Abstract
Human adenoviruses (HAdVs) have been related to several waterborne diseases such as acute gastroenteritis, conjunctivitis, and respiratory illness, and it has been shown that an important human exposure pathway is through recreational waters. However, HAdV occurrence at recreational freshwater beaches has not been previously investigated. In this study, a total of 58 water samples were collected from two recreational beaches on Lake Michigan (i.e., Silver Beach and Washington Park Beach) during the summer of 2004. Occurrences of HAdVs in these lake samples were determined using two hexon-based real-time PCR assays (one for monitoring all 51 serotypes of HAdVs and another for specifically detecting F species HAdVs, i.e., serotypes 40 and 41) and compared to an integrated cell culture (ICC) PCR method. The real-time PCR results showed that 8 of 30 Silver Beach samples and 6 of 28 Washington Park Beach samples contained HAdVs, and F species HAdVs were detected in three of these positive samples. The concentrations of HAdVs ranged from (1.7 ± 0.7) × 101 to (3.4 ± 0.8) × 102 and from (7 ± 2) × 100 to (3.8 ± 0.3) × 103 virus particles/liter for Silver Beach and Washington Park Beach, respectively. F species HAdVs were detected at levels ranging from (4.8 ± 0.8) × 101 to (4.6 ± 1.5) × 102 virus particles/liter. Approximately 60% of the ICC-PCR analyses agreed with the real-time PCR results. This study revealed the occurrence of HAdVs at Lake Michigan recreational beaches. Given the potential health risks, further assessment regarding sources, virus transport, and survival is needed to improve the safety of the region.
Recently, there has been much attention given to emerging viruses because of their low infectious dose, survival in water, and considerable health impacts (from diarrhea to death). Human adenoviruses (HAdVs) have been considered critical emerging viruses since the potential health risks associated with their waterborne transmission were noticed by the scientific community (18, 48). The HAdV serotypes 40 and 41 (HAdV40 and HAdV41, respectively) are critical etiological agents of viral gastroenteritis in children (12, 51). Adenoviruses are currently included in the contaminant candidate list of the U.S. Environmental Protection Agency. Waterborne outbreaks caused by or associated with HAdVs (e.g., acute gastroenteritis and conjunctivitis) have been documented (mostly in recreational swimming pools) (35, 37, 50, 54). Yet the occurrence of HAdVs in freshwater at recreational lake beaches has not been widely examined.
The use of reliable methods to precisely detect HAdVs is essential, particularly when human risk of exposure is assessed. Molecular methods have been applied to the detection of viruses due to their advantages of high speed, specificity, and sensitivity. Because HAdVs (especially types 40 and 41) do not produce consistent cytopathic effects in cell cultures, the use of cultivation-independent molecular techniques is even more crucial. Many PCR primer pairs have been designed and used to target the hexon gene of HAdVs in diverse water sources using conventional, nested, and/or multiplex PCR (2, 3, 5, 6, 13, 24, 32, 33, 34, 41, 44, 46, 53, 57). In addition, integrated cell culture PCR (ICC-PCR) techniques have been used to detect infectious HAdV (7, 9, 21, 29, 43). However, these methods do not allow direct quantification of HAdVs, impeding their relevance to health risk assessment (10). The latest advancement in molecular methods is the development of quantitative real-time PCR that relies upon the detection and quantification of a fluorescent reporter (e.g., 6-carboxyfluorescein [FAM], hexachloro-6-carboxyfluorescein, or 6-carboxytetramethylrhodamine) for which the signal intensity increase is proportional to the amount of PCR product amplified in the reaction. Until now, several hexon-based real-time PCR assays have been developed to detect generic HAdVs or particular HAdV groups (11, 16, 22, 23, 24, 26, 33). However, only a limited number of the presented assays have been applied to examine environmental water samples (22, 23, 24).
