Abstract
Bacterial leaf streak, caused by Xanthomonas oryzae pv. oryzicola, is an important disease of rice. Transposon-mediated mutational analysis of the pathogen with a quantitative assay revealed candidate virulence factors including genes involved in the pathogenesis of other phytopathogenic bacteria, virulence factors of animal pathogens, and genes not previously associated with virulence.
Bacterial leaf streak is an important disease of rice (Oryza sativa) for which control measures are limited (22). In particular, no simply inherited gene for resistance to the disease has been reported. The disease is caused by Xanthomonas oryzae pv. oryzicola, a member of the gamma subdivision of the class Proteobacteria. The pathogen enters through leaf stomata or wounds and colonizes the parenchyma apoplast, causing interveinal lesions that appear water soaked initially and then develop into translucent, yellow-to-white streaks. Leaf streak is prevalent in Asia and parts of Africa, where it can decrease yield by as much as 30%. In the United States, the pathogen is quarantined and has been designated a select agent under the Agricultural Bioterrorism Act of 2002. To lay the groundwork for disease prevention and control strategies based on interference with bacterial virulence, transposon-mediated mutational analysis of X. oryzae pv. oryzicola was carried out to identify candidate virulence factors.
Identification and characterization of reduced-virulence mutants.
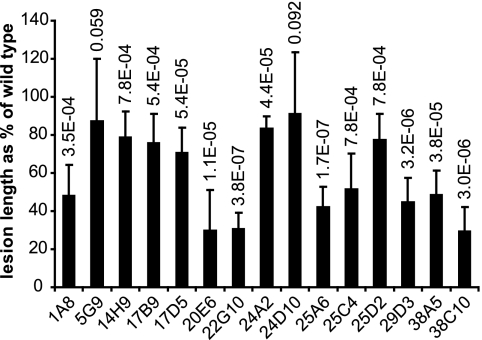
Strain BLS303 of X. oryzae pv. oryzicola (C. Vera-Cruz, International Rice Research Institute) was mutagenized by using the EZ::TN <R6Kγori/KAN-2> Tn5 insertion kit (Epicentre Biotechnologies), which generates random, stable insertions. BLS303 cells were transformed by electroporation as described previously (30). Insertion mutants were selected on glucose yeast extract agar (18) containing 25 μg/ml kanamycin and then cultured overnight in liquid glucose yeast extract with kanamycin. Cells were washed twice and resuspended in sterile water to an optical density at 600 nm of 0.5 and used to spot infiltrate, in duplicate, leaves of 4-week-old rice plants of Indica variety IRBB10 with a needleless syringe. Plants were grown and maintained in a PGC-105 growth chamber (1,000 μmol/m2/s; Percival Scientific, Inc., Perry, IA) under a cycle of 12 h of light at 28°C and 12 h of dark at 25°C with relative humidity at 75 to 80%. Symptoms were observed after 4 days. Ten thousand mutants were screened. For 153 mutants, symptoms appeared reduced relative to the wild type or were absent. These were characterized further with a more stringent quantitative assay (Fig. 1). In this assay, leaves of 8-week-old rice plants were inoculated with a mutant on one side of the midrib and the wild type directly opposite on the other. For each mutant, five replicate, paired inoculations on each of two leaves were made. After 10 days, lesion lengths were measured for each paired inoculation, and a paired, two-tailed Student t test was performed across all replicates. By this test, 21 mutants were confirmed as being virulence impaired (P < 0.1). Of these, 6 were completely nonvirulent and 15 (Fig. 2) were reduced in virulence.
FIG. 1.
Quantitative virulence assay. X. oryzae pv. oryzicola cells suspended in water to an optical density at 600 nm of 0.5 were used to spot infiltrate leaves of 8-week-old rice plants with a needleless syringe. Mutants were inoculated on one side of the midrib, and the wild type (WT) was inoculated directly opposite on the other. For each mutant, five replicate, paired inoculations were done and lesion lengths were measured after 10 days and compared by using a paired, two-tailed Student t test. Shown are representative lesions caused by mutant 38C10 (see text) and the wild type.
FIG. 2.
Lengths of lesions in rice leaves caused by reduced-virulence mutants of X. oryzae pv. oryzicola as percentages of the wild type inoculated side by side. Mutants that caused no lesions are not shown. Error bars represent the standard deviation of 10 replicate inoculations. P values resulting from a paired, two-tailed Student t test for each mutant are shown at the top.
