022, NC_002166; Phi 80, X04051.
Tm, thermal denaturation midpoint temperature. Data are presented only for the primers designed for this study.
STEC strains (463 isolates) were previously obtained as part of a long-term study that examined both horizontal transmission (44, 45) and longitudinal carriage (26) of STEC on several farms in Cheshire, United Kingdom. All STEC strains had been typed with respect to their carriage of stx1 or stx2 genes. These strains were cultured in Luria broth (optical density at 600 nm of 0.45 to 0.55) and treated with norfloxacin (1 μg ml−1) at 37°C for 1 h to induce endogenous prophages (20, 22, 28). The cultures were diluted 10-fold in phage buffer (PB) (Luria broth supplemented with 10 mM CaCl2) and allowed to recover at 37°C for 2 h. The culture liquor was serially diluted and subjected to plaque assay on mid-exponential phase cultures of indicator host strain E. coli WG5rif+ (optical density at 600 nm of 0.5 to 0.6) grown in PB at 37°C with shaking (at 200 rpm). WG5rif+ is a rifampin-resistant derivative of the E. coli strain WG5 (17), an E. coli C strain that possesses an attenuated host restriction modification system and provides increased phage sensitivity (29). WG5rif+ was created through passage in increasing concentrations of rifampin (5 to 500 μg ml−1) (22), and its identity was confirmed by pulsed-field gel electrophoresis. Indicator host (100 μl) was incubated at 37°C with 450 μl of serially diluted broth from the norfloxacin-treated STEC cultures. After 25 min, 5 ml of PB with 0.4% (wt/vol) Difco agar and 300 μg ml−1 rifampin was added to the infection mixture and poured onto PB with 1.5% (wt/vol) Difco agar. The plates were then incubated overnight at 37°C and examined for plaques. A single semiconfluent plaque plate was flooded with PB (5 ml) and incubated for 4 h at 4°C to enable the phage to diffuse from the top agar into the buffer. The resultant phage preparation was used as a template in a stx-targeted PCR. Naïve WG5rif+ was also used as a control in DNA amplifications to rule out the identification of remnant prophage genes from across its genome and to serve as an additional control for the carryover of host DNA.
The presence of Stx phages in the preparations was determined using a novel primer set (Table 1) capable of amplifying all known stx genes (2). Of the 463 STEC isolates screened in this manner, it was possible to detect inducible phage in 89% of the isolates and stx genes in 101 of the phage preparations, indicating a 22% carriage rate of inducible Stx phages in the screened STEC population. Of these 101 strains that possessed inducible Stx phages, a subset of 70 STEC strains was chosen to produce the validation of the multilocus typing strategy. The excluded 31 STEC isolates possessed inducible Stx phages that either were not possible to reproducibly induce or were labile upon overnight storage at 4°C, a general problem that has been reported before (31).
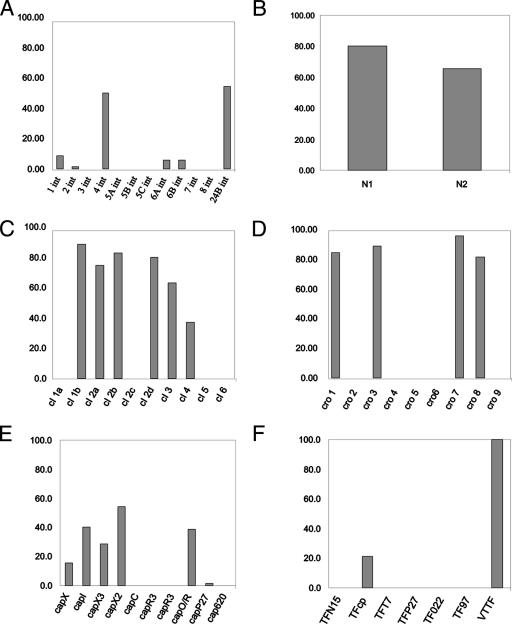
When this collection of 70 STEC strains was screened with the primers comprising the multilocus typing scheme, ∼50% of the phage preparations possessed an int gene identical to that carried by the sequenced Stx phage 933W (34) (Fig. 2A), ∼54% of the induced phage preparations possessed an int gene with homology to the gene associated with the Stx phage Φ24B (3, 16, 46), and 19% possessed both int genes. The cro gene, a key element in the regulation of lysis and lysogeny, required nine oligonucleotide primer pairs (Table 1) to cover the known genetic variation among phages. Four cro gene types (cro 1, cro 3, cro 8, and cro 9) dominated those identified in the phage preparations (Fig. 2D). Two of these loci (cro 1 and cro 3) have been described in other Stx phages and STEC strains and were identified here in >80% of the phage preparations. The cro gene