Although HAdVs have frequently been identified in various environments such as wastewater (19, 22, 39, 41), drinking waters (20, 30, 31, 55), groundwater (28, 40), surface waters (7, 15, 17, 25, 42, 44), and pool recreational waters (35, 38, 50, 54), quantitative information for HAdV occurrence at public lake beaches is still insufficient. The overall goal of this study was to gain insight into HAdV abundance in freshwaters at two popular recreational beaches on Lake Michigan using quantitative PCR assays. This investigation provides HAdV data for quantifying exposures and thus could be used in the future for assessing risks associated with recreational activities at Great Lakes beaches, with the long-term aim of preventing transmission of HAdV-related diseases.
MATERIALS AND METHODS
Sampling location.
Two recreational beaches (i.e., Silver Beach and Washington Park Beach) located on Lake Michigan were selected to assess the occurrence of HAdVs. Washington Park Beach is located at Michigan City, IN, with a public beach area of approximately 1,100 m wide. Beach water samples were taken at three sites (approximately 120 m apart) within a 360-m stretch that experienced the most foot traffic. Silver Beach is located at St. Joseph, MI. The public beach is approximately 600 m wide. Sampling was carried out at three positions inside a stretch of approximately 270 m which had the most foot traffic.
Sample collection, concentration, and elution.
For HAdV analysis, a total of 58 samples were collected from the two recreational beaches based on a concentration-elution approach (14, 52). Each sample represented a composite of lake water from three spatial locations. At each spatial location, 80 to 120 liters of lake water was first pumped into a clean 20-gal plastic barrel (disinfected previously with chlorine and neutralized with sodium thiosulfate), and the sample's pH was adjusted to a neutral level of 7.0 to 7.5 using 5 N hydrochloric acid. The beach waters from the three sampling sites were then sequentially filtered through a 1MDS filer to give a total sample volume of 250 to 350 liters. All filtered viral samples were held on ice, transported to the Water Quality Laboratory at Michigan State University, and eluted within 72 h.
Virus elution and further concentration were carried out by organic flocculation as modified from the U.S. Environmental Protection Agency's virus monitoring protocol for the Information Collection Requirements Rule (52). Briefly, the filters were backwashed twice with 0.5 liters of beef extract solution (1.5% [wt/vol] beef extract, 0.05 M glycine, pH 9.0 to 9.5) to elute absorbed viral particles. Subsequently, the eluates were flocculated by adding ferric chloride to a final concentration of 2.5 mM and by lowering the solution pH to 3.5 (38, 52). The flocs were collected by centrifugation at 2,500 × g for 15 min and resuspended in 30 ml of 0.15 M sodium phosphate (final pH of 9.0). The redissolved precipitates were centrifuged at 10,000 × g for 10 min. Finally, the supernatants (approximately 30 ml) were collected (pellet was discarded), neutralized (pH 7.0 to 7.5) with 1 M HCl, supplemented with 100 units of penicillin, 100 μg of streptomycin, and 0.25 μg of fungizone, and stored in aliquots at −80°C until analyses were conducted.
Nucleic acid extraction.
Viral nucleic acids were extracted from the concentrated lake samples and from the infected cell culture using a QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA) based on the spin protocol listed in the manufacturer's handbook. Several HAdV-positive samples were reextracted using two modified methods to attempt to eliminate environmental inhibitors with applications of extra washes for lysates prior to nucleic acid elution or for viral samples embedded in low-melting-point (LMP) agarose before lysis. Briefly, the lysates were further washed with 500 ml of 99.9% ethanol twice after the regular washing steps of the QIAamp protocol. Alternatively, the concentrated lake samples were embedded in 1 volume (i.e., 330 μl) of 1.6% (wt/vol) LMP agarose according to the conceptual approach of a previous publication (36). The solidified agarose blocks were washed twice with 1 ml of 1× Tris-EDTA buffer by shaking horizontally at 100 rpm for 1 h, and then the QIAamp protocol to extract viral nucleic acids was used. In addition, several selected extracts (which tested positive in real-time PCR) were further treated with ethanol precipitation (45), InhibitEX tablets (QIAamp DNA Stool mini kit; Qiagen, Valencia, CA), and a nucleic acid binding matrix solution (FastDNA kit; Qbiogene Inc., Morgan Irvine, CA) in order to eliminate possible environmental inhibitors that might adversely effect real-time PCR efficiency. All the extracts were stored in a freezer at −20°C for further detection.
