Abstract
The 2.1-kb cryptic plasmid pCIBAO89 from Bifidobacterium asteroides harbors a 1.4-kb segment which is sufficient for its autonomous replication. The segment is divided into two parts, the presumed replication origin, ori89, and the rep gene encoding the putative 41-kDa Rep89 replication initiation protein. This minimal replication region of pCIBAO89 was functionally dissected by transcriptional analyses as well as by DNA-binding studies, and the information obtained was exploited to create a number of Escherichia coli-Bifidobacterium shuttle vectors capable of transforming various bifidobacteria with an efficiency of up to 106 transformants/μg DNA.
Members of the genus Bifidobacterium are gram-positive, obligate anaerobic, non-spore-forming, nonmotile, rod-shaped bacteria with spatulate extremities (53). Their genomes have a relatively high GC content with values ranging from 55% to 67%. Furthermore, bifidobacteria are saccharolytic organisms, fermenting various hexoses exclusively by the fructose-6-phosphate pathway or by bifid shunt (44).
Currently the genus, which belongs to the phylum Actinomycetaceae of high-GC-content gram-positive bacteria (45), includes 34 species (58) which are mainly classified on the basis of their ecological origins and many of which have been isolated from fecal samples (for a review, see reference 59). Bifidobacteria, first described by Tissier (56), are widely used as probiotics. The concept of probiotic bacteria has evolved from that of a live culture that maintains a balanced and healthy gut microflora environment to one where specific beneficial effects are attributed to the microorganism (16). Particular therapeutic properties have been attributed to various strains of the Bifidobacterium genus (35), including prevention of enteric infection, reduction of serum cholesterol levels, immunostimulation, anticarcinogenic properties, alleviation of constipation, and treatment of lactose intolerance.
Despite these described beneficial properties and the widely growing commercial and consumer interest in probiotic members of this genus, bifidobacteria still remain poorly understood. The public release of the first complete genome sequence of a Bifidobacterium longum strain (46), along with ongoing efforts to sequence additional strains, has significantly contributed to advancing our knowledge of bifidobacterial genetics and metabolism. Such information will be a first step in providing a better understanding of the mode of action of a specific probiotic property. This type of analysis is currently hampered by a paucity of effective molecular tools such as cloning and expression vectors and plasmid-based gene disruption systems.
Plasmids in members of the genus Bifidobacterium were initially reported by Sgorbati et al. (48). Since then plasmids have been reported to have been found in six different species of bifidobacteria. To date, 12 plasmids from B. longum have been completely sequenced: pMB1 (40), pKJ36 (32), pKJ50 (33), pBLO1 (46), pNAC1, pNAC2, and pNAC3 (5), pTB6 (52), pDOJH10L and pDOJH10S (21), pNAL8 (12), and pB44 (GenBank accession no. NC004443). Furthermore, plasmids from other Bifidobacterium species have been sequenced as well: pVS809 from B. globosum (24), pCIBb1 (31) and pNBb1 (GenBank accession no. E17316) from B. breve, pBI from B. indicum (48), pBC1 from B. catenulatum (2), pAP1 from B. asteroides (GenBank accession no. Y11549), pASV479 (43) from B. pseudolongum subsp. globosum, and p4M from B. pseudocatenulatum (GenBank accession no. NC003527). The biological significance of these mostly small plasmids is unclear, as most of the open reading frames (ORF), apart from those that are predicted to specify replication functions, code for proteins of unknown function (5). An exception to this appears to be an as-yet-unsequenced B. bifidum plasmid that was reported to be responsible for the production of a bacteriocin named bifidocin B (63). Significant efforts have been made to exploit these native replicons for the construction of Escherichia coli-Bifidobacterium shuttle vectors. Successful applications of such vectors include the development of pMDY23, a reporter plasmid based on a B. longum cryptic plasmid and the E. coli gusA gene (18); expression of recombinant amylase (38); murine mucosal immunization with respect to Salmonella spp. through flagellin expression (51), and various tumor suppressor studies (8, 10, 28, 60, 61, 62). A hindrance to the widespread application of recombinant gene expression by Bifidobacterium-based shuttle vectors is the low transformation efficiency (3, 21, 39) complicated in some cases by segregational instability (8). In this study two B. asteroides strains were found to contain plasmids which, upon sequencing, were shown to be virtually identical. One of these plasmids (designated pCIBA089) was selected for analysis of its replication functions and for construction of E. coli-Bifidobacterium shuttle vectors.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Bifidobacteria were routinely cultured anaerobically at 37°C in either de Man, Rogosa and Sharpe medium (MRS; Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) (6) supplemented with 0.05% (wt/vol) cysteine-HCl or reinforced clostridial medium (Oxoid). Anaerobic conditions were maintained using a Merck Anaerocult oxygen-depleting system (Merck, Darmstadt, Germany). E. coli strains were cultivated in Luria-Bertani medium (27). Lactococcus lactis cultures were cultivated in M17 medium (Oxoid Ltd.) supplemented with 0.5% (wt/vol) glucose (GM17). Where necessary, antibiotics were added to the growth medium at the following final concentrations: for E. coli, ampicillin (Amp; 100 μg ml−1), chloramphenicol (Cm; 20 μg mg−1), and erythromycin (Em; 300 μg ml−1); for Bifidobacterium spp., Cm (5 μg ml−1) and Em (2 μg ml−1); and for L. lactis, Cm (5 μg ml−1).
TABLE 1.