type cro 7, which was identified in the highest number of phage preparations (96%), has been previously identified in the bacteriophage SfV from Shigella flexneri (1). A single oligonucleotide primer pair was able to amplify the cII gene in 90% of the phage preparations, while a single cIII-specific oligonucleotide primer pair (Table 1), possessing no degeneracies, amplified a product from all of the phage preparations. The gene responsible for encoding the bacteriophage repressor cI exhibits high levels of heterogeneity among lambdoid phages (Table 1) compared to other lambdoid regulatory genes such as cIII, cII, and Q. Ten primer sets were required to cover the known cI sequence diversity, but only six of these amplified products from the 70 induced phage preparations, indicating the presence of cI genes 1b, 2a, 2b, 2d, 3, and 4 (Fig. 2C). The cI gene 1b was detected most frequently (∼89%) and has been reported previously in the genomes of Stx phages 933W and VT2-Sa. The genes encoding the antiterminators involved in controlling the expression of either the early (N) or late (Q) genes were also examined. The N gene was readily identifiable by two primer sets yielding one of two products, N1 or N2, amplified in 80% and 66% of the samples, respectively (Fig. 2B), and both have been previously identified in Stx phages. The Q gene was detected by the use of one oligonucleotide primer pair, and this amplified the Q gene in all phage preparations.
FIG. 2.
Distribution of PCR targeted genes in phage preparations induced from 70 STEC strains. Panels present the percentage distributions of various gene types: int genes (A), N genes (B), cI genes (C), cro genes (D), capsid genes (E), and host recognition proteins (tail spikes/fibers) (F).
Phage structural genes were also analyzed. The capsid genes detected in the phage preparations had all been previously identified in prophages carried by the STEC isolate EDL933 (36) or Stx phage P27 (42); these included capX (present in 14% of phage preparations), capI (40%), capO/R (39%), and capP27 (∼2%) (Fig. 2E). The terminase genes associated with 933W, Term1 and Term2 (Table 1), involved in the packaging of DNA into the head, were present in 100 and 93% of the phage preparations, respectively. The bacteriophage host recognition factors (tail spike protein and/or tail fibers) described thus far for Stx phages were used to design oligonucleotide primer pairs (Table 1). These include the tail spike proteins from short-tailed bacteriophages (Podoviridae) such as 933W (37) and Φ24B (3, 22); long, noncontractile-tailed bacteriophages (Siphoviridae), e.g., H19B (33); and bacteriophages with complex contractile tails (Myoviridae) like P27 (42). DNA amplification using all of these primer pairs (Table 1) indicated that 100% of the samples were positive for the tail spike protein gene, previously described (47) as associated with the short-tailed Stx phages 933W and Φ24B (Fig. 2F). It was also possible to amplify sequences homologous to the tail spike protein gene of a remnant, long-tailed prophage (EcS1650) found within the Sakai O157:H7 genome; this gene was present in 21% of phage preparations (Fig. 2F). The high detection frequency of tail spike proteins associated with short-tailed Stx phages in the inducible phage preparations was not predicted, as there is considerable diversity in the tail fiber, base plate, or tail spike genes of coliphages, i.e., HK97, HK022 (25), and other phages such as N15 (41). The diversity of phage tails and their specific host recognition proteins has also been reported throughout the population of bacteriophages infecting the Mycobacteriaceae (18). Identification here of conservation of the tail spike protein in the inducible phages from STEC strains suggests selection pressure exerted by the conserved E. coli surface receptor for these phages (47).