Real-time PCR primers and probes.
The generic primers and TaqMan probe used for quantification of HAdVs were forward primer JTVXF (GGA-CGC-CTC-GGA-GTA-CCT-GAG), reverse primer JTVXR (ACI-GTG-GGG-TTT-CTG-AAC-TTG-TT), and probe JTVXP (6-FAM-CTG-GTG-CAG-TTC-GCC-CGT-GCC-A-BHQ) (26). This JTVXP assay amplifies 96 bp of hexon gene. Additionally, the hexon gene of F species HAdV (i.e., serotypes 40 and 41) was quantified with another real-time PCR assay using a primer/probe set modified from Jiang's (i.e., f-AD157/p-AD196/r-AD245) (24). The reverse primer and TaqMan probe were modified to enhance specificity according to the alignment of hexon gene sequences for the HAdV F species (GenBank accession numbers NC_001454, L19443, X51782, X51783, and D13781). The adapted primer/probe set is forward primer HAdV-F4041-hex157f (ACC-CAC-GAT-GTA-ACC-ACA-GAC), reverse primer (for serotype 40) HAdV-F40-hex245r (ACT-TTG-TAA-GAG-TAG-GCG-GTT-TC), reverse primer (for serotype 41) HAdV-F41-hex246r (CAC-TTT-GTA-AGA-ATA-AGC-GGT-GTC), and TaqMan probe HAdV-F4041-hex214rprobe (6-FAM-CGA-CKG-GCA-CGA-AKC-GCA-GCG-T-BHQ-1). This assay amplifies 88 to 89 bp of the hexon gene.
For sequence comparison in this study, 64 hexon gene sequences (Table 1) obtained from the National Center for Biotechnology Information GenBank were aligned using CLUSTAL X software (49). Sequences of seven published real-time PCR primer/probe sets (Table 2) were compared with these hexon gene sequences to assess their specificities. Furthermore, the published primers and probes were evaluated and/or modified according to the typical design guidelines provided by Applied Biosystems (Foster City, CA).
TABLE 1.
GenBank accession numbers for hexon genes used for sequence comparison in this study
| HAdV species | GenBank accession no. |
|---|---|
| A | NC_001460, AC_000005, DQ149610, DQ149611 |
| B | NC_001460, AY599834, AY594255, AF532578, DQ149612, AY601636, AY601633, AY737797, AC_000019, DQ149643 |
| C | NC_001405, AF534906, AC_000007, J01917, AF542118, AY224392, AY339865, AY601635, AF542130, AF542121, DQ149613 |
| D | NC_002067, DQ149614, AJ854486, DQ149615, DQ149616, DQ149617, DQ149618, DQ149619, DQ149620, DQ149621, DQ149622, DQ149623, DQ149624, DQ149625, DQ149626, DQ149627, DQ149628, DQ149629, DQ149630, DQ149631, DQ149632, DQ149633, DQ149634, DQ149635, DQ149636, DQ149637, DQ149638, DQ149639, DQ149640, DQ149641, DQ149642 |
| E | AY487947, AY594253, AY599837 |
| F | NC_001454, L19443, X51782, X51783, D13781 |
TABLE 2.
