Abstract
Listeria monocytogenes HrcA and CtsR negatively regulate class I and III stress response genes, respectively, while σB positively regulates the transcription of class II stress response genes. To define the HrcA regulon and identify interactions between HrcA, CtsR, and σB, we characterized newly generated L. monocytogenes ΔhrcA, ΔctsR ΔhrcA, and ΔhrcA ΔsigB strains, along with previously described ΔsigB, ΔctsR, and ΔctsR ΔsigB strains, using phenotypic assays (i.e., heat resistance, acid resistance, and invasion of human intestinal epithelial cells) and performed whole-genome transcriptome analysis of the ΔhrcA strain. The hrcA and sigB deletions had significant effects on heat resistance. While the hrcA deletion had no significant effect on acid resistance or invasion efficiency in Caco-2 cells, a linear regression model revealed a significant (P = 0.0493) effect of interactions between the hrcA deletion and the ctsR deletion on invasiveness. Microarray-based transcriptome analyses and promoter searches identified (i) 25 HrcA-repressed genes, including two operons (the groESL and dnaK operons, both confirmed as HrcA regulated by quantitative real-time PCR) and one gene directly repressed by HrcA, and (ii) 36 genes that showed lower transcript levels in the ΔhrcA strain and thus appear to be indirectly upregulated by HrcA. A number of genes were found to be coregulated by either HrcA and CtsR (2 genes), HrcA and σB (31 genes), or all three regulators (5 genes, e.g., gadCB). Combined with previous evidence that σB appears to directly regulate hrcA transcription, our data suggest that HrcA and σB, as well as CtsR, form a regulatory network that contributes to the transcription of a number of L. monocytogenes genes.
Listeria monocytogenes is a gram-positive food-borne pathogen that can cause severe invasive disease in humans, as well as in a number of different animal species (31). The capacity of L. monocytogenes to survive and multiply under a wide range of environmental stress conditions appears to be critical for the food-borne transmission of this pathogen (10). A number of transcriptional regulators (e.g., PrfA, σB, HrcA, and CtsR) that are important for the transcription of stress response and virulence genes have been identified in this organism (20, 25, 32, 37). While clear evidence for interactions between PrfA and σB has been reported (26, 40, 45), our understanding of interactions among other L. monocytogenes transcriptional regulators is limited. As no L. monocytogenes hrcA null mutant appears to have previously been reported, our understanding of the contributions of the negative regulator HrcA to stress response, transcriptional regulation, and regulatory networks is limited. In a number of gram-positive bacteria, including Bacillus subtilis, HrcA (heat regulation at CIRCE) has been found to repress the dnaK and groESL operons by binding to a region designated as the controlling inverted-repeat chaperone expression (CIRCE) element (38). Sequence analyses in L. monocytogenes also identified putative CIRCE elements upstream of hrcA (20) and of groES (13), which suggests that HrcA represses the dnaK operon (i.e., hrcA-grpE-dnaKJ-lmo1471-lmo1470) and the groESL operon in L. monocytogenes. Previous studies showed that insertional inactivation of L. monocytogenes dnaK resulted in decreased acid resistance and a growth defect at temperatures of 37°C or higher (19), suggesting contributions of HrcA to acid and temperature stress resistance. Transcription of the putative HrcA-dependent groESL was also found to be upregulated in L. monocytogenes exposed to selected stress conditions (e.g., heat) and in intracellular bacteria, further supporting the idea that groESL (and thus, HrcA) may contribute to L. monocytogenes stress response and virulence (13).
The alternative sigma factor σB directly regulates the transcription of a large regulon in L. monocytogenes (25) and appears to play a central role in regulating the transcription of L. monocytogenes stress response and virulence genes (25, 26, 30, 36). The characterization of sigB null mutants has also shown that L. monocytogenes σB contributes to survival under a variety of stress conditions, including acid and oxidative stresses and during carbon starvation (3, 10, 35). CtsR (class three stress gene repressor) is a transcriptional repressor which regulates at least 64 genes in L. monocytogenes, including directly repressing at least 10 genes (e.g., clpC, clpB, and clpP) (21, 37). Phenotypic characterization of different ctsR mutants also showed that L. monocytogenes CtsR contributes to virulence and survival under a variety of stress conditions, including heat, acid, oxidative, and high-pressure stress (6, 22, 23, 24).
In order to characterize the contributions of HrcA to L. monocytogenes stress response and virulence, including the potential interactions between HrcA and the HrcA regulon and other stress response regulators (i.e., CtsR and σB), we generated a series of isogenic single and double hrcA, sigB, and ctsR mutations in L. monocytogenes for phenotypic characterization and transcriptome analysis.
MATERIALS AND METHODS
Bacterial strains.
L. monocytogenes 10403S and isogenic mutant strains were used in this study (Table 1); ΔhrcA, ΔhrcA ΔsigB, and ΔctsR ΔhrcA strains have not previously been reported. The isogenic internal deletion mutants were constructed as previously detailed (7) by using the allelic exchange mutagenesis approach first reported by Smith and Youngman (46). Briefly, splicing by overlap extension PCR (see Table S1 in the supplemental material for primers) was used to construct an ΔhrcA allele with an in-frame 745-bp deletion within the hrcA open reading frame (ORF), which was cloned into pKSV7, yielding plasmid pBMB-28. This mutant allele was introduced into L. monocytogenes 10403S, as well as into previously constructed ΔsigB and ΔctsR strains, yielding ΔhrcA, ΔhrcA ΔsigB, and ΔctsR ΔhrcA strains (Table 1). The hrcA null mutation was confirmed in all strains by PCR amplification and direct sequencing of the PCR product (see Table S1 in the supplemental material).
TABLE 1.
L. monocytogenes strains used in this study
| Strain | Characteristicsa | Reference |
|---|---|---|
| 10403S | Serotype 1/2a parent strain | 4 |
| FSL A1-254 | 10403S ΔsigB | 51 |
| FSL H6-190 | 10403S ΔctsR | 21 |
| FSL H6-193 | 10403S ΔctsR ΔsigB | 21 |
| FSL B2-101 | 10403S ΔhrcA | This study |
| FSL H6-194 | 10403S ΔhrcA ΔsigB | This study |
| FSL H6-198 | 10403S ΔctsR ΔhrcA | This study |
The hrcA null mutation represents a 745-bp deletion within the hrcA ORF; this deletion allele encodes a 98-amino-acid nonsense protein that includes the first 17 and last 81 amino acids of the native HrcA protein.
The hrcA null mutant constructed here was designed to maintain the S2 transcriptional start site, which is located in the 3′ end of hrcA (20), to ensure that the hrcA deletion would not interfere with the transcription of the downstream genes grpE and dnaK. Maintenance of the S2 transcriptional start site (as well as 177 nucleotides upstream of S2) appears to be important, as an initial hrcA mutant that included a deletion of this region showed reduced growth at 37°C, which is the same phenotype previously reported for a dnaK insertional mutant (19). The hrcA mutant used here (FSL B2-101), which did not include a deletion of the S2 region, showed no reduction in growth at 37°C compared to the growth of the isogenic parent strain.
All phenotypic characterization experiments (see below) were performed using L. monocytogenes cells grown to early stationary phase, as initial quantitative real-time (qRT-PCR) showed that the HrcA-dependent genes dnaK and groES showed HrcA-dependent repression only in stationary-phase cells (see Results for details). Specifically, for all phenotypic characterization experiments, early-stationary-phase cells (i.e., bacterial cells incubated for an additional 1 h after they were grown to an optical density at 600 nm [OD600] of 0.8 at 37°C) were used to allow direct comparison with the results of previous phenotypic characterization experiments conducted in our laboratory (e.g., see reference 14), where L. monocytogenes cells were also grown to early stationary phase. For microarray experiments, on the other hand, stationary-phase cells (bacterial cells incubated for an additional 3 h after being grown to an OD600 of 1.0) were used to allow comparison with the results of other microarray studies (see, e.g., reference 39a).
Heat stress experiment.