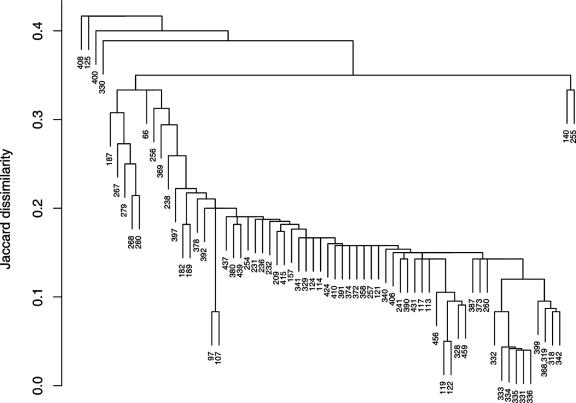
A dendrogram (Fig. 3) was produced to provide an indication of the level of heterogeneity identified in the induced phage preparations from the STEC strains. The Jaccard dissimilarity (12) between the inducible phage preparations from each pair of 70 strains was calculated using the presence and absence of genes in the multilocus characterization scheme. For a pair of strains (i, j), the Jaccard dissimilarity is the proportion of genes present in i and/or j that is not present in both i and j. Thus, the shared absence of a gene in two strains is not treated as indicating similarity between them, but the shared presence of a gene is scored. The dendrogram (Fig. 3.) was constructed using single-linkage hierarchical clustering (12), and the dissimilarity between a pair of strains is represented by the height at which they join a single group. R, version 2.4.0 (40), was used for these analyses and illustrates differences between the genetic profiles of these inducible phages for each STEC strain. The patterns are indicative of a fluid gene pool and also highlight the sensitivity of the multilocus characterization scheme to identify, quickly and accurately, differences among the inducible phages from each STEC strain. This analysis shows that only two STEC strains exhibited similar inducible prophage profiles (Fig. 3), demonstrating a remarkably fluidic population of inducible phage genes.
FIG. 3.
Jaccard dissimilarity dendrogram generated from data on the presence or absence of genes in each of the induced phage preparations from 70 STEC isolates. The numbers represent the laboratory identification of each phage preparation obtained from a single STEC isolate. A value of 0.1 indicates 10% shared genes.
The detection sensitivity of phage genes was assessed by PCR amplification of the Q gene from serial dilutions of a purified Φ24B phage preparation. The data (not shown) demonstrated that PCR was capable of detecting the presence of a single PFU in a reaction mixture, providing the same sensitivity of a more conventional plaque assay. It has also been reported in the literature that STEC strains harboring more than one phage upon induction do not necessarily produce equal numbers of the different phages (3, 43, 51); therefore, the overall sensitivity of our amplification reaction, using 4 μl of phage preparation, relies on the presence of 250 phage particles ml−1 (per phage genotype). Since induction rates of wild-type phages have been previously reported to be in the range of 5 × 104 to 2 × 109 ml−1 of STEC culture following induction (3), the technique reported here should be sufficiently sensitive to characterize inducible phages directly from a lysogen, even if they cannot be further propagated on laboratory host strains. Although a bacterial lysogen usually possesses multiple phage-related segments of DNA in its genome, often only one or two infectious phage types can be detected following induction/activation of the SOS response (3, 43, 53), so even though each of the phage preparations from this study is unlikely to consist of one pure phage, there will probably not be more than a few phages. This multilocus characterization scheme also has the potential to profile remnant bacteriophages within the bacterial host genome if patterns of inducible phage genes are compared to patterns of genes amplified from the lysogen directly. This approach to phage typing has advantages over DNA amplification typing techniques such as amplified fragment length polymorphism (50) or repetitive extragenic palindromic PCR (49). Amplified fragment length polymorphism (50) might provide quite diverse lengths of amplified products due to the levels of recombination among Stx and other lambdoid phages, and the levels of recombination might also mask identification of phages that are carrying similar or identical genes. Acquisition or loss of genes flanking the amplification target will obscure the presence of genes that control the biology of phage-host interactions. Bioinformatic analyses of the annotated genome sequences for 933W (NC_000924), Stx II bacteriophage II (NC>004914), VT2-Sa (NC_000902), P27 (NC_003356), and the raw genome sequence of Φ24B did not reveal complete repetitive extragenic palindromic or enterobacterial repetitive intergenic consensus sequences (49) as would be expected for DNA regions of ∼60 kb. The primer sets presented in this paper were designed from sequences that were first divided into clades and then subsequently checked for specificity; amplification products are therefore indicative of a specific type of gene and do not routinely require sequencing for identification of the amplification product. This level of gene identification has previously been possible only by gross sequencing and sequence alignment data. The PCR-based typing scheme described here will not distinguish single nucleotide polymorphisms but can identify an insertion or deletion encompassing a stretch of nucleotides, as has been reported for the tail spike protein (47). Not all of the primer sets amplified genes from the pool of 70 phage preparations, so these genes may be poorly represented or not present in the general Stx phage population. Therefore, the subset of primers that have amplified genes in the 70 induced preparations used here could represent the progenitor of a routinely applicable Stx phage scheme.