Rescue and sequence analysis of disrupted genes.
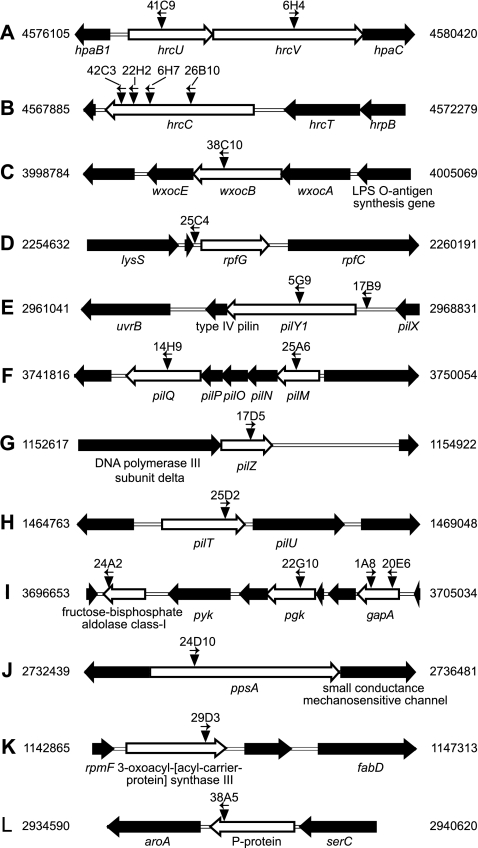
Each of the 21 mutants contained just one insertion, as determined by Southern blot hybridization of EcoRI-digested genomic DNA with the 1-kb XhoI/BamHI fragment of the transposon. To rescue the DNA containing the transposon, which carries the Pir protein-dependent origin of replication R6K and the nptII gene and lacks EcoRI sites, EcoRI-digested DNA was treated with T4 ligase and electroporated into Escherichia coli S17 λ pir. Transformants were selected on LB agar containing kanamycin (25 μg/ml). Nucleotide sequences flanking the transposon were determined with transposon-specific primers provided with the insertion kit. Insertions were mapped and oriented by aligning the sequences to the finished whole-genome sequence of X. oryzae pv. oryzicola strain BLS256 available through the Comprehensive Microbial Resource (www.tigr.org/cmr) and through the National Center for Biotechnology Information (GenBank accession no. AAQN01000001), with reference to the draft annotation for this genome, available through the Comprehensive Microbial Resource (Fig. 3). Insertions mapped to genes that encode components of the type III secretion system (T3SS), a lipopolysaccharide (LPS) synthesis enzyme, a two-component system response regulator, type IV pilus assembly proteins, enzymes involved in carbohydrate metabolism, and enzymes for fatty acid and aromatic amino acid synthesis (Table 1).
FIG. 3.
Genomic locations and orientations of transposon insertions in reduced-virulence mutants of X. oryzae pv. oryzicola (A to L). Each mutant carries only one insertion. Disrupted genes are represented by empty block arrows. Transposon insertions are represented by inverted triangles. The orientation of the nptII promoter in each insertion is shown by an arrow above the triangle. Mutant designations corresponding to each insertion are given at the top. Numbers to the left and right are genome coordinates in base pairs. Gene symbols or products are noted below disrupted genes and selected flanking genes.
TABLE 1.