A list of published hexon-based real-time PCR primer/probe sets used to detect different groups of HAdVs
| Primer and probe setsa | Target HAdV species (serotype[s]) | Reference(s) |
|---|---|---|
| Adeno.fwd/probe1,2/Adeno.rev | F | 33 |
| JTVXF/JTVXP/JTVXR | A, B, C, D, E, F | 26 |
| f-AD157/p-AD196/r-AD245 | F (AdV40) | 24 |
| AD3/ADP/AD2 | B(AdVs 3, 16, 21), C (AdVs 1, 2, 5), D (AdVs 9, 17, 19, 28, and 37), E, F | 22 |
| JHKXF/JHKXP/JHKXR | C (AdVs 1, 2, 5, 6), D (AdVs 8 and 19), F | 11, 27 |
| AQ2/AP/AQ1 | A, B, C, D, E, F | 23 |
| Ad:f/Ad:ACDEF,B/Ad:r | A, B, C, D, E, F | 16 |
Given in the order forward primer, probe, and reverse primer.
Quantitative real-time PCR assays.
A section of HAdV40 hexon gene (∼380 bp) was PCR amplified using a published primer set specific for HAdV40 (26). The amplicon was subsequently cloned into plasmid vector (pCR4-TOPO) based on the one-shot chemical transformation described in the manufacturer's instructions (TOPO TA Cloning Kit for Sequencing; Invitrogen, Carlsbad, CA). Plasmid DNA carrying the cloned HAdV40 hexon gene was purified using Wizard Plus SV Minipreps DNA Purification System (Promega, Madison, WI) and adjusted to 2 × 108 copies/μl to serve as a standard stock solution. Standards were diluted (with negative extracts to minimize the difference in matrix conditions between standards and lake extracts) in a range from 101 to 105 copies per PCR to calibrate concentrations of the target hexon gene detected in the HAdV generic real-time PCR assay and the PCR assay for HAdV F species (HAdV-F4041 assay).
In the HAdV generic assay, each PCR mix (total final volume of 20 μl) included 4 μl of 5× LightCycler TaqMan Master Mix, 0.5 μl of each 10 μM primer (final concentration, 250 nM), 0.3 μl of 10 μM TaqMan probe (final concentration, 150 nM), 9.7 μl of PCR-grade water, and 5 μl of DNA sample or standard. The real-time PCR running program (all thermocycles were performed at a temperature transition rate of 20°C/s) was 95°C for 15 min, followed by 45 cycles at 95°C for 10 s, 55°C for 30 s, and 72°C for 15 s, with a final step of 30 s at 40°C.
For the HAdV-F4041 assay, each 20-μl PCR mixture contained 4 μl of 5× LightCycler TaqMan Master Mix, 0.8 μl of 10 μM forward primer (final concentration, 400 nM), 0.4 μl of each 10 μM reverse primer (final concentration, 200 nM), 0.6 μl of 10 μM TaqMan probe (final concentration, 300 nM), 8.8 μl of PCR-grade water, and 5 μl of DNA sample or standard. The real-time PCR program (all thermocycles were performed at a temperature transition rate of 20°C/s) was 15 min at 95°C, followed by 45 cycles at 95°C for 15 s, 60°C for 30 s, and 72°C for 10 s, with a final step for 30 s at 40°C.
Both real-time PCR assays were performed in a Roche LightCycler 1.5 instrument (Roche Applied Sciences, Indianapolis, IN). The samples (i.e., viral DNA extracts) and standards were each run at least in triplicate. All PCR runs included a negative control reaction mixture (PCR-grade H2O without template) and a positive control reaction mixture. The crossing point (Cp) of each PCR was automatically determined by the LightCycler program, version 4.0, and was used to calculate the hexon gene concentration. The concentrations of HAdVs in the lake water samples from the studied recreational beaches were normalized from hexon gene copies per liter to virus particles per liter (lake beach water) using a ratio of one (i.e., each HAdV viral particle consists of one copy of the hexon gene) (47).
After the real-time PCR runs, the PCR products of positive samples were run in a gel to evaluate the integrities of the amplicons. Then, the target bands (i.e., ∼100 bp) were cut out, purified, and sent for sequencing. The sequences were compared with hexon gene sequences in the GenBank database using the BLAST (Basic Local Alignment Search Tool) program (4).