Heat survival experiments were conducted to evaluate the heat resistance of the parent strain 10403S and all six mutant strains (Table 1). For these experiments L. monocytogenes cells were grown at 37°C with shaking to early stationary phase in 5 ml of brain heart infusion (BHI) in 16- by 125-mm disposable borosilicate glass culture tubes. Exposure to 55°C was performed by placing the tubes with the stationary-phase L. monocytogenes cells into a 55°C water bath, followed by incubation in the 55°C water bath (without shaking) for 30 or 60 min; initial temperature measurements showed that the broth reaches a temperature of 55°C within 158 s (standard deviation, 17 s; four replicate measurements). Bacterial numbers (log CFU/ml) were determined prior to and after heat treatment by plating on BHI agar plates using a spiral plater (Autoplate 4000; Spiral Biotech, Inc., Norwood, MA). Survival was expressed as the log reduction in the survival rate, which was calculated by subtracting the bacterial numbers (in log CFU/ml) before heat treatment from the bacterial numbers (in log CFU/ml) after heat treatment. Three independent experiments were performed.
Acid stress experiment.
To evaluate the acid resistance of parent strain 10403S and all six mutant strains, early-stationary-phase cells were exposed to acid stress (by adjusting the BHI to pH 2.5 with 12 N HCl) for 1 and 2 h at 37°C with shaking (230 rpm). Bacterial numbers (in log CFU/ml) prior to and after acid treatment were determined by plating on BHI agar as described above. Survival was expressed as the log reduction in the survival rate, which was calculated by subtracting the bacterial numbers (in log CFU/ml) before acid exposure from the bacterial numbers (in log CFU/ml) after acid exposure. Three independent experiments were performed.
Invasion assay.
The invasiveness in human intestinal epithelial cells of the parent strain and all mutant strains was determined using Caco-2 cells (ATCC HTB-37) as previously described (39) and bacterial cells grown to early stationary phase. Confluent Caco-2 monolayers grown in a 24-well plate were inoculated with 10 μl L. monocytogenes cells (3 wells/strain). After incubation for 30 min, Caco-2 cells were washed three times with phosphate-buffered saline, and at 45 min postinoculation, fresh medium containing gentamicin (150 μg/ml) was added to kill extracellular bacteria. At 90 min postinoculation, Caco-2 cells were washed three times with phosphate-buffered saline and then lysed by the addition of ice-cold sterile distilled water and vigorous pipetting. Intracellular L. monocytogenes numbers were determined by plating lysed Caco-2 cell suspensions on BHI agar. The invasion efficiency was calculated as the number of intracellular bacteria (in log CFU) recovered relative to the bacterial numbers (in log CFU) used for the inoculation. Invasion efficiencies were normalized to those of the parent strain, for which invasion efficiency was set at 100%. Three independent invasion assays were performed for each L. monocytogenes strain tested.
Microarray.
To identify HrcA-regulated genes, whole-genome transcriptome analyses were performed to compare transcript levels between the parent strain and the ΔhrcA strain. Microarray construction, RNA isolation and purification, cDNA labeling, and hybridization were performed as previously described (5, 21, 39a). Briefly, RNA for microarrays was isolated from stationary-phase cells using RNAprotect bacteria reagent and an RNeasy Midi kit (QIAGEN, Valencia, CA). DNase treatment of RNA was performed essentially as described previously (25), using 40 U of DNase. Isolated RNA (in RNase-free water) was quantified and checked for purity using OD260 and OD280 measurements performed on a Nanodrop spectrophotometer (Nanodrop Technologies, Inc., Wilmington, DE). Agarose gel electrophoresis was used to verify RNA integrity.
Microarrays were constructed using 70-mer oligonucleotides targeting 2,857 L. monocytogenes ORFs (QIAGEN operon array-ready oligonucleotide sets) identified in the annotated genome sequence of L. monocytogenes EGD-e (16). 70-mer oligonucleotides targeting five Saccharomyces cerevisiae ORFs (act1, mfa1, mfa2, ras1, and ste3) were included on the microarray to serve as nonhybridizing controls, as described previously (52). L. monocytogenes strains EGD-e and 10403S both represent the same L. monocytogenes lineage (II), serotype (1/2a), and ribotype (DUP-1039C), and probes designed based on the EGD-e genome were thus expected to hybridize well with 10403S genes (5). As an unfinished genome sequence for strain 10403S has recently become available (1), we also verified cross-hybridization identities (CHI) between the EGD-e probes and the target genes in strain 10403S; totals of 2,107, 2,578, and 2,695 of the EGD-e probes showed CHI values of 100, ≥95, and ≥90; 45 probes showed CHI values of <90 (5). A total of 117 of the EGD-e-based microarray probes were not detected in the 10403S genome, likely reflecting either sequence divergence in 10403S, deletion of these genes in 10403S, or genes located in the gaps of the unfinished 10403S genome sequence (5). We thus were confident that the array used here would allow for comprehensive identification of differentially expressed genes in strain 10403S, with the possibility of some false negatives (i.e., for genes targeted by a probe with a low CHI or for genes present in the 10403S genome and absent in the EGD-e genome) (5). Mismatches to selected target genes would be unlikely to yield false positives (i.e., genes identified as differentially regulated even if they were not) since the same mismatches would occur with RNA from the parent strain and the ΔhrcA strain (5).
cDNA was synthesized and differentially labeled by using a SuperScript Plus indirect cDNA labeling system (Invitrogen, Carlsbad, CA). Briefly, cDNA was generated from 10 μg of RNA using random primers in an overnight reverse transcription reaction at 42°C. cDNA was purified by using a QIAGEN PCR purification kit prior to indirect labeling with Alexa Fluor 555 or Alexa Fluor 647 fluorescent dyes (performed overnight at room temperature). Labeled cDNA was purified by using a QIAGEN PCR purification kit to remove any unincorporated dye, and the labeled cDNA was quantified and checked for purity using OD260 and OD280 measurements. For each microarray, cDNA from the parent and the ΔhrcA strain were combined into one tube, dried, and then resuspended in hybridization buffer containing sodium dodecyl sulfate (SDS), sodium chloride-sodium citrate (SSC), salmon sperm DNA, dithiothreitol, and formamide. The combined cDNA target (in a 50-μl volume) was overlaid onto the microarray slides using mSeries LifterSlips (Erie Scientific, Portsmouth, NH). Following overnight hybridization at 42°C, slides were washed in 2× SSC-0.1% SDS (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 5 min at 42°C, followed by sequential room temperature washes in 2× SSC-0.1% SDS, 2× SSC, and 0.2× SSC for 5 min each. Slides were centrifuged to dry them and scanned with a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA). All microarray experiments were performed three times using three independent RNA isolations to provide true biological replicates.
Scanned microarray images were gridded using GenePix Pro 6.0 software. Raw image analysis data were preprocessed, and significant differences in gene expression patterns between strains of interest were determined using the LIMMA software package (47) from R/BioConductor (15). Following background correction with the normexp method, within-array normalization (print-tip loess) and between-array normalization (scale) were used to correct for spatial and intensity bias and to make the results comparable across arrays. The LIMMA software package was also used for differential expression analysis (48) to calculate moderated t and B statistics and P values (adjusted for multiple comparisons by controlling for the false discovery rate). Genes with an adjusted P value of <0.05 were considered statistically significant, and an n-fold change of ≥1.5 was used as the cutoff for the identification of differentially expressed genes. Genes that showed significantly different transcript levels between the parent and the ΔhrcA strain were considered putative HrcA-regulated genes.
HMM searches.
Potential HrcA binding sites were determined by Hidden Markov model (HMM) searches as previously described (25). The HMM training alignments included 57 unique HrcA binding sites (identified upstream of hrcA and groES in different gram-positive bacteria), which were retrieved from the Comprehensive Microbial Resource at the J. Craig Venter Institute (http://cmr.tigr.org). Forward and reverse HMM models were generated to search for HrcA binding sites on the plus and minus strands of the published EGD-e genome (16). Outputs were filtered, and only hits within 300 bp upstream of a start codon for an ORF, as annotated by Listilist (http://genolist.pasteur.fr/ListiList), and with an E value of <0.01 were considered to be putative HrcA binding sites.
qRT-PCR.