In conclusion, we report the use of a novel multilocus characterization scheme to simultaneously characterize the inducible phage preparations from a variety of farm environment-derived STEC lysogens. These data do not allow specific identification of the individual phages harvested from STEC strains, but they do enable description of the gene pool present in inducible phage preparations from any single bacterial lysogen. One aim is to identify prevalent phage genes that can be induced from STEC, thereby making it possible to identify the genetic background of inducible and/or noninducible prophages from clinically relevant or environmentally circulating STEC strains. Such data would highlight Stx phage-borne traits associated with clinical disease and/or particular farm environments and possibly identify more stable Stx phages that are likely to persist and disseminate stx genes. Analyses of these data by Jaccard dissimilarity (12) or other matrices together with a knowledge of the Stx phage-borne traits could identify phage profiles important in the transmission of stx or enable tracking of genes across sample study sites in order to gauge the rate of dissemination of a gene by bacteriophages. This multilocus characterization scheme can be further developed. Most Stx phages have larger genomes than the archetypal λ; 933W is 21% larger (2), and most of the additional genes have no designated function. Some of this additional DNA may influence the biology of the E. coli lysogen (e.g., fitness in the mammalian gut or colonization potential) and, therefore, be subject to positive selection (2, 9). As additional factors are identified, it will be possible to augment this multilocus typing scheme to generate further information on the genotypes of Stx phages and their epidemiological significance.
Acknowledgments
This research was funded by the Department for Environment, Food and Rural Affairs, United Kingdom, and the Biotechnology and Biological Sciences Research Council, United Kingdom.
We also acknowledge the free donation of bacteriophages and lysogens from David Friedman, University of Minnesota (EDL933); Graham Hatfull, University of Pittsburgh (HK620, N15); Eric Oswald, Ecole Nationale Vétérinaire de Toulouse, France (Sakai strain); Herbert Schmidt, Universität Hohenheim, Germany (P27); and Pauline Wang, University of Toronto (D3112). We are grateful to the staff of the Veterinary Science Department, University of Liverpool, for providing the collection of STEC isolates.
Footnotes
Published ahead of print on 19 October 2007.
REFERENCES
- 1.Allison, G. E., D. Angeles, N. Tran-Dinh, and N. K. Verma. 2002. Complete genomic sequence of SfV, a serotype-converting temperate bacteriophage of Shigella flexneri. J. Bacteriol. 184:1974-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, H. E. 2007. Stx-phages: drivers and mediators of the evolution of STEC and STEC-like pathogens. Future Microbiol. 2:165-174. [DOI] [PubMed] [Google Scholar]
- 3.Allison, H. E., M. J. Sergeant, C. E. James, J. R. Saunders, D. L. Smith, R. J. Sharp, T. S. Marks, and A. J. McCarthy. 2003. Immunity profiles of wild-type and recombinant Shiga-like toxin-encoding bacteriophages and characterization of novel double lysogens. Infect. Immun. 71:3409-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Balding, C., S. A. Bromley, R. W. Pickup, and J. R. Saunders. 2005. Diversity of phage integrases in Enterobacteriaceae: development of markers for environmental analysis of temperate phages. Environ. Microbiol. 7:1558-1567. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Brussow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, A. 1994. Comparative molecular biology of lambdoid phages. Annu. Rev. Microbiol. 48:193-222. [DOI] [PubMed] [Google Scholar]
- 9.Canchaya, C., G. Fournous, and H. Brussow. 2004. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 53:9-18. [DOI] [PubMed] [Google Scholar]
- 10.Casas, V., J. Miyake, H. Balsley, J. Roark, S. Telles, S. Leeds, I. Zurita, M. Breitbart, D. Bartlett, F. Azam, and F. Rohwer. 2006. Widespread occurrence of phage-encoded exotoxin genes in terrestrial and aquatic environments in southern California. FEMS Microbiol. Lett. 261:141-149. [DOI] [PubMed] [Google Scholar]
- 11.Casjens, S. R. 2005. Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 8:451-458. [DOI] [PubMed] [Google Scholar]
- 12.Chatfield, C., and A. J. Collins. 1980. Introduction to multivariate analysis. Chapman and Hall, London, United Kingdom.