Bacterial strains and plasmids used in this studya
| Bacterial strain or plasmid | Relevant properties | Source/reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL1-Blue | thi recA1 hsd17 gyrA94 αlacZ | Stratagene Ltd. |
| TOP10 | F−mcrA ϕ80lacZΔM15 ΔlacX74 recA1 araD galU rpsL endA1 nupG | Invitrogen |
| Bifidobacterium | ||
| B. asteroides DSM20089 | Source of pCIBA089 | Hindgut of honeybee |
| B. asteroides DSM20431 | Source of pCIBA431 | Hindgut of honeybee |
| B. animalis subsp. lactis Bb12 | C. Hansen, Hørsholm, Denmark | |
| B. breve UCC2003 | Electroporation host | UCC collection |
| B. dentium NCFB2843 | NCFB | |
| B. longum NCIMB8809 | UCC collection | |
| B. pseudocatenulatum LMG10505 | BCCM | |
| B. pseudolongum NCIMB2244 | UCC collection | |
| B. globosum JCM5820 | JCM | |
| Lactococcus | ||
| Lactococcus lactis NZ9000 | MG1363 pepN nisRK | 19 |
| Plasmids | ||
| pCR2.1-TOPO | 3.9-kb E. coli cloning vector; Ampr | Invitrogen |
| pBluescript KS(−) | 2.9-kb E. coli cloning vector; Ampr | Stratagene Ltd. |
| pNZ272 | 4.6 kb; promoterless gusA for promoter probe vector analysis; Cmr | 36 |
| pGC series | Set of promoter probe constructs based on pNZ272; Cmr | This study |
| pCIBA089 | Native 2.1-kb plasmid | B. asteroides |
| pCIBA431 | Native 2.1-kb plasmid | B. asteroides |
| pUC7C | 3.6-kb E. coli plasmid; Ampr, Cmr | 22 |
| pNZ8048 | 3 kb; nisin-inducible L. lactis expression vector; Cmr | 19 |
| pNZ44 | pNZ8048 containing constitutive P44 promoter from L. lactis | 26 |
| pNZrep89 | pNZ44 containing rep89 in sense orientation | This study |
| pBlueCm | 4.1 kb; Cmr cassette from pUC7C in BamHI site of pBluescript KS(−) | This study |
| pPKCm series | Set of E. coli-Bifidobacterium shuttle vectors based on pBlueCm | This study |
| pErythromycin | 2.7 kb; 1.6-kb PCR fragment of pBluescript KS(−), Emr gene of pGKV210 | This study |
| pSKEm | 4.4-kb E. coli-Bifidobacterium shuttle vector derived from pErythromycin | This study |
DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany; UCC, University College Cork, Cork, Ireland; BCCM, Belgian Coordinated Collections of Microorganisms, Universiteit Gent, Gent, Belgium; JCM, Japan Collection of Microorganisms, RIKEN BioResource Center, Wako, Japan; NCFB, National Collection of Food Bacteria, Reading, United Kingdom.
DNA isolation procedures.
E. coli plasmid DNA was routinely isolated using a QIAprep Spin plasmid Miniprep kit (Qiagen, Santa Clara, CA) according to the manufacturer's instructions. Bifidobacterial plasmid DNA was also isolated using this kit with the adaptation that buffer P1 was supplemented with 40 mg ml−1 lysozyme and, when used, was incubated at 37°C for 45 min. Small-scale total bifidobacterial DNA preparations were obtained as described by Mazé et al. (25).
DNA manipulations, sequence analysis, and electroporation.
Plasmids and PCR products were purified using a Qiagen rapid PCR purification kit (Qiagen). Restriction endonucleases, alkaline phosphatase, and T4 DNA ligase were used according to the manufacturer's instructions (Roche Diagnostic GmbH, Mannheim, Germany). Standard PCRs were carried out using Taq PCR MasterMix (Qiagen), while high-fidelity PCR was carried out using KOD polymerase (Novagen, Darmstadt, Germany). Oligonucleotide primers used in this study are listed in Table S1 in the supplemental material. DNA sequencing was performed by MWG-Biotech (Ebersberg, Germany). All sequence data assembly and analysis were performed using a DNASTAR software package (DNASTAR Inc., Madison, WI). Phylogenetic and multiple sequence alignment analyses were conducted using MEGA version 3.1 (20). Database searches were performed using BLAST (1) located at the National Center for Biotechnology Information web facility (http://www.ncbi.nlm.nih.gov). Electrotransformation of E. coli and L. lactis was performed as previously described by Sambrook et al. (42) using a Bio-Rad Gene Pulser. Electrotransformations of bifidobacteria were performed as described previously by MacConaill et al. (23).
Plasmid constructions.
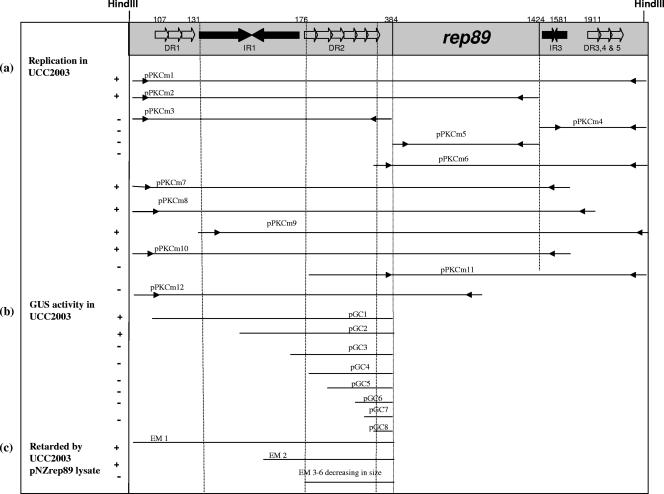
Plasmid pBlueCm (4.1 kb) was constructed by excising the chloramphenicol resistance cassette from pUC7C and cloning this into the BamHI site of pBluescript KS(−). The HindIII-linearized 2.1-kb pCIBA089 plasmid was cloned into the HindIII site of pBlueCm, resulting in pPKCm1 (Fig. 1a and Fig. 2). The vector pErythromycin (2.7 kb) was generated by ligation of a 1.6-kb fragment of pBluescript KS(−) (containing the E. coli ColE1 origin of replication, the intact pBluescript multiple cloning site [MCS], and the αlacZ gene) to the Em resistance (Emr) gene from pGKV210. The construction of pSKEm (Fig. 1b) was achieved by inserting a 1.7-kb PCR-amplified fragment of pCIBA089 (primers pCIBFA and pCIBRA) into the SphI site of pErythromycin. Eight different fragments covering various parts of the promoter region upstream of the replication protein-encoding gene from pCIBAO89 were generated by PCR (using one of eight forward primers [GusF1 through GusF8] in combination with GusRev). The resultant amplicons were digested with BglII and PstI and ligated into similarly restricted pNZ272 to generate plasmids pGC1, pGC2, pGC3, pGC4, pGC5, pGC6, pGC7, and pGC8 by use of L. lactis NZ9000 as the cloning host (Fig. 2). To create plasmid pNZrep89, the rep89 gene was amplified as an AflIII/HindIII fragment (primers rep44f and rep44r) and ligated into the NcoI/HindIII sites of the L. lactis pNZ44 vector (26). Integrity of all plasmids was verified by restriction analysis and subsequent DNA sequencing. In order to determine the minimum region of pCIBA089 necessary to support replication in Bifidobacterium spp., various segments of pCIBA089 were PCR amplified as HindIII fragments and ligated into pBlueCm (the cloned sections of pCIBA089 and their corresponding names are displayed in Fig. 2a). These constructs, designated pPKCm1 through pPKCm12, were subsequently introduced into B. breve UCC2003 by electroporation, and constructs with the ability to replicate in UCC2003 were identified by their capacity to confer chloramphenicol-resistance, which was subsequently verified by restriction analysis of plasmid DNA isolated from randomly selected transformants.