RNA isolation for TaqMan qRT-PCR was performed as described previously (30). qRT-PCR primers and probes for dnaK and groES (see Table S2 in the supplemental material) were designed using PrimerExpress. qRT-PCR with these primer/probe sets, as well as primer and probe sets for the housekeeping genes rpoB and gap (45, 49), was performed using RNA isolated from the L. monocytogenes parent and ΔhrcA strain (grown to stationary phase) to confirm HrcA-dependent repression of these two genes. qRT-PCR was performed by using an ABI prism 7000 sequence detection system (Applied Biosystems, Foster City, CA) as detailed previously (30, 49). Reverse transcriptase negative control reactions, DNA standard curves, and analysis of qRT-PCR were also performed as described previously (30). Relative cDNA copy numbers were calculated as the log cDNA copy numbers for the target gene relative to the geometric mean of the cDNA copy numbers for the housekeeping genes rpoB and gap {i.e., log10 target gene − [(log10 rpoB + log10 gap)/2]}. qRT-PCR was repeated three times using three independent RNA isolations from cells grown on different days.
Statistical analysis.
All statistical analyses were performed using SAS (SAS onlineDoc8 version 8, SAS Institute, Inc., Cary, NC). qRT-PCR data for the parent strain and the ΔhrcA strain were analyzed by using a t test. Phenotypic data (heat survival, acid survival, and invasion efficiency) were analyzed using one-way analysis of variance with Duncan's multiple comparison procedure. In addition, the phenotypic data were also analyzed with linear regression models to test the effects of the three categorical variables (presence/absence of hrcA, ctsR, and sigB) and their interactions on a given phenotype; the final model was phenotype = hrcA + ctsR + sigB, where “phenotype” represents heat survival, acid survival, or invasion efficiency. For all tests, statistical significance was established as a P value of <0.05; significant P values are reported as the actual value, unless P is <0.0001.
Microarray data accession numbers.
Raw and normalized microarray data in MIAME format are available at the NCBI Gene Expression Omnibus (GEO) data repository under accession number GSE7517.
RESULTS
An hrcA deletion affects L. monocytogenes heat resistance.
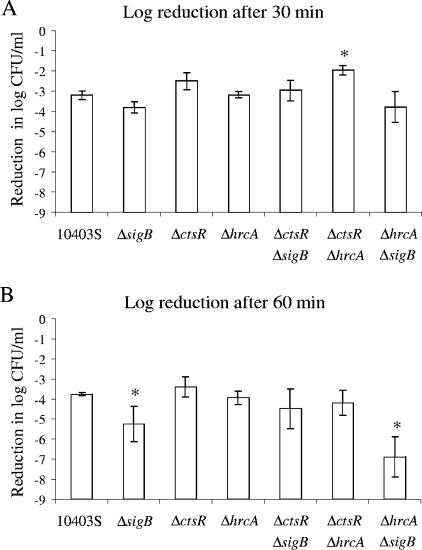
Analysis of heat survival rates for all strains after 30 and 60 min using a linear regression model (as detailed in Materials and Methods) showed (i) significant effects of the terms “ctsR deletion” and “sigB deletion” on survival after 30 min (P = 0.0003 and P = 0.0161, respectively) and (ii) significant effects of the terms “sigB deletion” and “hrcA deletion” on survival after 60 min (P < 0.0001 and P = 0.0110, respectively). The significant effect of the hrcA deletion on survival after 60 min, despite the observation that the ΔhrcA strain showed no significant difference in its survival rate compared to that of the parent strain (Fig. 1), likely reflects the fact that both ΔhrcA double mutants (i.e., the ΔctsR ΔhrcA and ΔhrcA ΔsigB strains) showed numerically reduced rates of survival compared to the survival rate of the paired null mutants with the hrcA wild-type allele (i.e., the ΔctsR and ΔsigB strains), which suggests that an hrcA deletion is associated with increased heat sensitivity in L. monocytogenes.
FIG. 1.
Heat survival of the L. monocytogenes parent strain (10403S) and the ΔsigB, ΔctsR, ΔhrcA, ΔctsR ΔsigB, ΔctsR ΔhrcA, and ΔhrcA ΔsigB strains after (A) 30 min or (B) 60 min of exposure to 55°C. Strains were grown to early stationary phase before exposure to 55°C. Survival was expressed as the log reduction, which was calculated by subtracting the bacterial numbers (in log CFU/ml) before heat exposure from the bacterial numbers (in log CFU/ml) after heat exposure. The data shown represent the averages of the results of three independent experiments; error bars indicate standard deviations. Overall analysis of variance showed a significant effect of the factor “strain” on heat survival after both 30 and 60 min. Duncan's multiple-comparison procedure was used to determine whether heat survival differed between specific strains. An asterisk indicates a strain with a log reduction in the survival rate that differed significantly (P < 0.05) from the survival rate of the parent strain; in addition, the ΔctsR ΔhrcA strain also showed a significantly higher survival rate than the ΔhrcA ΔsigB and the ΔsigB strains after 30 min of heat stress. After 60 min, the survival rate of the ΔhrcA ΔsigB strain was significantly lower than those of all other strains, and the survival rate of the ΔctsR strain was significantly higher than that of the ΔctsR ΔsigB strain.
The significant effect of the sigB null mutation on survival rates after 30 and 60 min appears to reflect increased heat sensitivity associated with this deletion, as the sigB null mutant strains consistently showed reduced survival compared to the survival rates of the paired strains without an sigB deletion, including significantly lower survival of the ΔsigB strain, compared to the survival rate of the parent strain after 60 min of exposure to 55°C (Fig. 1B). The significant effect of the ctsR null mutation on survival after 30 min appears to reflect increased heat resistance associated with this deletion, as the ctsR null mutant strains consistently showed higher survival rates than the paired strains without a ctsR deletion. These findings are consistent with those of other studies (22, 23), which also showed increased heat resistance of ctsR null mutant strains.
The linear regression model did not show any evidence for effects of interactions between the three factors (i.e., “hrcA deletion,” “ctsR deletion,” and “sigB deletion”) on heat survival.
An hrcA deletion shows no effect on L. monocytogenes acid resistance.
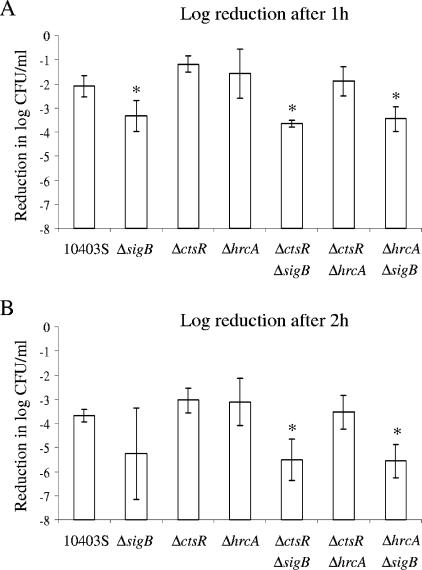
Analyses of L. monocytogenes survival after 1 and 2 h of exposure to pH 2.5 (Fig. 2) using a linear regression model showed no significant effect of either the term “hrcA deletion” or the term “ctsR deletion” on survival, indicating that acid resistance in early-stationary-phase cells is not affected by either the hrcA or the ctsR deletion. The term “sigB deletion,” on the other hand, had a significant effect on acid stress survival after both 1 and 2 h (P < 0.0001 and P = 0.0003, respectively), and the sigB null mutant strains consistently showed reduced acid stress survival rates compared to the survival rates of the paired strains without a sigB deletion (Fig. 2). These results are consistent with the well-documented importance of sigB in L. monocytogenes acid resistance (10, 11, 51).
FIG. 2.
Acid stress survival of the L. monocytogenes parent strain (10403S) and the ΔsigB, ΔctsR, ΔhrcA, ΔctsR ΔsigB, ΔctsR ΔhrcA, and ΔhrcA ΔsigB strains after (A) 1 h or (B) 2 h of exposure to acid stress (pH 2.5). Strains were grown to early stationary phase before exposure to pH 2.5. Duncan's multiple-comparison procedure was used to determine whether acid survival differed between specific strains; an asterisk indicates a strain with acid stress survival that differed significantly (P < 0.05) from that of the parent strain. Survival was expressed as the log reduction, which was calculated by subtracting the bacterial numbers (in log CFU/ml) before acid exposure from the bacterial numbers (in log CFU/ml) after acid exposure. The data shown represent the averages of the results of three independent experiments; error bars indicate standard deviations. A linear regression model to test the effects of the three categorical variables (presence/absence of hrcA, ctsR, and sigB) and their interactions on acid stress survival indicated a significant effect of the sigB deletion on acid stress survival (P value of <0.0001 and P value of 0.0003 for survival after 1 and 2 h, respectively).