- 13.Copeland, N. G., N. A. Jenkins, and D. L. Court. 2001. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2:769-779. [DOI] [PubMed] [Google Scholar]
- 14.Dobrindt, U. 2005. (Patho-)genomics of Escherichia coli. Int. J. Med. Microbiol. 295:357-371. [DOI] [PubMed] [Google Scholar]
- 15.Dundas, S., W. T. Todd, A. I. Stewart, P. S. Murdoch, A. K. Chaudhuri, and S. J. Hutchinson. 2001. The central Scotland Escherichia coli O157:H7 outbreak: risk factors for the hemolytic uremic syndrome and death among hospitalized patients. Clin. Infect. Dis. 33:923-931. [DOI] [PubMed] [Google Scholar]
- 16.Fogg, P. C. M., S. M. Gossage, D. L. Smith, J. R. Saunders, A. J. McCarthy, and H. E. Allison. 2007. Identification of multiple integration sites for Stx phage Φ24B in the E. coli genome, description of a novel integrase and evidence for a functional anti-repressor. Microbiology. doi: 10.1099/mic.0.2007/011205. [DOI] [PubMed]
- 17.Grabow, W. O., and P. Coubrough. 1986. Practical direct plaque assay for coliphages in 100-ml samples of drinking water. Appl. Environ. Microbiol. 52:430-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatfull, G. F., M. L. Pedulla, D. Jacobs-Sera, P. M. Cichon, A. Foley, M. E. Ford, R. M. Gonda, J. M. Houtz, A. J. Hryckowian, V. A. Kelchner, S. Namburi, K. V. Pajcini, M. G. Popovich, D. T. Schleicher, B. Z. Simanek, A. L. Smith, G. M. Zdanowicz, V. Kumar, C. L. Peebles, W. R. Jacobs, Jr., J. G. Lawrence, and R. W. Hendrix. 2006. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet. 2:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 20.Herold, S., J. Siebert, A. Huber, and H. Schmidt. 2005. Global expression of prophage genes in Escherichia coli O157:H7 strain EDL933 in response to norfloxacin. Antimicrob. Agents Chemother. 49:931-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 22.James, C. E., K. N. Stanley, H. E. Allison, H. J. Flint, C. S. Stewart, R. J. Sharp, J. R. Saunders, and A. J. McCarthy. 2001. Lytic and lysogenic infection of diverse Escherichia coli and Shigella strains with a verocytotoxigenic bacteriophage. Appl. Environ. Microbiol. 67:4335-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johannessen, G. S., C. E. James, H. E. Allison, D. L. Smith, J. R. Saunders, and A. J. McCarthy. 2005. Survival of a Shiga toxin-encoding bacteriophage in a compost model. FEMS Microbiol. Lett. 245:369-375. [DOI] [PubMed] [Google Scholar]
- 24.Johansen, B. K., Y. Wasteson, P. E. Granum, and S. Brynestad. 2001. Mosaic structure of Shiga-toxin-2-encoding phages isolated from Escherichia coli O157:H7 indicates frequent gene exchange between lambdoid phage genomes. Microbiology 147:1929-1936. [DOI] [PubMed] [Google Scholar]
- 25.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 26.Kemp, R. 2005. The epidemiology of VTEC O157, non-O157 VTEC and Campylobacter spp. in a 100 km2 dairy farming area in northwest England. University of Liverpool, Liverpool, United Kingdom.
- 27.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsushiro, A., K. Sato, H. Miyamoto, T. Yamamura, and T. Honda. 1999. Induction of prophages of enterohemorrhagic Escherichia coli O157:H7 with norfloxacin. J. Bacteriol. 181:2257-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muniesa, M., and J. Jofre. 2004. Factors influencing the replication of somatic coliphages in the water environment. Antonie Leeuwenhoek 86:65-76. [DOI] [PubMed] [Google Scholar]
- 30.Muniesa, M., F. Lucena, and J. Jofre. 1999. Comparative survival of free Shiga toxin 2-encoding phages and Escherichia coli strains outside the gut. Appl. Environ. Microbiol. 65:5615-5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muniesa, M., R. Serra-Moreno, and J. Jofre. 2004. Free Shiga toxin bacteriophages isolated from sewage showed diversity although the stx genes appeared conserved. Environ. Microbiol. 6:716-725. [DOI] [PubMed] [Google Scholar]
- 32.Murphy, F., C. Fauquet, D. Bishop, S. Ghabrial, A. Jarvis, G. Martelli, M. Mayo, and M. Summers. 1995. Virus taxonomy: the classification and nomenclature of viruses. The sixth report of the International Committee on Taxonomy of Viruses. Springer-Verlag, Vienna, Austria.