FIG. 1.
(a) Schematic representation of pPKCm. Ampr, ampicillin resistance gene. Cmr, chloramphenicol resistance cassette from pUC7C. (b) Schematic representation of pSKEm. Emr, erythromycin resistance gene. pBluescript, pBluescript KS(−). MCS / LacZ, MCS and αlacZ gene.
FIG. 2.
Structural features of pCIBA089. (a) Schematic representation of deletion analysis of pCIBAO89; black arrows indicate primers used for PCR amplification (see Table S1 in the supplemental material), rep89 represents the rep89 ORF, + indicates the fragments that support plasmid replication in B. breve UCC2003, and − indicates the fragments that do not support plasmid replication in UCC2003. (b) GUS fragments generated for cloning into the reporter vector pNZ272. + indicates GUS activity generated in B. breve UCC2003, and − indicates that the GUS activity generated in UCC2003 was at same level as that of the control vector. (c) Fragments generated for EMSA analysis. + indicates fragments whose mobility was retarded by B. breve UCC2003 pNZrep89 lysate, and − indicates fragment whose mobility was not retarded by B. breve UCC2003 pNZrep89 lysate. Note: diagram is not to scale; IR2 results are not shown.
Plasmid stability studies.
B. breve UCC2003 colonies containing individual pPKCm constructs were first cultured in MRS broth containing 4 μg ml−1 Cm. Cells were then subcultured in fresh MRS broth without antibiotic selection every 12 h for a total of 200 generations. Vector segregation stability was monitored by plating for isolated colonies every 25 generations, spot inoculating 100 colonies onto reinforced clostridial agar plates with and without 4 μg ml−1 Cm, and incubating at 37°C for 24 h. The percentage of loss of the test plasmid in the population was then calculated.
GUS assay.
Intracellular β-glucuronidase (β-GUS) activity was determined essentially as described by Sangrador-Vegas et al. (43).
Transcriptional start site analysis.
Total RNA was isolated from cells grown to an optical density at 600 nm (OD600) of 0.6 using the macaloid acid method as described by Ventura et al. (57) and then treated with DNase (Roche). A 10-μg volume of total RNA was used to determine the transcriptional start site for the gene carrying rep89 by use of a First Choice RNA ligase-mediated rapid amplification of cDNA ends (RACE) kit (Ambion, Cambridgeshire, United Kingdom) essentially as described by Han et al. (13) with the following adaptations. Reverse transcription for cDNA was mediated by thermostable Superscript reverse transcriptase (Invitrogen). The nested PCR contained 10 pmol of gene-specific outer primer pRLMout in combination with 10 pmol of 5′ RACE outer primer (Ambion). The 5′ inner PCR was carried out with 10 pmol of gene-specific inner primer pRLMin plus 10 pmol of 5′ RACE inner primer (Ambion; for primer sequences, see Table S1 in the supplemental material). PCR products were cloned in pCR2.1-TOPO vector (Invitrogen), and multiple clones were sequenced (MWG Biotech).
Gel mobility shift DNA binding assays.
Six DNA fragments were generated by PCR (using one of six forward primers [em1 to em6] in combination with emrev) covering decreasing sections of the pCIBAO89 promoter region. The resultant amplicons were purified and then labeled using [33P]dATP and T4 polynucleotide kinase (New England Biolabs). In order to obtain a crude lysate containing the presumed replication protein, B. breve UCC2003 containing pNZrep89 was inoculated (2%) in MRS supplemented with 4 μg ml−1 Cm, grown at 37°C until an OD600 of approximately 0.5 was reached, and then harvested by centrifugation. The cell pellet was resuspended in 500 μl of 20 mM Tris (pH 7.4) supplemented with 10 mM dithiothreitol, and this cell suspension was then bead beaten with glass beads and centrifuged twice at 7,000 rpm for 5 min. Binding studies were carried out using 20-μl reaction volumes containing 20 mM Tris-HCl (pH 8.0), 8.7% (vol/vol) glycerol, 1 mM EDTA (pH 8.0), 5 mM MgCl2, 100 mM KCl, 0.5 mM dithiothreitol, labeled DNA fragment (3,000 cpm), and increasing volumes of crude B. breve UCC2003 pNZrep89 cell lysate. As a negative control, cell lysates were used which had been obtained from B. breve UCC2003 harboring pNZ44. Bovine serum albumin (1 μg) and poly(dI-dC) (Amersham Pharmacia Biotech) were added to the reaction mixtures in order to increase the specificity of the binding reaction. After incubation for 30 min at 37°C, samples were loaded onto a 4% polyacrylamide gel. Electrophoresis was performed at 150 V for 2.5 h. Gels were dried and used for autoradiography at −80°C with Kodak XAR-5 film and intensifying screens.
Relative plasmid copy number determination.
Two oligonucleotide primer sets for quantitative real-time PCR (qPCR) were designed to have a predicted melting temperature of ∼56°C and to generate amplicons of ∼300 bp in length. One primer set was designed to target the replication gene of pCIBAO89 (primers pPlasF and pPlasR), while the second set of primers was based on the sequence of the unique amylopullulanase (apuB) gene (primer combination pAmuF and pAmuR) on the chromosome of B. breve UCC2003 (41). qPCRs were conducted using a FastStart DNA master SYBR green 1 kit (Roche). Serial dilutions (10−1 to 10−3) of total DNA isolated from B. breve UCC2003 containing pPKCm1, grown under conditions of chloramphenicol selection, were analyzed using 0.3 μM of the relevant forward and reverse primer pair. Three independent qPCR trials were conducted, and in each trial triplicate samples of the template were analyzed. Copy numbers were calculated essentially as described by Providenti et al. (37), where the mean threshold cycle value of the amplicons-targeting apuB gene (single-copy reference) was compared to the amplicon of the plasmid using the formula Nrelative = (1 + E)ΔCT, where E is the amplification efficiency of target and reference genes, and ΔCT is the difference between the threshold cycle number (CT) of the apuB reaction and that of the rep89 reaction.
Nucleotide sequence accession number.
The GenBank accession number for the sequences of pCIBAO89 is EU030683.
RESULTS
Identification and sequence analysis of two bifidobacterial plasmids.