The linear regression model did not show any evidence for effects of interactions between the three factors (i.e., “hrcA deletion,” “ctsR deletion,” and “sigB deletion”) on acid stress survival.
An hrcA deletion has limited effects on L. monocytogenes invasion efficiency.
Statistical analyses of Caco-2 cell invasion efficiencies for the L. monocytogenes parent strain and the six mutant strains using a linear regression model showed no significant effect of the term “hrcA deletion” but a significant effect of the terms “ctsR deletion” (P = 0.0154) and “sigB deletion” (P < 0.0001) on invasion efficiency. The sigB null mutant strains always showed significantly lower invasion efficiencies than the paired strains without a sigB deletion (i.e., the ΔsigB, ΔsigB ΔctsR, and ΔsigB ΔhrcA strains showed significantly lower invasion efficiencies than the parent, ΔctsR, and ΔhrcA strains, respectively) (Fig. 3), consistent with the well-documented role of sigB in Caco-2 cell invasion (14, 27). While the ΔctsR and parent strains showed similar invasion efficiencies, the ΔsigB ΔctsR and ΔctsR ΔhrcA strains showed lower invasion efficiencies than the corresponding mutant strains without a ctsR deletion (i.e., the ΔsigB and ΔhrcA strains) (Fig. 3), suggesting that a ctsR deletion is associated with reduced invasion efficiency, consistent with the results of previous reports (6).
FIG. 3.
Invasion efficiency in Caco-2 cells of the L. monocytogenes parent strain (10403S) and the ΔsigB, ΔctsR, ΔhrcA, ΔctsR ΔsigB, ΔctsR ΔhrcA, and ΔhrcA ΔsigB strains that were grown to early stationary phase in BHI at 37°C. The invasion efficiency was calculated as the number of intracellular bacteria (in CFU) recovered relative to the bacterial numbers (in CFU) used for the inoculation; invasion efficiencies were normalized to the invasion efficiency of one of the two parent strains included in each experiment (which was set at 100%). The data shown represent the averages of the results of three independent experiments; error bars indicate standard deviations. Duncan's multiple-comparison procedure was used to determine whether the invasion efficiency differed between specific strains; an asterisk indicates a strain with a percent invasion that differed significantly (P < 0.05) from that of the parent strain. A linear regression model used to test the effects of the three categorical variables (presence/absence of hrcA, ctsR, and sigB) and their interactions on invasion efficiency showed significant effects on invasion efficiency for the ctsR deletion (P = 0.0154), the sigB deletion (P < 0.0001), and the interaction between the hrcA and ctsR deletions (P = 0.0493).
The linear regression model showed significant (P = 0.0493) evidence for interactions between the factors “hrcA deletion” and “ctsR deletion” affecting invasiveness but showed no evidence for other interactions affecting it. The invasion efficiency of the ΔctsR ΔhrcA strain (83%) was numerically lower than the invasion efficiencies of both the ΔctsR and the ΔhrcA strain (102 and 109%, respectively), supporting contributions of hrcA and ctsR to L. monocytogenes' invasion efficiency.
Whole-genome microarray analysis identified 61 HrcA-regulated genes.
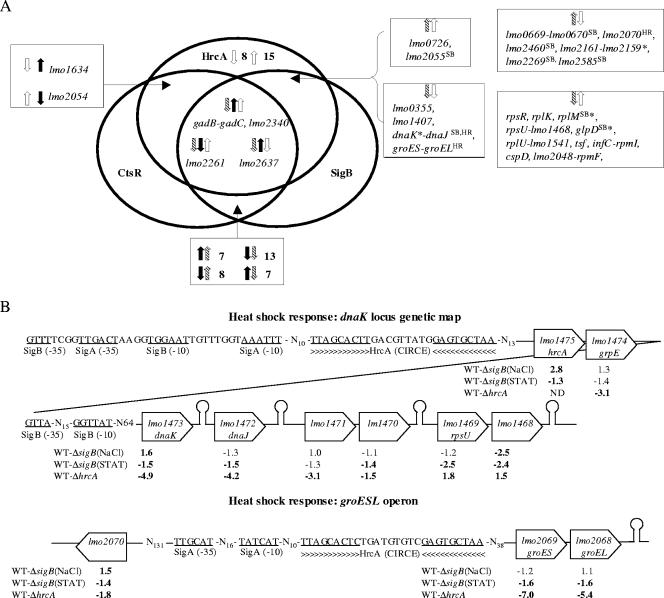
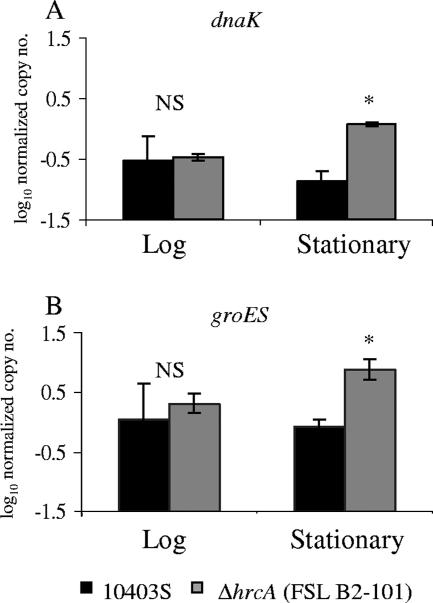
Microarray experiments comparing transcript levels in the L. monocytogenes parent strain and the ΔhrcA strain grown to stationary phase identified a total of 61 HrcA-regulated genes, including (i) 25 genes negatively regulated by HrcA (i.e., genes showing higher transcript levels in the ΔhrcA strain) (Table 2) and (ii) 36 genes that showed lower transcript levels in the ΔhrcA strain (Table 3). The “duplicateCorrelation” function in LIMMA was used to estimate gene-specific correlations across arrays; the overall estimate of the correlation across gene correlations was computed as 0.895 (this correlation was taken into account when fitting a linear model for each gene). While all genes that showed lower transcript levels in the ΔhrcA strain (indicating positive regulation by HrcA) are likely indirectly regulated by HrcA (as HrcA is a negative regulator), genes that show higher transcript levels in the ΔhrcA strain could be directly or indirectly repressed by HrcA. An HMM search identified HrcA binding sites upstream of two operons and one gene (see Fig. 5) that showed higher transcript levels in the ΔhrcA strain, representing a total of nine genes that are likely to be directly repressed by HrcA. The two operons identified as directly repressed by HrcA include the groES-groEL and the hrcA-grpE-dnaK-dnaJ-lmo1471-lmo1470 operon, which had both previously been hypothesized to be HrcA dependent in L. monocytogenes based on the identification of putative upstream HrcA binding sites by using DNA sequence analyses (13, 20). Interestingly, lmo2070, which was shown to be repressed by HrcA in the microarray data, is directly adjacent to the directly HrcA-repressed groES-groEL (lmo2069-lmo2068) operon and transcribed in the opposite direction; lmo2070 thus may share the HrcA binding site upstream of groES-groEL and may consequently be directly repressed by HrcA. HrcA-dependent repression of dnaK and groES was also confirmed by qRT-PCR (Fig. 4); both dnaK and groES showed significantly higher transcript levels in the ΔhrcA strain than in the parent strain in stationary-phase cells, while no significant differences in dnaK and groES transcript levels were found in ΔhrcA and parent strain cells grown to log phase (Fig. 4).
TABLE 2.