- 33.Neely, M. N., and D. I. Friedman. 1998. Arrangement and functional identification of genes in the regulatory region of lambdoid phage H-19B, a carrier of a Shiga-like toxin. Gene 223:105-113. [DOI] [PubMed] [Google Scholar]
- 34.O'Brien, A. D., and G. D. LaVeck. 1983. Purification and characterization of a Shigella dysenteriae 1-like toxin produced by Escherichia coli. Infect. Immun. 40:675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 36.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 37.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pringle, C. R. 1998. The universal system of virus taxonomy of the International Committee on Virus Taxonomy (ICTV), including new proposals ratified since publication of the Sixth ICTV Report in 1995. Arch. Virol. 143:203-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pringle, C. R. 1999. Virus taxonomy at the XIth International Congress of Virology, Sydney, Australia, 1999. Arch. Virol. 144:2065-2070. [DOI] [PubMed] [Google Scholar]
- 40.R Development Core Team. 2006. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 41.Ravin, V., N. Ravin, S. Casjens, M. E. Ford, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 299:53-73. [DOI] [PubMed] [Google Scholar]
- 42.Recktenwald, J., and H. Schmidt. 2002. The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage φP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect. Immun. 70:1896-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rietra, P. J., G. A. Willshaw, H. R. Smith, A. M. Field, S. M. Scotland, and B. Rowe. 1989. Comparison of Vero-cytotoxin-encoding phages from Escherichia coli of human and bovine origin. J. Gen. Microbiol. 135:2307-2318. [DOI] [PubMed] [Google Scholar]
- 44.Robinson, S. E., P. E. Brown, E. John Wright, M. Bennett, C. A. Hart, and N. P. French. 2005. Heterogeneous distributions of Escherichia coli O157 within naturally infected bovine faecal pats. FEMS Microbiol. Lett. 244:291-296. [DOI] [PubMed] [Google Scholar]
- 45.Robinson, S. E., E. J. Wright, C. A. Hart, M. Bennett, and N. P. French. 2004. Intermittent and persistent shedding of Escherichia coli O157 in cohorts of naturally infected calves. J. Appl. Microbiol. 97:1045-1053. [DOI] [PubMed] [Google Scholar]
- 46.Saunders, J. R., H. Allison, C. E. James, A. J. McCarthy, and R. Sharp. 2001. Phage-mediated transfer of virulence genes. J. Chem. Technol. Biotechnol. 76:1-5. [Google Scholar]
- 47.Smith, D. L., C. E. James, M. J. Sergeant, Y. Yaxian, J. R. Saunders, A. J. McCarthy, and H. E. Allison. 2007. Short-tailed Stx phages exploit the conserved YaeT protein to disseminate Shiga toxin genes amongst enterobacteria. J. Bacteriol. 189:7223-7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, et al. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner, P. L., D. W. K. Acheson, and M. K. Waldor. 1999. Isogenic lysogens of diverse Shiga toxin 2-encoding bacteriophages produce markedly different amounts of Shiga toxin. Infect. Immun. 67:6710-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, J., M. Sarov, J. Rientjes, J. Fu, H. Hollak, H. Kranz, W. Xie, A. F. Stewart, and Y. Zhang. 2006. An improved recombineering approach by adding RecA to lambda Red recombination. Mol. Biotechnol. 32:43-53. [DOI] [PubMed] [Google Scholar]
- 53.Willshaw, G. A., H. R. Smith, S. M. Scotland, A. M. Field, and B. Rowe. 1987. Heterogeneity of Escherichia coli phages encoding Vero cytotoxins: comparison of cloned sequences determining VT1 and VT2 and development of specific gene probes. J. Gen. Microbiol. 133:1309-1317. [DOI] [PubMed] [Google Scholar]