ICC-PCR.
Viruses were cultured on an animal cell line (the Buffalo green monkey [BGM] kidney cells) using the total culturable virus method described in the virus monitoring protocol for the Information Collection Requirements rule (52), with minor modifications. Briefly, the BGM cells were grown in flasks until at least 70 to 90% confluence was obtained. Virus concentrates were added to the flasks and incubated at 36.5 ± 1°C for 2 h with occasional rocking to ensure complete contact between the cells and viral particles. After the growth medium was decanted and discarded, the cells were washed with Dulbecco's phosphate-buffered saline. Cells were maintained with minimum essential medium supplemented with l-glutamine, Earle's salts, and 2% fetal bovine serum. The developments of cytopathic effects (indicative of a viral infection) in the cell cultures were monitored for up to 14 days. Presence or absence of cytopathic effects was confirmed by performing a secondary passage for each flask. The cell culture was used to perform PCR screening for HAdVs using hexAA1885/hexAA1913 and nehexAA1893/nehexAA1905 primer sets (1, 3, 7, 41). The PCR mix consisted of 1 unit of Hotstart Taq polymerase, 1.5 mM MgCl2, 1× PCR buffer, 1× Q-solution, a 0.5 μM concentration of each primer, and a 0.5 mM concentration of each deoxynucleoside triphosphate. The PCR thermocycler settings were 95°C for 15 min, followed by 35 cycles at 95°C for 0.5 min, 57°C for 0.5 min, and 72°C for 0.5 min, with a final elongation step of 72°C for 5 min. The PCR products were separated on a 1.5% agarose gel stained with GelStar nucleic acid stain (BioWhittaker, Walkersville, MD) and viewed under UV light.
RESULTS
Evaluation of real-time PCR assays and inhibition control.
To date, several hexon-based real-time PCR assays have been reported for detecting HAdVs using TaqMan probes (Table 2). Primers and probes used in these published assays were evaluated based on typical guidelines for primer/probe design in order to identify suitable assays for this study. Results showed that the primer/probe set of JTVXF/JTVXP/JTVXR (forward primer, probe, and reverse primer, respectively) (26) fulfilled the most criteria. This primer/probe set has been suggested as being capable of amplifying the target HAdV hexon gene and can detect generic HAdVs (all 51 serotypes) (26).
For specifically quantifying F species of HAdVs (serotypes 40 and 41), the TaqMan probe and reverse primer of a published primer/probe set (f-AD157/p-AD196/r-AD245) (24) were intentionally modified to enhance specificity and to overcome a few of the original biases (e.g., mismatches, short nucleotide length, and/or low annealing temperature for the probe). The adapted primers and probe satisfied nearly all the criteria and were assessed to be highly specific for HAdV40 and HAdV41 (no mismatch with the HAdV40 and HAdV41 hexon genes but having at least 5 to 16 mismatches with the other HAdVs at these primer and probe sites).
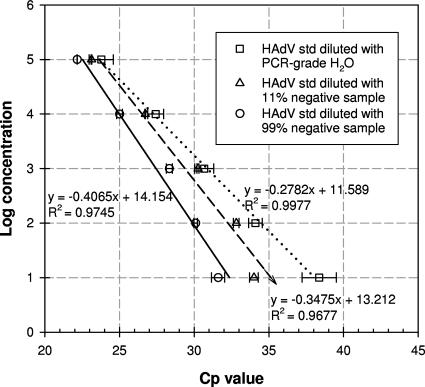
Based on observation of PCR amplification history (fluorescence curves accumulated during real-time PCR runs), significant inhibitory effects on amplification efficiency were noticed for both real-time PCR assays when the lake sample extracts were analyzed (data not shown). Specifically, fluorescence increases were much sharper for standards (diluted with PCR-grade water) than for lake samples. When standard solutions were diluted with negative samples (lake extracts without target templates showing no amplification signals in preliminary real-time PCR runs) to imitate matrix conditions of the lake extracts, the amplification efficiency decreased, and the fluorescence accumulation profile changed (data not shown), leading to shifts in Cp values toward the lower end (Fig. 1). The shifts in Cp values increased as more negative extracts were added. Moreover, the inhibitory effects on the low-level standards (10 copies per PCR) seemed more significant than effects on the high-level standards (fluorescence increase was retarded more for the lower-level standards) (Fig. 1). Concentrations of the target hexon gene were determined using calibration curves generated from the standards in negative extracts to account for matrix effects. The modified standard curve gave enumerated hexon gene concentrations at least 1 order of magnitude lower than those obtained by the original standard curve (prepared with PCR-grade water).