Genes identified by microarray analysis as downregulated by HrcAa
| Geneb | Differential expression in L. monocytogenes parent strain vs ΔhrcA strain
|
Protein function (specific gene name) | |
|---|---|---|---|
| Adjusted P value | Fold changec | ||
| lmo0117 | 0.0003 | −1.8 | LmaB, antigen B (lmaB) |
| lmo0124 | 0.0058 | −1.5 | Unknown |
| lmo0355 | 0.0013 | −1.6 | Similar to flavocytochrome C fumarate reductase chain A |
| lmo0641 | 0.0005 | −1.9 | Similar to heavy metal-transporting ATPase |
| lmo0669 | 0.0001 | −2.0 | Similar to oxidoreductase |
| lmo0670 | 0.0005 | −1.7 | Unknown |
| lmo1257 | 0.0010 | −2.8 | Unknown |
| lmo1293 | 0.0003 | −1.9 | Similar to glycerol 3-phosphate dehydrogenase (glpD) |
| lmo1407 | 0.0003 | −1.8 | Pyruvate formate lyase-activating enzyme (pflC) |
| lmo1474 | 0.0017 | −3.1 | Heat shock protein (grpE) |
| lmo1473 | 0.0017 | −4.9 | Heat shock protein (dnaK) |
| lmo1472 | 0.0003 | −4.2 | Heat shock protein (dnaJ) |
| lmo1471 | 0.0001 | −3.1 | Similar to ribosomal protein L11 methyltransferase |
| lmo1470 | 0.0027 | −1.5 | Unknown |
| lmo1634 | 0.0071 | −1.9 | Similar to alcohol-acetaldehyde dehydrogenase |
| lmo2069 | 0.0003 | −7.0 | Heat shock protein (groES) |
| lmo2068 | 0.0003 | −5.4 | Heat shock protein (groEL) |
| lmo2070 | 0.0185 | −1.8 | Unknown |
| lmo2161 | 0.0006 | −1.5 | Unknown |
| lmo2159 | 0.0010 | −1.6 | Similar to oxidoreductase |
| lmo2269 | 0.0039 | −1.6 | Unknown |
| lmo2460 | 0.0134 | −1.5 | Similar to B. subtilis CggR hypothetical transcriptional regulator |
| lmo2459 | 0.0046 | −1.5 | Glyceraldehyde 3-phosphate dehydrogenase (gap) |
| lmo2585 | 0.0004 | −1.6 | Similar to B. subtilis YrhD protein |
| lmo2637 | 0.0014 | −2.1 | Conserved lipoprotein |
All genes that showed lower transcript levels in the parent strain (i.e., an adjusted P value of <0.05 and n-fold change of ≤−1.5) in the microarray comparison are listed; these genes are either directly or indirectly downregulated by HrcA.
Putative operons are in bold font; gene names correspond to the gene designations for L. monocytogenes strain EGD-e (as listed on the ListiList server http://genolist.pasteur.fr/ListiList/).
A negative n-fold change indicates that the gene has lower transcript levels in the L. monocytogenes parent strain.
TABLE 3.
Genes identified by microarray analysis as showing lower transcript levels in the ΔhrcA straina
| Geneb | Differential expression in L. monocytogenes parent strain vs ΔhrcA strain
|
Protein function (specific gene name) | |
|---|---|---|---|
| Adjusted P value | Fold changec | ||
| lmo0001 | 0.0134 | 1.5 | Chromosomal replication initiation protein (dnaA) |
| lmo0044 | 0.0162 | 1.5 | Ribosomal protein S6 (rpsF) |
| lmo0045 | 0.0003 | 1.8 | Single-strand binding protein (ssb) |
| lmo0046 | 0.0042 | 1.5 | Ribosomal protein S18 (rpsR) |
| lmo0223 | 0.0366 | 1.5 | Cysteine synthase (cysK) |
| lmo0248 | 0.0004 | 1.9 | Ribosomal protein L11 (rplK) |
| lmo0249 | 0.0005 | 2.1 | Ribosomal protein L1 (rplA) |
| lmo0726 | 0.0012 | 1.9 | Unknown |
| lmo0727 | 0.0004 | 1.9 | Similar to l-glutamine-d-fructose-6-phosphate aminotransferase |
| lmo1059 | 0.0501 | 1.5 | Unknown |
| lmo1268 | 0.0003 | 1.7 | ATP-dependent Clp protease ATP-binding subunit ClpX (clpX) |
| lmo1469 | 0.0036 | 1.8 | 30S ribosomal protein S21 (rpsU) |
| lmo1468 | 0.0016 | 1.5 | Unknown |
| lmo1523 | 0.0006 | 1.5 | Similar to (p)ppGpp synthetase (relA) |
| lmo1542 | 0.0017 | 1.6 | Ribosomal protein L21 (rplU) |
| lmo1541 | 0.0273 | 1.8 | Unknown |
| lmo1657 | 0.0042 | 1.6 | Translation elongation factor (tsf) |
| lmo1658 | 0.0209 | 1.5 | 30S ribosomal protein S2 (rpsB) |
| lmo1683 | 0.0036 | 1.8 | Similar to transcription regulators (Fur family), PerR in B. subtilis |
| lmo1785 | 0.0008 | 1.7 | Translation initiation factor IF-3 (infC) |
| lmo1784 | 0.0017 | 1.6 | Ribosomal protein L35 (rpmI) |
| lmo1859 | 0.0021 | 1.6 | Similar to transcriptional regulator, PilB family |
| lmo1879 | 0.0209 | 1.5 | Similar to cold shock protein (cspD) |
| lmo1921 | 0.0079 | 1.6 | Unknown |
| lmo1956 | 0.0003 | 2.1 | Transcriptional regulator, Fur family (fur) |
| lmo2048 | 0.0006 | 1.7 | Unknown |
| lmo2047 | 0.0013 | 1.6 | Ribosomal protein L32 (rpmF) |
| lmo2055 | 0.0007 | 1.6 | Unknown |
| lmo2054 | 0.0032 | 1.5 | Unknown |
| lmo2261 | 0.0042 | 1.6 | Unknown |
| lmo2340 | 0.0002 | 1.6 | Similar to Erwinia chrysanthemi IndA protein |
| lmo2362 | 0.0032 | 1.8 | Amino acid antiporter (gadC) |
| lmo2363 | 0.0010 | 2.3 | Glutamate decarboxylase (gadB) |
| lmo2426 | 0.0032 | 1.7 | Unknown |
| lmo2478 | 0.0005 | 1.6 | Thioredoxin reductase (trxB) |
| lmo2597 | 0.0048 | 1.9 | Ribosomal protein L13 (rplM) |
All genes that showed evidence for lower transcript levels in the ΔhrcA strain in the microarray comparison are listed (adjusted P value of <0.05; n-fold change of ≥1.5); as HrcA is a negative regulator, genes downregulated in the ΔhrcA strain are most likely indirectly affected by HrcA.
Putative operons are in bold font; gene names correspond to the gene designations for L. monocytogenes strain EGD-e (as listed on the ListiList server http://genolist.pasteur.fr/ListiList/).
Positive changes indicate genes that have higher transcript levels in the L. monocytogenes parent strain.
FIG. 5.
Genes and operons regulated by HrcA, CtsR, and σB. Genes that showed significant and >1.5-fold difference in transcript levels were considered to be affected by a given regulator. (A) Venn diagram summarizing genes and operons that showed transcript levels that were found in microarray experiments to be significantly affected by either the hrcA deletion (this study), a ctsR deletion (21), or a sigB deletion (39a); ↑ and ↓ indicate genes that are up- or downregulated by a given regulator; color and shading of broad arrows indicate genes regulated by different regulators (CtsR [black], HrcA [white], and σB [hatched]). Genes preceded by a σB consensus promoter or a consensus HrcA binding site are marked with superscript SB or HR, respectively. Asterisks indicate genes that showed different transcription patterns in the two microarray comparisons conducted with the ΔsigB strain (i.e., comparisons conducted with log-phase cells exposed to 0.3 M NaCl for 10 min and comparisons conducted with cells grown to stationary phase); for these genes, the trends shown represent those found in L. monocytogenes grown to stationary phase (e.g., dnaK, rplM, and glpD were found to be downregulated by σB in stationary-phase cells but were found to be upregulated by σB in log-phase cells exposed to 0.3 M NaCl; the lmo2161-lmo2159 operon is upregulated by σB in stationary-phase cells but downregulated by σB in log-phase cells exposed to 0.3 M NaCl). (B) Selected genes and operons regulated by both HrcA and σB. Data for σB genes were taken from reference 39a. Putative σB-dependent promoters and HrcA binding sites, as well as putative terminators (shown as stem-loop structures), are indicated. Transcript ratios determined in the microarray experiments are indicated below a given gene; severalfold differences in transcript levels between parent strain (wild type [WT]) and ΔsigB strain were determined either in log-phase cells exposed to salt [WT-ΔsigB(NaCl)] or in stationary-phase cells [WT-ΔsigB(STAT)]. Positive numbers indicate that transcript levels were higher in the parent strain (indicating positive regulation by σB), while negative numbers indicate that transcript levels were lower in the parent strain (indicating negative regulation by σB). For HrcA, severalfold differences in transcript levels were determined between the parent strain (WT) and the ΔhrcA strain grown to stationary phase (WT-ΔhrcA); negative numbers indicate that transcript levels were lower in the parent strain (indicating negative regulation by HrcA), while positive numbers indicate that transcript levels were higher in the parent strain, indicating indirect positive regulation by HrcA. n-fold differences with a P value of <0.05 are in bold font. ND, not determined (as hrcA is absent in the ΔhrcA strain).