X. oryzae pv. oryzicola transposon insertion mutants affected in virulence
| Mutant | Locus | Product | Gene | Virulencea | Insertion positionb | Orientationc |
|---|---|---|---|---|---|---|
| 41C9 | XOCORF_4443 | Type III secretion protein HrcU | hrcU | − | 554 | R |
| 6H4 | XOCORF_4444 | Type III secretion protein HrcV | hrcV | − | 1398 | F |
| 42C3 | XOCORF_4434 | Type III secretion protein HrcC | hrcC | − | 1693 | F |
| 22H2 | XOCORF_4434 | Type III secretion protein HrcC | hrcC | − | 1580 | F |
| 6H7 | XOCORF_4434 | Type III secretion protein HrcC | hrcC | − | 1411 | F |
| 26B10 | XOCORF_4434 | Type III secretion protein HrcC | hrcC | − | 728 | F |
| 38C10 | XOCORF_3864 | Putative glycosyltransferase | wxocB | + | 544 | F |
| 25C4 | XOCORF_2264 | Two-component system response regulator RpfG | rpfG | ++ | −90 | R |
| 5G9 | XOCORF_2899 | Type IV fimbrial biogenesis protein | pilY1 | +++ | 1380 | F |
| 17B9 | XOCORF_2899 | Type IV fimbrial biogenesis protein | pilY1 | +++ | −115 | F |
| 14H9 | XOCORF_3636 | Fimbrial assembly protein | pilQ | +++ | 1595 | F |
| 25A6 | XOCORF_3640 | Type IV pilus assembly protein | pilM | ++ | 685 | F |
| 17D5 | XOCORF_1181 | Type IV pilus assembly protein | pilZ | +++ | 151 | F |
| 25D2 | XOCORF_1487 | Twitching mobility protein | pilT | +++ | 986 | F |
| 24A2 | XOCORF_3585 | Fructose-bisphosphate aldolase class I | +++ | 900 | F | |
| 22G10 | XOCORF_3589 | Phosphoglycerate kinase | pgk | + | 530 | F |
| 1A8 | XOCORF_3592 | Glyceraldehyde-3-phosphate dehydrogenase, type I | gapA | ++ | 638 | F |
| 20E6 | XOCORF_3592 | Glyceraldehyde-3-phosphate dehydrogenase, type I | gapA | + | 303 | R |
| 24D10 | XOCORF_2661 | Phosphoenolpyruvate synthase | ppsA | +++ | 540 | F |
| 29D3 | XOCORF_1172 | 3-Oxoacyl-[acyl-carrier-protein] synthase III | ++ | 825 | F | |
| 38A5 | XOCORF_2877 | P protein | ++ | 960 | F |
Virulence is rated as lesion length relative to wild-type lesion length (percent). Symbols: −, 0%; +, 10 to 30%; ++, 31 to 60%; +++, 61 to 95%.
Relative to the first nucleotide of the open reading frame.
Orientation of the nptII promoter in the transposon relative to the orientation of the disrupted gene. F, forward; R, reverse.
BLS256 and BLS303 are both Philippine isolates and are similar in their degrees of virulence. With three exceptions, the genetic context of each insertion shown in Fig. 3 is conserved in the genome of X. oryzae pv. oryzae strain MAFF311018 (GenBank accession no. NC_007705) and in most cases in Xanthomonas genomes less closely related to X. oryzae pv. oryzicola (not shown). The gene content and organization at these loci in BLS303 therefore are likely to be similarly conserved if not identical to those in BLS256. The first exception is the LPS biosynthesis locus (Fig. 3C), which is variable across Xanthomonas genomes, in some cases at the interstrain level (P. Patil and R. Sonti, personal communication). However, long-range, nested PCR amplification of the locus yielded products identical in size for BLS303 and BLS256 (L. Chen and A.J.B., unpublished data), indicating that the locus arrangement in BLS256 is likely shared by BLS303. The second exception is a small open reading frame upstream of rpfG (Fig. 3D) in the annotation of BLS256 that is absent from the other Xanthomonas genomes, as annotated. The final exception is the locus containing the pgk and gapA genes (see Fig. 3I). In other Xanthomonas genome sequences, including that of MAFF311018, upstream of pgk several conserved hypothetical protein-encoding genes replace gapA and nearby open reading frames, and gapA localizes elsewhere. In MAFF311018, gapA is surrounded by insertion sequence elements. Thus, although BLS256 and BLS303 are likely syntenic at this locus, further characterization of the insertion in gapA in BLS303, including its potential effect on genes downstream, will require cloning the locus from this strain.
Type III secretion genes.
Six insertions mapped to the hrp/hrc gene cluster, which encodes proteins involved in the regulation and assembly of the T3SS, a macromolecular, syringe-like complex that delivers effector proteins into host cells. In many plant pathogens, the T3SS is essential for eliciting the host hypersensitive reaction and for pathogenicity (5, 13). In mutant strains 41C9 and 6H4, the insertions occurred in hrcU and hrcV, respectively, which encode inner membrane components of the complex. The insertions in mutant strains 42C3, 22H2, 6H7, and 26B10 reside in hrcC, whose product is a key outer membrane component. As expected, and shown previously for hrcC in BLS303 (18), each of the six mutants was nonpathogenic (not shown).