From a total of 55 strains of bifidobacteria of human, animal, and insect origin, two strains were found to contain plasmid DNA (data not shown). The identified plasmids were designated pCIBA089 (from B. asteroides DSM20089) and pCIBA431 (from B. asteroides DSM20431). Sequence analysis of pCIBA089 and pCIBA431 revealed circular molecules of 2,111 bp with an overall GC content of 52.3%. Further analysis showed that these two plasmids are highly homologous to each other and to pAP1, a plasmid previously isolated from B. asteroides DSM20089 (49; GenBank accession no. Y11549) (>99% identity), but that at the same time they also exhibit some interesting differences. The most striking of these was the occurrence of an extra cytosine residue in position 1262 present in both pCIBA089 and pCIBA431 but absent from the deposited pAP1 sequence. This residue extends the size of the only identified ORF (designated rep in pAP1, rep89 in pCIBAO89, and rep31 in pCIBA431) in these plasmids from 894 nucleotides in pAP1 to 1,041 nucleotides in pCIBA089 and pCIBA431. The second observed difference was an imperfect repeated sequence of 31 nucleotides present in pAP1 but absent from pCIBA089 and pCIBA431. This region is located between positions 1470 and 1502 on pAP1, immediately downstream of rep.
A BLAST search (1) revealed that Rep89 (and therefore Rep31) exhibits various degrees of amino acid sequence similarity, ranging from 25 to 58%, to putative replication proteins identified on other plasmids, most of which were isolated from high-GC-content gram-positive bacteria (see Table S2 in the supplemental material). It shows the highest level of homology to predicted plasmid replication proteins from the B. longum RW041 plasmid pNAC3 (58% identity) (5) and the B. longum DJO10A plasmid pDOJH10L (53% identity to the repC gene) (21) as well as 35% identity to an as-yet-unidentified replicase from B. adolescentis ATCC 15703 (GenBank accession no. YP_910104). Excluding the putative bifidobacterial replication proteins, Rep89 also shows similarity to the replication proteins of a number of broad-host-range plasmids (detailed in Table S2 in the supplemental material). Therefore, the rep89 protein product is predicted to act as the plasmid replication protein.
The sequence upstream of the rep89 gene contains a number of inverted and direct repeats (DRs) (Fig. 2 and 3) which may represent features required for plasmid replication. Analysis of the sequence between positions 131 and 172 revealed the presence of an inverted repeat (IR), designated IR1 (ΔG = −22.5 kcal mol−1), with the potential to form a stem-loop structure. Two further inverted repeats with the potential to form stem-loops were also identified between positions 325 and 352 (designated IR2; ΔG = −9.2 kcal mol−1) and positions 1581 and 1602 (designated IR3; ΔG = −7.7 kcal mol−1). A number of DRs were also identified in the putative promoter region. The sequence TATAGG (positions 107 to 112) repeated three times just upstream of IR1 was designated DR1, while a second DR beginning four bp downstream of IR1 and containing six imperfect 10-bp DRs with an ATATTTAAAG consensus sequence was designated DR2. In some cases DR2 contains a 5-bp repeat extension (TACAC) which is present in three of the six DR2 sequences, one of which overlaps the putative −10 site (Fig. 3). Finally, an additional cluster of three DRs (designated DR3, DR4, and DR5) is located downstream of IR3 between positions 1911 and 1980. DRs are typically present in both rolling circle and theta replicating plasmids, where they constitute so-called iterons required for the initiation of replication (34). Such DR sequences have been shown to function as binding sites for Rep-like proteins (15).
FIG. 3.
Structural features of the pCIBAO89 promoter region. The first base pair of the single HindIII site was designated bp 1 and is underlined. The ribosome binding sites (−10 and −35 hexamers) are boxed. Letters in bold denote the transcriptional and translational start sites; arrows indicate the significant repeat regions.
Construction of the shuttle vector pPKCm.
In order to test whether the replication functions of pCIBAO89 can be used to construct an E. coli-Bifidobacterium shuttle vector, the entire 2.1-kb pCIBA089 was ligated into the HindIII site of pBlueCm, resulting in pPKCm1 (see Materials and Methods). This 6.2-kb construct was found to be capable of transforming B. breve UCC2003 with an average transformation efficiency of 3.8 × 106 transformants per μg of DNA. All tested chloramphenicol-resistant B. breve transformants were found to contain the expected plasmid, and restriction analysis confirmed that no obvious alterations in the DNA sequence had occurred. To investigate whether pPKCm1 could replicate in related hosts, a selection of bifidobacterial strains was tested. Plasmid pPKCm1 could be introduced into a diverse range of bifidobacterial strains, including B. animalis subsp. lactis (101) (the numbers of transformants obtained per microgram of DNA are given in parentheses), B. longum NCIMB8809 (102), B. pseudolongum NCIMB2244 (102), B. globosum JCM5820 (103), B. pseudocatenulatum LMG10505 (103) and B. dentium NCFB2843 (104) (see Table 1 for specifics on strains). However, it failed to generate Cmr transformants in L. lactis NZ9000, suggesting that the host range of the vector may be limited to certain gram-positive bacteria.
Determination of the minimal replicon of pCIBAO89.
In order to determine the minimal region of pCIBA089, deletion analysis of its replication region in pPKCm1 was performed as schematically illustrated in Fig. 2a. The smallest of these constructs found to be capable of replicating in B. breve UCC2003 (designated pPKCm2; 5.5 kb) contains a 1.4-kb pCIBA089 fragment encompassing DR1, IR1, DR2, and rep89. This suggests that the 0.7-kb DNA sequence following the ORF is not essential for pCIBA089 replication. The pPKCm6 construct containing a 1.9-kb pCIBA089 PCR fragment lacking the IR1 and DR2 elements but containing rep89, its promoter, and a ribosome binding site failed to generate Cmr transformants of B. breve UCC2003. pPKCm11 (differing from pPKCm6 only in that it also contains DR2) also failed to produce transformants in B. breve UCC2003. This implies that the IR1 sequences are essential components for pCIBAO89 replication. As expected, rep89 also appears to be vital for pCIBA089 replication, as pPKCm3 (containing a 360-bp pCIBA089 PCR fragment encoding IR1, DR2, and the rep89 promoter) and pPKCm12 (encoding all potential promoter elements but with the final 122 bp of rep89 removed by XhoI digestion and religation) failed to produce Cmr transformants when introduced into B. breve UCC2003. The deletion of the various DR sequences at the end of the pCIBA089 sequence (pPKCm8; 6.0 kb) did not appear to interfere with replication in UCC2003. When analyzed by agarose gel electrophoresis, no apparent copy number variations among the pPKCm constructs were observed (data not shown). To establish the segregational stability of the various pPKCm constructs a plasmid stability assay was performed over 200 generations. All six constructs capable of replication in B. breve UCC2003 (Fig. 2a) were found to exhibit considerable stability (96.6% ± 3%) in the absence of selective pressure (data not shown).