FIG. 4.
Transcript levels for dnaK and groES in the parent strain (10403S) and the ΔhrcA strain grown to (A) log phase (OD600 of 0.4) or (B) stationary phase in BHI at 37°C. Transcript levels were determined by qRT-PCR and are expressed as the log cDNA copy numbers normalized to the geometric mean of cDNA copy numbers for the housekeeping genes rpoB and gap (i.e., log10 target gene − [(log10 rpoB + log10 gap)/2], indicated as “log10 normalized copy no.”). The values shown represent the averages of the results of qRT-PCR assays performed on three independent RNA collections; error bars show standard deviations. For a given gene, an asterisk indicates transcript levels of the ΔhrcA strain that differed significantly (P < 0.05) from those of the parent strain by t test. NS, no significant difference between the parent and ΔhrcA strains for the given gene's transcript levels.
Genes that appear to be indirectly repressed by HrcA (i.e., genes without a putative upstream HrcA binding site) included at least three operons (lmo0669-lmo0670, lmo2460-lmo2459, and lmo2161-lmo2159) (Table 2), as well as a number of individual genes. Interestingly, genes indirectly repressed by HrcA include lmo2460 (encoding a putative transcriptional regulator with 75.4% similarity to B. subtilis CggR) and a number of putative stress response genes, including the PerR-regulated gene lmo0641, as well as lmo0669 and lmo2159, which encode proteins with 62.0% and 55.7% similarity to the Oceanobacillus iheyensis oxidoreductase OB1186 and the Geobacillus kaustophilus oxidoreductase GK2114, respectively, suggesting a role for HrcA in regulating oxidative stress response. In addition, a number of HrcA-repressed genes encode proteins that contribute to metabolism, including lmo1293 (glpD, encoding a putative glycerol-3-phosphate dehydrogenase with 74.2% amino acid similarity to B. subtilis GlpD), lmo1407 (pflC, encoding a putative pyruvate formate lyase-activating enzyme), and lmo1634 (encoding a putative alcohol-acetaldehyde dehydrogenase). Finally, lmaB, which is located in the lmaDCBA operon and encodes L. monocytogenes antigen B (17, 43), as well as one downstream gene (lmo0124), also showed a higher transcript level in the ΔhrcA strain; overall, all genes located in this region (i.e., lmo0117 to lmo0129) showed higher transcript levels in the ΔhrcA strain, although not meeting the criterion of a P value of <0.05 and/or the ≥1.5-fold-change cutoff, suggesting that this operon may also be indirectly repressed by HrcA.
The 36 genes that appeared to be indirectly upregulated by HrcA (as supported by lower transcript levels in the ΔhrcA strain) include 10 genes encoding ribosomal proteins (rpsF, rpsR, rplK, rplA, rpsU, rplU, rpsB, rpmI, rpmF, and rplM) (Table 3), as well as a number of genes encoding proteins contributing to DNA replication, transcription, or translation (dnaA, ssb, infC, and tsf) (Table 3). Additional genes that appear to be indirectly upregulated by HrcA encode proteins contributing to virulence and stress response (relA and perR, as well as the PerR-regulated fur and trxB), acid stress resistance (gadC and gadB), heat stress (clpX), and cold stress response (lmo1879 [cspD]), as well as proteins with unknown functions (Table 3).
Transcription of 38 HrcA-regulated genes is coregulated by σB or CtsR or both.
Analysis of the HrcA-regulated genes identified here in conjunction with the recently completed characterization of the σB regulon (39a) and the CtsR regulon (21) identified 38 genes as coregulated by either HrcA and CtsR (2 genes), HrcA and σB (31 genes), or all three regulators (5 genes) (Fig. 5). The five genes indirectly regulated by HrcA, σB, and CtsR include the gadC-gadB operon, which encodes a glutamate transporter and decarboxylase, respectively, that are important for acid resistance (8) (Fig. 5 and 6).
FIG. 6.
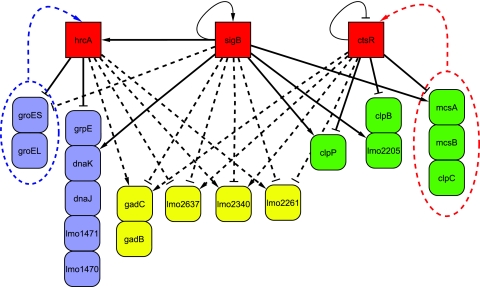
Partial HrcA, σB, and CtsR interaction network. The network is based on HrcA microarray data presented here, σB microarray data (39a), and CtsR microarray data (21). Solid lines indicate direct regulation of the gene by a given regulator as determined by the presence of an HrcA operator site, CtsR operator site, or σB promoter; dashed lines represent indirect regulation. Target arrows (↓) indicate positive regulation by a given regulator (as indicated by higher transcript levels in the parent strain than in the mutant strain); target stops (⊥) indicate negative regulation by a given regulator (as indicated by lower transcript levels in the parent strain than in the mutant strain). Loops indicate autoregulation. Color coding was used to identify (i) genes solely regulated by HrcA or dually regulated by HrcA and σB (blue), (ii) genes solely regulated by CtsR or dually regulated by CtsR and σB (green), and (iii) genes regulated by all three regulators (yellow). Genes arranged in vertical columns represent operons. The blue arrow targeting HrcA indicates posttranscriptional regulation of HrcA by proteins encoded by the groESL operon, based on evidence reported for B. subtilis (33). The red arrow targeting CtsR indicates posttranscriptional regulation of CtsR by McsA, McsB, and ClpC, based on evidence reported for B. subtilis (28, 29).
The observations that the whole dnaK operon appears to be directly repressed by HrcA (as supported by microarray data and the identification of an HrcA binding site upstream of hrcA [Fig. 5]) and that σB appears to directly regulate the transcription of the 5′ portion of the dnaK operon (i.e., hrcA-grpE-dnaK) (Fig. 5) in L. monocytogenes exposed to salt stress (as supported by microarray data and the identification of σB consensus promoter sequences upstream of hrcA and dnaK [S. Raengpradub, M. Wiedmann, and K. J. Boor, submitted]) provide evidence for a regulatory network involving these two regulators. Interestingly, σB-dependent regulation of hrcA was apparent only in log-phase L. monocytogenes cells exposed to 0.3 M NaCl, while in stationary-phase cells, the hrcA transcript levels were 1.3-fold higher in a ΔsigB null mutant (adjusted P < 0.05) (39a), suggesting growth phase-dependent effects on σB-dependent regulation of hrcA. Overlaps and interactions between σB- and HrcA-dependent regulation are further supported by the observation that 31 genes (including dnaK and dnaJ) (Fig. 5) are affected in their transcription by both hrcA and sigB deletions. While one would hypothesize that σB- and HrcA-dependent transcription patterns would follow the same trend in all genes if the only interaction between these two regulons is σB-dependent transcription of hrcA, genes whose transcription was found to be affected by both hrcA and sigB deletions represented (i) 14 genes positively regulated by HrcA and negatively regulated by σB, (ii) 7 genes negatively regulated by HrcA and negatively regulated by σB, (iii) 2 genes positively regulated by HrcA and positively regulated by σB, and (iv) 8 genes negatively regulated by HrcA and positively regulated by σB (including five genes with a putative σB-dependent promoter). These findings suggest complex interactions between the HrcA and σB regulons, including growth phase- and σB-dependent transcription of hrcA, as well as direct regulation by σB of other HrcA-regulated genes. Interestingly, the 14 genes positively regulated by HrcA and negatively regulated by σB predominantly encode ribosomal proteins (rpsR, rplK, rplM, rpsU, rplU, rpmI, and rpmF) and translation factors (tsf and infC), while the 8 genes negatively regulated by HrcA and positively regulated by σB include genes encoding proteins that potentially contribute to oxidative stress response (lmo0699 and lmo2159), as well as CggR, a transcriptional regulator highly similar to the B. subtilis regulator of central glycolytic genes (Fig. 5).