LPS synthesis gene.
LPS is a component of the bacterial cell surface thought to protect against environmental stresses and antimicrobial compounds by restricting membrane permeability. It comprises three covalently linked components: an outer membrane-bound moiety called lipid A, a core oligosaccharide, and an outermost polysaccharide known as the O chain (23). The core and lipid A without the O chain constitute lipooligosaccharide. In mutant 38C10, the transposon disrupts the gene wxocB, which encodes a predicted member of the rhamnose-glucose polysaccharide assembly protein F (RgpF) family (31). Members of the RgpF family are involved in the assembly of the O chain, but immediately downstream of wxocB resides wxocE, which is predicted to be involved in the synthesis of the core. So the insertion, if polar, might block the assembly of even lipooligosaccharide. LPS has been implicated previously in plant pathogenesis owing to the isolation of reduced-virulence mutants that exhibited LPS deficiencies. Plants recognize certain LPSs as microbe-associated molecular patterns, triggering innate defense responses (17, 26). In the Rhizobium-legume symbiosis, structural changes in the O chain take place during nodulation, suggesting an adaptive role (15). Changes in LPS structure affect the efficiency of the T3SS of Shigella (30a) and the expression or function of adhesin, phospholipase A, and other virulence factors in Yersinia (3a). Rice leaves inoculated with 38C10 developed lesions dramatically reduced in length relative to the wild type. Although structural consequences of the mutation in this strain and the reason for its reduced virulence remain to be elucidated, this observation implies that LPS plays a central role in the virulence of X. oryzae pv. oryzicola.
Two-component regulatory gene rpfG.
In mutant 25C4, the transposon disrupts rpfG, which encodes the regulatory component of the RpfG/RpfC two-component system that has been shown to positively regulate the synthesis of virulence factors and dispersal of biofilms in X. campestris pv. campestris in response to cell-to-cell signaling mediated by a diffusible signal factor (25, 29). The mutant caused lesions in rice leaves about half the length of those caused by the wild type.
Type IV pilus assembly and twitching motility genes.
Twitching motility in bacteria is movement independent of flagella that occurs by extension, tethering, and retraction of type IV pili (16). Twitching motility plays an important role in host colonization by several animal pathogens (20). Reports on plant-associated bacteria are fewer and, to our knowledge, limited to bacteria that colonize the vascular system (6, 14, 21). Six of the mutant strains reported here carry insertions in type IV pilus assembly and twitching motility genes. In mutants 5G9 and 17B9, the transposons disrupt the coding and promoter regions, respectively, of pilY1. Disruption of pilY1 in Xylella fastidiosa caused a reduction, but not a complete loss, of type IV pili and twitching motility (16). Mutants 14H9 and 25A6 carry insertions in pilQ and pilM, respectively, which reside at either end of an operon conserved across several species and required for pilus assembly, twitching motility, and phage sensitivity (10, 19, 24). The transposons in mutants 17D5 and 25D2 integrated within pilZ and pilT, respectively. PilZ is a predicted receptor for the secondary messenger bis-(3′-5′)-cyclic dimeric GMP, which regulates processes such as biofilm formation, twitching motility, photosynthesis, and virulence (1, 7, 11). pilT encodes a putative hexameric ATPase required for type IV pilus retraction (2). With the exception of 25A6, which showed a relatively severe (greater than 50%) reduction in virulence, the impairment of the virulence of each of these mutants was slight to moderate. Nevertheless, to our knowledge, this is the first time that type IV pili (and twitching motility) have been implicated in nonvascular plant pathogenesis. The mechanistic basis of this involvement, whether attachment, motility, biofilm formation, or some combination, remains to be determined.
Carbohydrate synthesis genes.