Transcriptional analysis of the rep89 gene in pCIBAO89.
The transcriptional start site of rep89 was identified as a thymine at position 312 just downstream of sequences resembling the consensus −10 (TACACG) and −35 (CTGCTA) sequences (Fig. 3). In order to assess whether this promoter region contains additional regulatory elements that control the promoter activity other than the identified −10 and −35 sequences, a number of transcriptional fusions were constructed using various rep89 promoter fragments (Fig. 2b) and the promoterless gusA reporter gene in the promoter probe vector pNZ272 (36). B. breve UCC2003 transformants containing each of the relevant constructs were assayed for β-GUS activity and OD600; a representative result at an OD600 value of 0.5 is presented in Table S3 in the supplemental material. The largest construct, pGC1, containing the entire putative promoter region consistently exhibited the highest level of GUS activity. Significant GUS activity, although at a lower level than that seen with pGC1, was detected for pGC2, which contains all but the first half of IR1. In contrast, the remaining constructs pGC3 through pGC8, from which IR1 was removed and containing successively smaller segments of the promoter region, displayed only background activity similar to that obtained with the control pNZ272 vector.
In order to investigate the effect of the presence of the Rep89 protein on the activity of the rep89 promoter, B. breve UCC2003 transformants containing each of the relevant GUS constructs described above were cotransformed with pSKEm (Fig. 1b), an erythromycin-based shuttle vector which contains the replication region of pCIBAO89 and therefore was expected to produce the Rep89 protein. The presence of both plasmids was confirmed by dual antibiotic selection (2 μg ml−1 Cm, 1 μg ml−1 Em) as well as restriction profiling. GUS assays were then performed to assess the effect of the presence of Rep89 on the activity of the promoter fragments. A 10-fold decrease in GUS activity was observed for pGC1 and pGC2 (not shown), suggesting that Rep89 represses its own promoter activity, possibly (as shown below) through binding to the IR1 sequence.
Rep89 binds to a putative origin of replication.
One approach to identify the origin of replication of rep-containing plasmids is to elucidate the exact region of DNA with which the replication protein interacts as a prerequisite for plasmid replication. The ability of Rep89 to bind to its promoter and upstream region was examined by gel electrophoretic mobility shift assays (EMSAs). EMSA analysis (Fig. 4a) clearly showed that the presence of a crude lysate of B. breve UCC2003 (pNZrep89) retarded the mobility of the extended rep89 promoter region (PCR fragment EM1 generated using primers em1fwd/emerv; Fig. 2c). This binding was shown to be specific, as a control lysate of B. breve UCC2003 containing pNZ44, which does not contain and therefore cannot express rep89, was shown to be incapable of binding to the promoter region of pCIBAO89 (lanes 2 in Fig. 4a and b). Furthermore, the mobility of a control DNA fragment identical in size to the pCIBA089 promoter region was not retarded by the presence of pNZrep89 lysate (lanes 1 in Fig. 4a and b). To establish which part of the promoter region is important for the observed interaction with Rep89, various shortened versions of the rep89 promoter region were used for EMSA analysis. DNA fragments were generated by PCR (using EM2fwd, -3fwd, -4fwd, -5fwd, or -6fwd in combination with EMrev; see Table S1 in the supplemental material) dissecting the pCIBAO89 promoter region. The mobility of probe EM2 (primers em2fwd and emrev; Fig. 2c) was also retarded, although its mobility appeared less severely affected than that of EM1 (Fig. 4b), possibly reflecting binding of Rep89 to multiple independent binding sites. The mobility of probes which did not contain IR1 were not affected under similar experimental conditions (data not shown), highlighting the importance of IR1 for Rep89 interaction and its involvement in plasmid replication.
FIG. 4.
(a) Lane 1: 3,000 cpm labeled 300-bp control DNA fragment plus B. breve UCC2003 pNZrep89 lysate. Lanes 2 to 6 contain 3,000 cpm labeled EM1 fragment. Lane 2, control B. breve pNZ44 lysate; lanes 3 to 6, increasing volumes of crude pNZrep89 lysate. (b) Lane 1, 3,000 cpm labeled 300-bp control DNA fragment plus B. breve UCC2003 pNZrep89 lysate. Lanes 2 to 6 contain 3,000 cpm labeled EM2 fragment. Lane 2, control B. breve pNZ44 lysate; lanes 3 to 6, increasing volumes of crude pNZrep89 lysate.
Copy number determination of pPKCm1 in B. breve UCC2003.
To determine the relative copy numbers of the pPKCm1 plasmid, qPCR analysis using total DNA from exponentially growing B. breve UCC2003 harboring pPKCm1 was used in combination with primer pairs designed for targets on both the chromosome and plasmid (see Materials and Methods). Assigning the copy number of the chromosomal gene apuB a value of 1, the relative ratios of chromosome/plasmid levels were determined based on the crossing values achieved. In template-free negative controls, no amplicons were generated. The assay was repeated in triplicate, and variations in the results can be attributed to the quality of the total DNA and small differences in the growth phases of the cultures analyzed. This method produced a copy number of 4 ± 1.26 per copy of chromosome for pPKCm1. This finding compares well with the copy number obtained for the recombinant plasmid pCRY4-Rep (to which Rep89 shows 29% amino acid sequence similarity), which was calculated at three plasmid copies per chromosome in Corynebacterium glutamicum (55), indicating that both of these plasmids copy at low numbers in their respective hosts.
DISCUSSION
In order to link strain-specific probiotic activities to the expression of specific genes it will be necessary to perform genome-wide or gene-specific functional analyses. For bifidobacteria such analyses are currently very difficult, mainly due to a paucity of effective molecular tools such as cloning, expression, and mutagenesis vectors. This work addresses this scientific deficiency through the comprehensive characterization of the cryptic, self-replicating bifidobacterial plasmid pCIBAO89 and the construction of shuttle vectors exploiting its replication functions.