DISCUSSION
Initial evidence has shown that L. monocytogenes σB (class II stress response gene regulator), CtsR (class III stress response gene regulator), and PrfA interact and form regulatory networks important for the transcription of stress response and virulence genes in this food-borne pathogen (21, 26, 40, 45), suggesting that regulatory networks are likely to be critical for the appropriate expression of stress response and virulence genes in L. monocytogenes. However, our understanding of the contribution of the class I stress response gene regulator (HrcA) to transcriptional regulation and regulatory networks in L. monocytogenes is limited. To investigate the role of HrcA and the interactions between HrcA, CtsR, and σB in stress response and the expression of virulence and stress response genes in L. monocytogenes, we characterized a series of isogenic L. monocytogenes mutants, including nonpolar ΔsigB, ΔctsR, ΔhrcA, ΔctsR ΔsigB, ΔctsR ΔhrcA, and ΔhrcA ΔsigB strains, using phenotypic assays as well as microarray and qRT-PCR methods. Our data show that (i) while a total of 61 genes are regulated by HrcA, either directly or indirectly, including genes important for stress response and virulence, hrcA deletion has limited effects on heat and acid stress resistance or on invasion efficiency and that (ii) in addition to an overlap between the HrcA, CtsR, and σB regulons, σB also appears to contribute to direct regulation of hrcA transcription, indicating that HrcA and the HrcA regulon are part of an integrated network of transcriptional regulators contributing to stress response systems in L. monocytogenes.
While a total of 61 genes are regulated by HrcA, an hrcA deletion has limited effects on heat and acid stress resistance or on invasion efficiency.
Transcriptome analyses of the ΔhrcA null mutant, combined with HMM analyses, identified 61 genes as regulated by HrcA, including 9 genes directly repressed by HrcA. To our knowledge, this is the first whole-genome transcriptome study defining the HrcA regulon in any bacterium. While previous studies on HrcA-regulated genes in different gram-positive bacteria generally identified only the groES-EL and the dnaK operons as HrcA regulated (e.g., in B. subtilis [18, 44]), we identified an additional gene that appears to be directly regulated by HrcA (lmo2070, transcribed divergently from the groES-EL operon), as well as a number of additional genes whose transcription is affected by an hrcA null mutation and which appear to be indirectly regulated by HrcA. For example, genes indirectly repressed by HrcA (i.e., genes with higher transcript levels in the ΔhrcA strain, but with no apparent HrcA binding sites) include genes encoding proteins that contribute to oxidative stress response (i.e., lmo0669, lmo2159, and lmo0641). Genes indirectly activated by HrcA include genes encoding proteins that contribute to cold (cspD) (2), acid (gadC-B) (8), and, potentially, to salt (cysK) stress response (9). HrcA-regulated genes also encode a number of ribosomal proteins (RpsF, RpsR, RplK, RplA, RpsU, RplU, RpsB, RpmI, RpmF, and RplM), a single-strand binding protein (SSB), a chromosomal replication initiation protein (DnaA), translation initiation factor IF-3 (InfC), and a translation elongation factor (Tsf), suggesting that the adaptive expression of proteins contributing to DNA metabolism, transcription, and translation is necessary under certain stress conditions, including in stationary-phase cells (12).
While our data suggest that HrcA regulates the transcription of genes involved in responses to a variety of stresses in addition to heat stress, phenotypic characterization revealed a clear effect of the hrcA deletion only on L. monocytogenes heat resistance, with lower heat resistance associated with the hrcA deletion. In contrast, a B. subtilis ΔhrcA strain showed higher thermotolerance than the parent strain (44), suggesting possible differences in the overall regulation of HrcA and HrcA-dependent genes between L. monocytogenes and B. subtilis. While the hrcA deletion showed only a limited effect on invasion efficiency (i.e., interactions between the hrcA and ctsR deletions affected invasion efficiencies), HrcA was found to regulate genes with apparent roles in virulence, including relA, perR, and fur, which appear to contribute to the survival and growth of L. monocytogenes in a mouse model of infection (42, 50). Further evaluation of the isogenic hrcA null mutants using animal models will thus be necessary to pinpoint the specific contributions of HrcA to L. monocytogenes virulence.
HrcA and the HrcA regulon are part of an integrated network of transcriptional regulators contributing to stress response systems in L. monocytogenes.
Our data indicate that HrcA and the HrcA regulon are part of an integrated network of transcriptional regulators contributing to stress response systems in L. monocytogenes (Fig. 5 and 6). In particular, our data indicate considerable interactions and overlap between HrcA- and σB-dependent transcriptional regulation, including apparent σB-dependent transcription of hrcA by σB. These interactions appear complex, though as (i) both sigB (3, 40) and hrcA (20) also appear to autoregulate their own transcription, (ii) σB appears to directly regulate some HrcA-dependent genes, and (iii) the activities of both σB and HrcA are regulated posttranscriptionally (7, 33). For example, in B. subtilis, GroE, encoded by the HrcA-dependent groESL operon, appears to modulate HrcA activity (33). In addition, previous studies have also reported that certain genes may show σB-dependent transcription only under specific environmental stress conditions, including a number of genes that showed σB-dependent transcription only in L. monocytogenes exposed to salt stress, but not in stationary-phase bacteria (25). Overall, σB- and HrcA-dependent regulatory mechanisms thus appear to interact at transcriptional and posttranscriptional levels; these interactions are likely required to assure the appropriate expression of stress response systems, including heat shock response, in L. monocytogenes grown under different environmental conditions.
While we found only limited overlap between the HrcA and CtsR regulons, previous data have shown a considerable overlap between the CtsR regulon and the σB regulon, including regulation by σB of the mcsA-mscB-clpC operon (39a) which encodes proteins contributing to the posttranscriptional regulation of CtsR (28). Thus, σB appears to have a strong linkage to both the HrcA and CtsR regulons; these linkages appear not only to be important for heat resistance but also to contribute to the regulation of other stress response genes, including, for example, acid response, as supported by the fact that the transcription of the gadCB operon appears to be affected by CtsR, HrcA, and σB.
Interestingly, in addition to connections between HrcA and the HrcA regulon and σB- and CtsR-dependent transcriptional regulation, we also found initial evidence for linkages between HrcA-dependent regulation and other regulatory pathways, including the RelA, PerR, and CggR regulatory pathways. For example, perR and the PerR-dependent genes fur, trxB, and lmo0641 (41) showed HrcA-dependent transcription; PerR appears to regulate genes important for L. monocytogenes' response to oxidative stress and ethanol and alkaline pH stress resistance, as well as virulence (34, 42). HrcA also appears to affect the transcription of a gene encoding a protein highly similar to CggR, as well as putative CggR-dependent genes (i.e., gapA, which was found to be CggR dependent in B. subtilis [53]). While previous studies have shown significant overlaps and interactions between the PrfA regulon and σB, we identified only three PrfA-dependent genes that were also regulated by HrcA, including the lmo0669-lmo0670 operon, which encodes a putative oxidoreductase (lmo0669) and is also regulated by σB. While regulatory networks involving HrcA thus appear to be predominantly important for the regulation of responses to non-host-associated stress conditions (e.g., heat), HrcA also appears to contribute to the regulation of genes and networks with possible roles in L. monocytogenes virulence.
Supplementary Material
Acknowledgments
S. Raengpradub was supported by a USDA National Needs Fellowship grant (no. 2002-38420-11738 to K.J.B.). This work was also supported by a USDA National Research Initiative grant (no. 2005-35201-15330 to K.J.B.).
We thank B. Bowen for help with mutant construction.
Footnotes
Published ahead of print on 26 October 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anonymous. 2006. Listeria monocytogenes sequencing project. Broad Institute of Harvard and MIT. http://www.broad.mit.edu.