Five mutants carry insertions in genes that encode enzymes for sugar metabolism. In mutant 24A2, the disrupted gene encodes fructose-bisphosphate aldolase (class I). In mutant 22G10, the transposon resides in pgk, which encodes phosphoglycerate kinase. Mutants 1A8 and 20E6 both carry an insertion in the gapA gene for type 1 glyceraldehyde-3-phosphate dehydrogenase. Each of these insertions resulted in moderately to dramatically reduced lesion lengths in inoculated rice leaves. Interestingly, the affected genes colocalize in a cluster involved in glycolysis and gluconeogenesis (10). Products of gap, pgk, and another gene in the cluster, pyk, were identified as candidate virulence factors in Yersinia pestis by proteomic analysis of the low calcium response (9). In Streptococcus pneumoniae the gapA gene product is both cytoplasmic and cell wall associated and contributes to virulence through a role in plasminogen binding, recruiting proteolytic activity to the bacterial cell surface important for invasiveness (4). gapA also plays a role in adhesion and invasiveness in Paracoccidioides brasiliensis, a fungal pathogen of humans, possibly by binding to host surface matrix components (3). A role for the gapA product in plant pathogenesis has not been reported previously. Whether it recruits protease activity to the X. oryzae pv. oryzicola cell surface, binds to rice cell surfaces, or contributes in some other way remains to be determined. Although functional analysis remains to be done, isolation of these four mutants suggests that some housekeeping and metabolic proteins play more complex roles than previously thought. In the fifth mutant of this class, 24D10, the disrupted gene is ppsA, which encodes phosphoenolpyruvate synthase A, a key enzyme in gluconeogenesis. ppsA was reported as important for virulence in X. campestris pv. campestris (28). In rice leaves inoculated with 24D10, a slight reduction in lesion length relative to the wild type was observed.
Other genes.
Two genes identified among the reduced-virulence mutants encode enzymes for basic metabolism. In mutant 29D3, the transposon is in a gene that encodes 3-oxoacyl-[acyl-carrier-protein] synthase III, an enzyme involved in fatty acid and phospholipid biosynthesis. A homolog of this gene regulates virulence factors in the tobacco pathogen Pseudomonas syringae pv. tabaci, potentially through an effect on the synthesis of acyl homoserine lactones that are involved in quorum sensing (27). Acyl homoserine lactone-mediated quorum sensing has not been reported in any xanthomonads, however. In mutant 38A5, the disrupted gene encodes the P protein involved in the prephenate pathway for aromatic amino acid biosynthesis. Aromatic amino acid auxotrophic mutants of other plant and animal pathogens also have attenuated virulence (8, 12). The moderate reduction in the virulence of mutants 29D3 and 38A5 likely reflects general defects in cellular physiology.
Conclusion.
Transposon-mediated mutagenesis has been used to identify virulence factors in many plant-pathogenic bacteria. With our screen for virulence-impaired transposon insertion mutants of X. oryzae pv. oryzicola, we identified several factors associated with virulence in other plant pathogens, including the T3SS, the rpfG/rpfC two-component regulatory system, LPS, and type IV pili. Importantly, to our knowledge, ours is the first reported indication of a virulence function for type IV pili in a nonvascular plant pathogen. Also, we identified factors not previously associated with plant pathogenesis but important or implicated in the virulence of animal pathogens, namely, selected enzymes for sugar metabolism, of which some appear also to play roles in binding host substrates at the cell surface. Finally, we isolated mutants affected in fatty acid and aromatic amino acid synthesis, processes that likely contribute to virulence through their roles in basic metabolism.
In all, despite the large scale of the screen, only 21 mutants were confirmed as virulence impaired. Ten thousand mutants represent roughly twofold coverage for a targeted insertion rate of 1 per 1,000 bp (the genome is just under 5 × 106 bp), so the screen likely was not saturating. Also, leaf-to-leaf variability and the early time point used for scoring in the initial screen might have precluded the capture of subtle virulence deficiencies. These factors were accounted for in the more stringent quantitative assay used for further characterization of mutants. Another possibility is that strains with mutations that have a marked effect on colony size or morphology (e.g., loss of EPS) may have been overlooked. Finally, functional redundancy of some virulence genes may have precluded their isolation.
The genes identified will require confirmation by genetic complementation, since the reduced-virulence phenotypes may be due to polar effects of insertions on downstream genes or to spontaneous ectopic mutations. However, we have observed the transposon to be a nonpolar mutagen when in the forward orientation (Fig. 3; L.W. and A.J.B., unpublished), ostensibly because of the outreading promoter of the terminator-less nptII gene that it contains. Also, for several genes or gene classes, we isolated multiple independent insertions, strongly supporting the conclusions that the insertions are the cause of the virulence deficiencies and that the genes to which they localize are the relevant virulence factors.