The sequences of pCIBAO89, pCIBA431, and the publicly available pAP1 differ due to the presence of an extra cytosine base at position 1262 in the former two plasmids which extends the size of the only identified ORF from 894 nucleotides to 1,041. If this difference is due to a sequence error or mutation it may explain the failure to create a functional shuttle vector by use of the presumed replication region of pAP1 (47). The 2.1-kb pCIBA089 plasmid contains the rep89 gene, which encodes the presumed plasmid replication protein (Rep89). It exhibits similarities to replication proteins of plasmids isolated from a wide variety of bacterial species, but the most significant homologies are to replication proteins encoded by two B. longum plasmids of approximately 10 kb. Interestingly, the replication protein from pCIBAO89/pCIBA431 is also similar to mostly putative Rep proteins encoded by various large plasmids (see Table S2 in the supplemental material) such as pIPO2T (54), pBFp1 (4), pMOL98 (11), pCRY4 from C. glutamicum (55), pSa from E. coli (30), and pNL1 (GenBank accession no. NC_002033) from Novosphingobium aromaticivorans as well as to a Rep protein from pRM21, a small Rhodothermus marinus plasmid (7). The latter plasmid was found to possess a region containing three 19-bp DRs, dnaA boxes, and an A+T-rich region, which are characteristic of the replication origins of certain theta replicating plasmids. This suggests that, despite their limited sizes, pCIBAO89 and pCIBA431 may replicate via a theta-type mechanism. Based on sequence similarities of their replication proteins, they may form a subgroup of the IncW plasmids. A number of other small plasmids have been previously shown to replicate by a theta-type mechanism, e.g., the 2.6-kb pRJF1 from Butyrivibrio fibrisolvens (14) and the 3.8-kb plasmid pWV02 from L. lactis (17).
Upon investigation of the pCIBA089 sequence, a number of direct and inverted repeats were discovered upstream of rep89, including six 10-bp DRs which we propose represent the iterons required for the initiation of replication of this plasmid (Fig. 3). This organization closely resembles the IncW plasmid pSa (to which Rep89 shows 27% identity) except that the iterons in pSa consist of 17-bp repeats and are separated by 6 bp compared to approximately 10 bp for Rep89. For iteron-controlled plasmid replication it is suggested that binding of the corresponding replication protein to the DRs in the ori is a key step in the initiation of plasmid replication. However, replication proteins can also bind to inverted repeats in order to repress plasmid replication. The specificity of the binding appears to be dependent on whether the replication protein is present in a monomeric or dimeric form where the monomer binds to DRs, while the dimeric form associates with the IRs (9). The requirement for such a segment for plasmid replication is to allow a response to the intracellular concentration of the replication protein. From our DNA-binding studies we can attribute such a function to IR1, and the failure to demonstrate binding to the downstream iterons can be attributed to the dimeric nature of the replication protein used in the EMSA, as replication proteins obtained under laboratory conditions are known to predominately exist as dimers (50).
To establish the function of the upstream regulatory regions, the transcriptional start site was determined and subsequently the contribution of the various repeat sequences was experimentally achieved using the promoter probe vector pNZ272. Constructs containing IR1 (pGC1 and pGC2) gave consistently high GUS activity levels while removal of IR1 eliminated GUS activity to background levels. This would suggest that IR1 acts as a transcription-enhancing element, possibly mediated by a host-encoded factor. Inverted repeats compensating for ori may function as an enhancer element, as has previously been reported by Ohkubo and Yamaguchi (29). When cultures containing each of the GUS constructs (pGC1 to pGC8) were cotransformed with the Em-selective shuttle vector pSKEm, the presence of Rep89 had an inhibitory effect on the activity of the reporter gusA gene. We propose that this effect was due to Rep89 binding to IR1, which interferes with the transcription enhancement of this region, thereby representing an autoregulatory loop to control replication and consequent copy numbers of the plasmid.
A further goal of this work was to create a shuttle vector with improved cloning features, which was achieved with the construction of pSKEm. The vector contains a single selectable antibiotic resistance marker functional in both E. coli and Bifidobacterium spp. To the best of our knowledge, pSKEm is the first described E. coli-Bifidobacterium shuttle vector harboring the αlacZ gene and MCS of pBluescript, which allows the useful blue-white screening of recombinant constructs in E. coli.
The host range of bifidobacterial shuttle vectors is not fully known, although pPKCm1 could not be introduced into L. lactis; this is consistent with other studies on replication of plasmids of bifidobacterial origins (21). However, pPKCm1 was successfully electroporated into a diverse range of bifidobacterial strains, including the commercially relevant strain B. animalis subsp. lactis Bb12, clearly demonstrating its usefulness as a shuttle vector for a wide range of species within this genus.
Though several Bifidobacterium-E. coli shuttle vectors have been constructed based on cryptic plasmids from bifidobacteria, this work represents the first in-depth molecular characterization of the replication functions of a native bifidobacterial plasmid. This characterization, combined with the relatively high transformation efficiency and host range capabilities of the developed shuttle vectors, represents an important step for future functional genomic analyses of this commercially and medically significant gut inhabitant.
Supplementary Material
Acknowledgments
This work was financially supported by the Higher Education Authority Programme for Research in Third-Level Institutions and by the Science Foundation Ireland-funded Alimentary Pharmabiotic Centre located at University College Cork.