- 2.Bayles, D. O., B. A. Annous, and B. J. Wilkinson. 1996. Cold stress proteins induced in Listeria monocytogenes in response to temperature downshock and growth at low temperatures. Appl. Environ. Microbiol. 62:1116-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, L. A., M. S. Cetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 5.Chan, Y. C., S. Raengpradub, K. J. Boor, and M. Wiedmann. 2007. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl. Environ. Microbiol. 73:6484-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaturongakul, S., and K. J. Boor. 2004. RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 70:5349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotter, P. D., C. G. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 9.Duche, O., F. Tremoulet, P. Glaser, and J. Labadie. 2002. Salt stress proteins induced in Listeria monocytogenes. Appl. Environ. Microbiol. 68:1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreira, A., C. P. O'Byrne, and K. J. Boor. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira, A., D. Sue, C. P. O'Byrne, and K. J. Boor. 2003. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Folio, P., P. Chavant, I. Chafsey, A. Belkorchia, C. Chambon, and M. Hebraud. 2004. Two-dimensional electrophoresis database of Listeria monocytogenes EGDe proteome and proteomic analysis of mid-log and stationary growth phase cells. Proteomics 4:3187-3201. [DOI] [PubMed] [Google Scholar]
- 13.Gahan, C. G., J. O'Mahony, and C. Hill. 2001. Characterization of the groESL operon in Listeria monocytogenes: utilization of two reporter systems (gfp and hly) for evaluating in vivo expression. Infect. Immun. 69:3924-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garner, M. R., B. L. Njaa, M. Wiedmann, and K. J. Boor. 2006. Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect. Immun. 74:876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 17.Gohmann, S., M. Leimeister-Wachter, E. Schiltz, W. Goebel, and T. Chakraborty. 1990. Characterization of a Listeria monocytogenes-specific protein capable of inducing delayed hypersensitivity in Listeria-immune mice. Mol. Microbiol. 4:1091-1099. [DOI] [PubMed] [Google Scholar]
- 18.Grandvalet, C., G. Rapoport, and P. Mazodier. 1998. hrcA, encoding the repressor of the groEL genes in Streptomyces albus G, is associated with a second dnaJ gene. J. Bacteriol. 180:5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanawa, T., M. Fukuda, H. Kawakami, H. Hirano, S. Kamiya, and T. Yamamoto. 1999. The Listeria monocytogenes DnaK chaperone is required for stress tolerance and efficient phagocytosis with macrophages. Cell Stress Chaperones 4:118-128. [PMC free article] [PubMed] [Google Scholar]
- 20.Hanawa, T., M. Kai, S. Kamiya, and T. Yamamoto. 2000. Cloning, sequencing, and transcriptional analysis of the dnaK heat shock operon of Listeria monocytogenes. Cell Stress Chaperones 5:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, Y., S. Raengpradub, U. Schwab, C. Loss, R. Orsi, M. Wiedmann, and K. J. Boor. 12 October 2007. Phenotypic and transcriptomic analyses demonstrate interactions between the transcriptional regulators CtsR and sigma B in Listeria monocytogenes. Appl. Environ. Microbiol. doi: 10.1128/AEM.01085-07. [DOI] [PMC free article] [PubMed]
- 22.Joerger, R. D., H. Chen, and K. E. Kniel. 2006. Characterization of a spontaneous, pressure-tolerant Listeria monocytogenes Scott A ctsR deletion mutant. Foodborne Pathog. Dis. 3:196-202. [DOI] [PubMed] [Google Scholar]
- 23.Karatzas, K. A., and M. H. Bennik. 2002. Characterization of a Listeria monocytogenes Scott A isolate with high tolerance towards high hydrostatic pressure. Appl. Environ. Microbiol. 68:3183-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karatzas, K. A., J. A. Wouters, C. G. Gahan, C. Hill, T. Abee, and M. H. Bennik. 2003. The CtsR regulator of Listeria monocytogenes contains a variant glycine repeat region that affects piezotolerance, stress resistance, motility and virulence. Mol. Microbiol. 49:1227-1238. [DOI] [PubMed] [Google Scholar]
- 25.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kazmierczak, M. J., M. Wiedmann, and K. J. Boor. 2006. Contributions of Listeria monocytogenes σB and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology 152:1827-1838. [DOI] [PubMed] [Google Scholar]
- 27.Kim, H., K. J. Boor, and H. Marquis. 2004. Listeria monocytogenes σB contributes to invasion of human intestinal epithelial cells. Infect. Immun. 72:7374-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirstein, J., D. Zuhlke, U. Gerth, K. Turgay, and M. Hecker. 2005. A tyrosine kinase and its activator control the activity of the CtsR heat shock repressor in B. subtilis. EMBO J. 24:3435-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kruger, E., D. Zuhlke, E. Witt, H. Ludwig, and M. Hecker. 2001. Clp-mediated proteolysis in Gram-positive bacteria is autoregulated by the stability of a repressor. EMBO J. 20:852-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGann, P., M. Wiedmann, and K. J. Boor. 2007. The alternative sigma factor σB and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins. Appl. Environ. Microbiol. 73:2919-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milohanic, E., P. Glaser, J. Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 33.Mogk, A., G. Homuth, C. Scholz, L. Kim, F. X. Schmid, and W. Schumann. 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16:4579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mongkolsuk, S., and J. D. Helmann. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 45:9-15. [DOI] [PubMed] [Google Scholar]
- 35.Moorhead, S. M., and G. A. Dykes. 2004. Influence of the sigB gene on the cold stress survival and subsequent recovery of two Listeria monocytogenes serotypes. Int. J. Food Microbiol. 91:63-72. [DOI] [PubMed] [Google Scholar]
- 36.Nadon, C. A., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nair, S., I. Derre, T. Msadek, O. Gaillot, and P. Berche. 2000. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 35:800-811. [DOI] [PubMed] [Google Scholar]
- 38.Narberhaus, F. 1999. Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31:1-8. [DOI] [PubMed] [Google Scholar]
- 39.Nightingale, K. K., K. Windham, K. E. Martin, M. Yeung, and M. Wiedmann. 2005. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 71:8764-8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Raengpradub, S., M. Wiedmann, and K. J. Boor. Comparative analysis of the σB-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 40.Rauch, M., Q. Luo, S. Muller-Altrock, and W. Goebel. 2005. SigB-dependent in vitro transcription of prfA and some newly identified genes of Listeria monocytogenes whose expression is affected by PrfA in vivo. J. Bacteriol. 187:800-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rea, R., C. Hill, and C. G. Gahan. 2005. Listeria monocytogenes PerR mutants display a small-colony phenotype, increased sensitivity to hydrogen peroxide, and significantly reduced murine virulence. Appl. Environ. Microbiol. 71:8314-8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rea, R. B., C. G. Gahan, and C. Hill. 2004. Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect. Immun. 72:717-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaferkordt, S., and T. Chakraborty. 1997. Identification, cloning, and characterization of the lma operon, whose gene products are unique to Listeria monocytogenes. J. Bacteriol. 179:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulz, A., and W. Schumann. 1996. hrcA, the first gene of the Bacillus subtilis dnaK operon, encodes a negative regulator of class I heat shock genes. J. Bacteriol. 178:1088-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwab, U., B. Bowen, C. Nadon, M. Wiedmann, and K. J. Boor. 2005. The Listeria monocytogenes prfAP2 promoter is regulated by sigma B in a growth phase dependent manner. FEMS Microbiol. Lett. 245:329-336. [DOI] [PubMed] [Google Scholar]
- 46.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 47.Smyth, G. K. 2005. Limma: linear models for microarray data. In R. Gentleman, V. Carey, S. Dudoit, R. Irizarry, and W. Huber (ed.), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY.
- 48.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:1-25. [DOI] [PubMed] [Google Scholar]
- 49.Sue, D., D. Fink, M. Wiedmann, and K. J. Boor. 2004. σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150:3843-3855. [DOI] [PubMed] [Google Scholar]
- 50.Taylor, C. M., M. Beresford, H. A. Epton, D. C. Sigee, G. Shama, P. W. Andrew, and I. S. Roberts. 2002. Listeria monocytogenes relA and hpt mutants are impaired in surface-attached growth and virulence. J. Bacteriol. 184:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zorrilla, S., T. Doan, C. Alfonso, E. Margeat, A. Ortega, G. Rivas, S. Aymerich, C. A. Royer, and N. Declerck. 2007. Inducer-modulated cooperative binding of the tetrameric CggR repressor to operator DNA. Biophys. J. 92:3215-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.