X. oryzae pv. oryzicola is a pathogen of emerging importance that constrains production of the world's most important food crop. Because rice is also an important biological model, bacterial leaf streak can serve as a representative system for understanding nonvascular pathogenesis in other plants. The identification of the candidate virulence factors reported here is an important first step toward elucidating molecules and mechanisms important in disease that may be targeted for the development of novel means of control and prevention. It should be noted that the inoculation technique used to identify these factors bypasses survival on the leaf surface and entry into the apoplast. Screening of the mutant library, by alternative methods, for mutants affected in these processes is likely to reveal yet additional virulence factor candidates.
Acknowledgments
We are grateful to J. Helgerson, Z. Sayre, E. Flemmig, J. Paulson, and J. Lorence for assistance with mutant screening, to K. Vogel for technical assistance, and to D. Meyer and D. Niño-Liu for critical reading of the manuscript.
This work was supported by award 0227357 from the Plant Genome Research Program of the National Science Foundation.
Footnotes
Published ahead of print on 2 November 2007.
REFERENCES
- 1.Amikam, D., and M. Y. Galperin. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3-6. [DOI] [PubMed] [Google Scholar]
- 2.Aukema, K. G., E. M. Kron, T. J. Herdendorf, and K. T. Forest. 2005. Functional dissection of a conserved motif within the pilus retraction protein PilT. J. Bacteriol. 187:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbosa, M. S., S. N. Bao, P. F. Andreotti, F. P. de Faria, M. S. Felipe, L. dos Santos Feitosa, M. J. Mendes-Giannini, and C. M. Soares. 2006. Glyceraldehyde-3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect. Immun. 74:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Bengoechea, J. A., H. Najdenski, and M. Skurnik. 2004. Lipopolysaccharide O antigen status of Yersinia enterocolitica O:8 is essential for virulence and absence of O antigen affects the expression of other Yersinia virulence factors. Mol. Microbiol. 52:451-469. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, S., M. Rohde, and S. Hammerschmidt. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect. Immun. 72:2416-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogdanove, A. J., S. V. Beer, U. Bonas, C. Boucher, A. Collmer, D. L. Coplin, G. R. Cornelis, H.-C. Huang, S. W. Hutchenson, N. J. Panopoulos, and F. Van Gijsegem. 1996. Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol. Microbiol. 20:681-683. [DOI] [PubMed] [Google Scholar]
- 6.Bohm, M., T. Hurek, and B. Reinhold-Hurek. 2007. Twitching motility is essential for endophytic rice colonization by the N2-fixing endophyte Azoarcus sp. strain BH72. Mol. Plant-Microbe Interact. 20:526-533. [DOI] [PubMed] [Google Scholar]
- 7.Brouillette, E., M. Hyodo, Y. Hayakawa, D. K. Karaolis, and F. Malouin. 2005. 3′,5′-Cyclic diguanylic acid reduces the virulence of biofilm-forming Staphylococcus aureus strains in a mouse model of mastitis infection. Antimicrob. Agents Chemother. 49:3109-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cersini, A., A. M. Salvia, and M. L. Bernardini. 1998. Intracellular multiplication and virulence of Shigella flexneri auxotrophic mutants. Infect. Immun. 66:549-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chromy, B. A., M. W. Choi, G. A. Murphy, A. D. Gonzales, C. H. Corzett, B. C. Chang, J. P. Fitch, and S. L. McCutchen-Maloney. 2005. Proteomic characterization of Yersinia pestis virulence. J. Bacteriol. 187:8172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folster, J. P., V. Dhulipala, R. A. Nicholas, and W. M. Shafer. 2007. Differential regulation of ponA and pilMNOPQ expression by the MtrR transcriptional regulatory protein in Neisseria gonorrhoeae. J. Bacteriol. 189:4569-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 12.Goel, A. K., L. Rajagopal, and R. V. Sonti. 2001. Pigment and virulence deficiencies associated with mutations in the aroE gene of Xanthomonas oryzae pv. oryzae. Appl. Environ. Microbiol. 67:245-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang, Y., H. Liu, S. Genin, M. A. Schell, and T. P. Denny. 2002. Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 46:427-437. [DOI] [PubMed] [Google Scholar]