Footnotes
Published ahead of print on 26 October 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez-Martín, P., A. B. Florez, and B. Mayo. 2007. Screening for plasmids among human bifidobacteria species: sequencing and analysis of pBC1 from Bifidobacterium catenulatum L48. Plasmid 57:165-174. [DOI] [PubMed] [Google Scholar]
- 3.Argnani, A., R. J. Leer, N. van Luijk, and P. H. Pouwels. 1996. A convenient and reproducible method to genetically transform bacteria of the genus Bifidobacterium. Microbiology 142:109-114. [DOI] [PubMed] [Google Scholar]
- 4.Bergström, M., M. Hermansson, and C. Dahlberg. 2004. Isolation and sequencing of the replication region of plasmid pBFp1 isolated from marine biofilm. Plasmid 51:179-184. [DOI] [PubMed] [Google Scholar]
- 5.Corneau, N., E. Emond, and G. LaPointe. 2004. Molecular characterization of three plasmids from Bifidobacterium longum. Plasmid 51:87-100. [DOI] [PubMed] [Google Scholar]
- 6.de Man, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 7.Ernstsson, S., S. H. Bjornsdottir, Z. O. Jónsson, S. H. Thorbjarnardottir, G. Eggertsson, and A. Palsdottir. 2003. Identification and nucleotide sequence analysis of a cryptic plasmid, pRM21, from Rhodothermus marinus. Plasmid 49:188-191. [DOI] [PubMed] [Google Scholar]
- 8.Fu, G. F., X. Li, Y. Y. Hou, Y. R. Fan, W. H. Liu, and G. X. Xu. 2005. Bifidobacterium longum as an oral delivery system of endostatin for gene therapy on solid liver cancer. Cancer Gene Ther. 12:133-140. [DOI] [PubMed] [Google Scholar]
- 9.Fueki, T., and K. Yamaguchi. 2001. The structure and function of the replication initiator protein (Rep) of pSC101: an analysis based on a novel positive-selection system for the replication-deficient mutants. J. Biochem. (Tokyo) 130:399-405. [DOI] [PubMed] [Google Scholar]
- 10.Fujimori, M. 2006. Genetically engineered bifidobacterium as a drug delivery system for systemic therapy of metastatic breast cancer patients. Breast Cancer 13:27-31. [DOI] [PubMed] [Google Scholar]
- 11.Gstalder, M., M. Faelen, N. Mine, E. M. Top, M. Mergeay, and M. Couturier. 2003. Replication functions of new broad host range plasmids isolated from polluted soils. Res. Microbiol. 154:499-509. [DOI] [PubMed] [Google Scholar]
- 12.Guglielmetti, S., M. Karp, D. Mora, I. Tamagnini, and C. Parini. 2007. Molecular characterization of Bifidobacterium longum biovar longum NAL8 plasmids and construction of a novel replicon screening system. Appl. Microbiol. Biotechnol. 74:1053-1061. [DOI] [PubMed] [Google Scholar]
- 13.Han, S. O., H. Yukawa, M. Inui, and R. H. Doi. 2005. Molecular cloning and transcriptional analysis of engO, encoding a new noncellulosomal family 9 enzyme, from Clostridium cellulovorans. J. Bacteriol. 187:4884-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hefford, M. A., R. M. Teather, and R. J. Forster. 1993. The complete nucleotide sequence of a small cryptic plasmid from a rumen bacterium of the genus Butyrivibrio. Plasmid 29:63-69. [DOI] [PubMed] [Google Scholar]
- 15.Inui, M., J. H. Roh, K. Zahn, and H. Yukawa. 2000. Sequence analysis of the cryptic plasmid pMG101 from Rhodopseudomonas palustris and construction of stable cloning vectors. Appl. Environ. Microbiol. 66:54-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isolauri, M. 2004. Dietary modification of atopic disease: use of probiotics in the prevention of atopic dermatitis. Curr. Allergy Asthma Rep. 4:270-275. [DOI] [PubMed] [Google Scholar]
- 17.Kiewiet, R., S. Bron, K. do Jonge, G. Venema, and J. F. Seegers. 1993. Theta replication of the lactococcal plasmid pWVO2. Mol. Microbiol. 10:319-327. [PubMed] [Google Scholar]
- 18.Klijn, A., D. Moine, M. Delley, A. Mercenier, F. Arigoni, and R. D. Pridmore. 2006. Construction of a reporter vector for the analysis of Bifidobacterium longum promoters. Appl. Environ. Microbiol. 72:7401-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuipers, O. P., P. G. G. A. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 20.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 21.Lee, J. H., and D. J. O'Sullivan. 2006. Sequence analysis of two cryptic plasmids from Bifidobacterium longum DJO10A and construction of a shuttle cloning vector. Appl. Environ. Microbiol. 72:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindner, C., R. Nijland, M. van Hartskamp, S. Bron, L. W. Hamoen, and O. P. Kuipers. 2004. Differential expression of two paralogous genes of Bacillus subtilis encoding single-stranded DNA binding protein. J. Bacteriol. 186:1097-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacConaill, L. E., G. F. Fitzgerald, and D. van Sinderen. 2003. Investigation of protein export in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 69:6994-7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattarelli, P., B. Biavati, A. Alessandrini, F. Crociani, and V. Scardovi. 1994. Characterization of the plasmid pVS809 from Bifidobacterium globosum. Microbiology 17:327-331. [PubMed] [Google Scholar]
- 25.Mazé, A., M. O'Connell-Motherway, G. F. Fitzgerald, J. Deutscher, and D. van Sinderen. 2007. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73:545-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGrath, S., G. F. Fitzgerald, and D. van Sinderen. 2001. Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl. Environ. Microbiol. 67:608-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics, p. 433. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Nakamura, T., T. Sasaki, M. Fujimori, K. Yazawa, Y. Kano, J. Amano, and S. Taniguchi. 2002. Cloned cytosine deaminase gene expression of Bifidobacterium longum and application to enzyme/pro-drug therapy of hypoxic solid tumours. Biosci. Biotechnol. Biochem. 66:2362-2366. [DOI] [PubMed] [Google Scholar]
- 29.Ohkubo, S., and K. Yamaguchi. 1995. Two enhancer elements for DNA replication of pSC101, par and a palindromic binding sequence of the Rep protein. J. Bacteriol. 177:558-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okumura, M. S., and C. I. Kado. 1992. The region essential for efficient autonomous replication of pSa in Escherichia coli. Mol. Gen. Genet. 235:55-63. [DOI] [PubMed] [Google Scholar]
- 31.O'Riordan, K., and G. F. Fitzgerald. 1999. Molecular characterisation of a 5.75-kb cryptic plasmid from Bifidobacterium breve NCFB2258 and determination of a mode of replication. FEMS Microbiol. Lett. 174:285-294. [DOI] [PubMed] [Google Scholar]
- 32.Park, M. S., D. W. Shin, K. H. Lee, and G. E. Ji. 1997. Isolation and characterisation of two plasmids from Bifidobacterium longum. Lett. Appl. Microbiol. 25:5-7. [DOI] [PubMed] [Google Scholar]
- 33.Park, M. S., D. W. Shin, K. H. Lee, and G. E. Ji. 1999. Sequence analysis of plasmid pKJ50 from Bifidobacterium longum. Microbiology 145:585-592. [DOI] [PubMed] [Google Scholar]
- 34.Paulsson, J., and D. K. Chattoraj. 2006. Origin inactivation in bacterial DNA replication control. Mol. Microbiol. 61:9-15. [DOI] [PubMed] [Google Scholar]
- 35.Picard, C., J. Fioramonti, A. Francois, T. Robinson, F. Neant, and C. Matuchansky. 2005. Review article: bifidobacteria as probiotic agents—physiological effects and clinical benefits. Aliment. Pharmacol. Ther. 22:495-512. [DOI] [PubMed] [Google Scholar]
- 36.Platteeuw, C., G. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Providenti, M. A., J. M. O'Brien, R. J. Ewing, E. S. Paterson, and M. L. Smith. 2006. The copy-number of plasmids and other genetic elements can be determined by SYBR-Green-based quantitative real-time PCR. J. Microbiol. Methods 65:476-487. [DOI] [PubMed] [Google Scholar]
- 38.Rhim, S. L., M. S. Park, and G. E. Ji. 2005. Expression and secretion of Bifidobacterium adolescentis amylase by Bifidobacterium longum. Biotechnol. Lett. 23:163-168. [DOI] [PubMed] [Google Scholar]
- 39.Rossi, M., P. Brigidi, and D. Matteuzzi. 1997. An efficient transformation system for Bifidobacterium spp. Lett. Appl. Microbiol. 24:33-36. [Google Scholar]
- 40.Rossi, M., P. Brigidi, R. A. Gonzalez Vara, and D. Matteuzzi. 1996. Characterisation of the plasmid pMB1 from Bifidobacterium longum and its use for shuttle vector construction. Res. Microbiol. 147:133-143. [DOI] [PubMed] [Google Scholar]
- 41.Ryan, S., G. F. Fitzgerald, and D. van Sinderen. 2006. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl. Environ. Microbiol. 72:5289-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Sangrador-Vegas, A., C. Stanton, D. van Sinderen, G. F. Fitzgerald, and R. P. Ross. 2007. Characterisation of plasmid pASV479 from Bifidobacterium pseudolongum subsp. globosum and its use for expression vector construction. Plasmid 58:140-147. [DOI] [PubMed] [Google Scholar]
- 44.Scardovi, V., B. Sgorbati, and G. Zani. 1971. Starch gel electrophoresis of fructose-6-phosphate phophoketolase in the genus Bifidobacterium. J. Bacteriol. 106:1036-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scardovi, V. 1986. The genus Bifidobacterium Orla-jensen 1924, 472AL, p. 1418-1434. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systemic bacteriology, vol. 2. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 46.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schurch, C. 2002. Development of a novel DNA transformation system for bifidobacteria. Ph.D. thesis. Swiss Federal Institute of Technology, Zurich (ETHZ), Switzerland.