- 15.Lerouge, I., and J. Vanderleyden. 2002. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol. Rev. 26:17-47. [DOI] [PubMed] [Google Scholar]
- 16.Li, Y., G. Hao, C. D. Galvani, Y. Meng, L. De Le Fuente, H. C. Hoch, and T. J. Burr. 2007. Type I and type IV pili of Xylella fastidiosa affect twitching motility, biofilm formation and cell-cell aggregation. Microbiology 153:719-726. [DOI] [PubMed] [Google Scholar]
- 17.Mackey, D., and A. J. McFall. 2006. MAMPs and MIMPs: proposed classifications for inducers of innate immunity. Mol. Microbiol. 61:1365-1371. [DOI] [PubMed] [Google Scholar]
- 18.Makino, S., A. Sugio, F. F. White, and A. J. Bogdanove. 2006. Inhibition of resistance gene mediated defense in rice by Xanthomonas oryzae pv. oryzicola. Mol. Plant-Microbe Interact. 19:240-249. [DOI] [PubMed] [Google Scholar]
- 19.Martin, P. R., A. A. Watson, T. F. McCaul, and J. S. Mattick. 1995. Characterization of a five-gene cluster required for the biogenesis of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 16:497-508. [DOI] [PubMed] [Google Scholar]
- 20.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 21.Meng, Y., Y. Li, C. D. Galvani, G. Hao, J. N. Turner, T. J. Burr, and H. C. Hoch. 2005. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J. Bacteriol. 187:5560-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niño-Liu, D. O., P. C. Ronald, and A. J. Bogdanove. 2006. Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7:303-324. [DOI] [PubMed] [Google Scholar]
- 23.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rumszauer, J., C. Schwarzenlander, and B. Averhoff. 2006. Identification, subcellular localization and functional interactions of PilMNOWQ and PilA4 involved in transformation competency and pilus biogenesis in the thermophilic bacterium Thermus thermophilus HB27. FEBS J. 273:3261-3272. [DOI] [PubMed] [Google Scholar]
- 25.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 103:6712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Silipo, A., A. Molinaro, L. Sturiale, J. M. Dow, G. Erbs, R. Lanzetta, M. A. Newman, and M. Parrilli. 2005. The elicitation of plant innate immunity by lipooligosaccharide of Xanthomonas campestris. J. Biol. Chem. 280:33660-33668. [DOI] [PubMed] [Google Scholar]
- 27.Taguchi, F., Y. Ogawa, K. Takeuchi, T. Suzuki, K. Toyoda, T. Shiraishi, and Y. Ichinose. 2006. A homologue of the 3-oxoacyl-(acyl carrier protein) synthase III gene located in the glycosylation island of Pseudomonas syringae pv. tabaci regulates virulence factors via N-acyl homoserine lactone and fatty acid synthesis. J. Bacteriol. 188:8376-8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang, D. J., Y. Q. He, J. X. Feng, B. R. He, B. L. Jiang, G. T. Lu, B. Chen, and J. L. Tang. 2005. Xanthomonas campestris pv. campestris possesses a single gluconeogenic pathway that is required for virulence. J. Bacteriol. 187:6231-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang, J. L., Y. N. Liu, C. E. Barber, J. M. Dow, J. C. Wootton, and M. J. Daniels. 1991. Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris. Mol. Gen. Genet. 226:409-417. [DOI] [PubMed] [Google Scholar]
- 30.Tsuge, S., A. Furutani, R. Fukunaka, Y. Kubo, and O. Horino. 2001. Growth complementation of hrpXo mutants of Xanthomonas oryzae pv. oryzae by virulent strains in rice cultivars resistant and susceptible to the parental strain. J. Gen. Plant Pathol. 67:51-57. [Google Scholar]
- 30a.West, N. P., P. Sansonetti, J. Mounier, R. M. Exley, C. Parsot, S. Guadagnini, M. C. Prevost, A. Prochnicka-Chalufour, M. Delepierre, M. Tanguy, and C. M. Tang. 2005. Optimization of virulence functions through glucosylation of Shigella LPS. Science 307:1313-1317. [DOI] [PubMed] [Google Scholar]
- 31.Yamashita, Y., Y. Tsukioka, K. Tomihisa, Y. Nakano, and T. Koga. 1998. Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J. Bacteriol. 180:5803-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]