- 48.Sgorbati, B., V. Scardovi, and D. J. LeBlanc. 1982. Plasmids in the genus Bifidobacterium. J. Gen. Microbiol. 128:2121-2131. [DOI] [PubMed] [Google Scholar]
- 49.Sgorbati, B., V. Scardovi, and D. J. LeBlanc. 1986. Related structures in the plasmid profiles of Bifidobacterium asteroides, B. indicum and B. globosum. Microbiology 9:443-456. [PubMed] [Google Scholar]
- 50.Sugiura, S., M. Tanaka, Y. Masamune, and K. Yamaguchi. 1990. DNA binding properties of purified replication initiator protein (Rep) encoded by plasmid pSC101. J. Biochem. 107:369-376. [DOI] [PubMed] [Google Scholar]
- 51.Takata, T., T. Shirakawa, Y. Kawasaki, S. Kinoshita, A. Gotoh, Y. Kano, and M. Kawabata. 2006. Genetically engineered Bifidobacterium animalis expressing the Salmonella flagellin gene for the mucosal immunization in a mouse model. J. Gene Med. 8:1341-1346. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka, K., K. Samura, and Y. Kano. 2005. Structural and functional analysis of pTB6 from Bifidobacterium longum. Biosci. Biotechnol. Biochem. 69:422-425. [DOI] [PubMed] [Google Scholar]
- 53.Tannock, G. 1999. The bowel microflora: an important source of urinary tract pathogens. World J. Urol. 17:339-344. [DOI] [PubMed] [Google Scholar]
- 54.Tauch, A., S. Schneiker, W. Selbitschka, A. Puhler, L. S. van Overbeek, K. Smalla, C. M. Thomas, M. J. Bailey, L. J. Forney, A. Weightman, P. Ceglowski, T. Pembroke, E. Tietze, G. Schroder, E. Lanka, and J. D. van Elsas. 2002. The complete nucleotide sequence and environmental distribution of the cryptic, conjugative, broad-host-range plasmid pIPO2 isolated from bacteria of the wheat rhizosphere. Microbiology 148:1637-1653. [DOI] [PubMed] [Google Scholar]
- 55.Tauch, A., A. Puhler, J. Kalinowski, and G. Thierbach. 2003. Plasmids in Corynebacterium glutamicum and their molecular classification by comparative genomics. J. Biotechnol. 104:27-40. [DOI] [PubMed] [Google Scholar]
- 56.Tissier, H. 1906. Traitement des infections intestinales par la méthode de transformations de la flore bactérienne de l'intestin. C. R. Soc. Biol. 60:359-361. [Google Scholar]
- 57.Ventura, M., D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie Leeuwenhoek 86:205-223. [DOI] [PubMed] [Google Scholar]
- 58.Ventura, M., C. Canchaya, A. D. Casale, F. Dellaglio, E. Neviani, G. F. Fitzgerald, and D. van Sinderen. 2006. Analysis of bifidobacterial evolution using a multilocus approach. Int. J. Syst. Evol. Microbiol. 56:2783-2792. [DOI] [PubMed] [Google Scholar]
- 59.Ventura, M., C. Canchaya, G. F. Fitzgerald, R. S. Gupta, and D. van Sinderen. 2007. Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Antonie Leeuwenhoek 91:351-372. [DOI] [PubMed] [Google Scholar]
- 60.Xu, Y. F., L. P. Zhu, B. Hu, G. F. Fu, H. Y. Zhang, J. J. Wang, and G. X. Xu. 2007. A new expression plasmid in Bifidobacterium longum as a delivery system of endostatin for cancer gene therapy. Cancer Gene Ther. 14:151-157. [DOI] [PubMed] [Google Scholar]
- 61.Yazawa, K., M. Fujimori, J. Amano, Y. Kano, and S. Taniguchi. 2000. Bifidobacterium longum as a delivery system for cancer gene therapy: selective localization and growth in hypoxic tumors. Cancer Gene Ther. 7:269-274. [DOI] [PubMed] [Google Scholar]
- 62.Yazawa, K., M. Fujimori, T. Nakamura, T. Sasaki, J. Amano, Y. Kano, and S. Taniguchi. 2001. Bifidobacterium longum as a delivery system for gene therapy of chemically induced rat mammary tumors. Breast Cancer Res. Treat. 66:165-170. [DOI] [PubMed] [Google Scholar]
- 63.Yildirim, Z., D. K. Winters, and M. G. Johnson. 1999. Purification, amino acid sequence and mode of action of bifidocin B produced by Bifidobacterium bifidum NCFB 1454. J. Appl. Microbiol. 86:45-54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.