FIG. 1.
Calibration curves generated using standards (std) diluted with PCR-grade water and 11% and 99% HAdV-negative lake extracts for quantifying the target hexon gene.
HAdV occurrence at the recreational beaches.
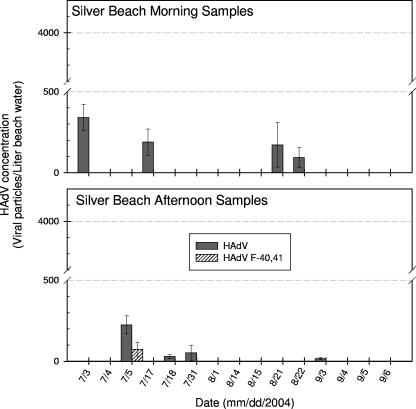
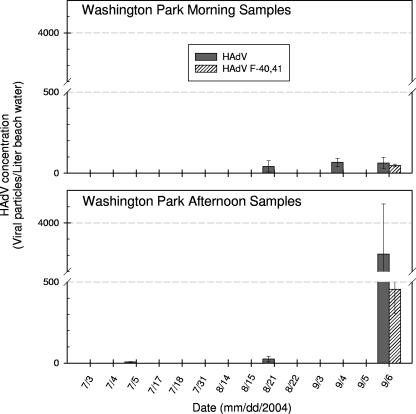
A total of 58 lake extracts were analyzed for overall HAdVs using the HAdV generic real-time PCR assay. Fourteen samples tested positive for HAdV (8 samples from Silver Beach and 6 from Washington Park Beach). The HAdV concentrations ranged from (1.7 ± 0.7) × 101 to (3.4 ± 0.8) × 102 and from (7 ± 2) × 100 to (3.8 ± 0.3) × 103 virus particles/liter for Silver Beach and Washington Park Beach, respectively (Fig. 2 and 3). Typically, HAdVs were detected at relatively low levels (<500 virus particles/liter) excluding a high peak (3.8 × 103 ± 0.3 × 103 virus particles/liter) observed for the sample taken from Washington Park Beach on 6 September 2004 during the afternoon.
FIG. 2.
HAdV abundances in the lake water samples taken from Silver Beach during the summer of 2004 and quantified by real-time PCR assays.
FIG. 3.
HAdV abundances in the lake water samples taken from Washington Park Beach during the summer of 2004 and quantified by real-time PCR assays.
The 14 HAdV-positive samples were further examined for F species of HAdVs using the modified HAdV-F4041 real-time PCR assay. The F species HAdVs were detected in 3 of the 14 positive samples (1 from Silver Beach and 2 from Washington Park Beach) with average concentrations of (7.3 ± 4.3) × 101, (4.8 ± 0.8) × 101, and (4.6 ± 1.5) × 102 virus particles/liter (Fig. 2 and 3). These F species HAdVs represented about 12 to 77% of overall HAdVs in the three lake samples. Sequencing results for the real-time PCR products purified from electrophoresis gel (with expected amplicon size of 88 to 89 bp) confirmed the presence of F species HAdVs in the positive samples.
In addition, all 58 lake samples were also analyzed by the ICC-PCR method. Using this assay, 24 samples were positive (16 from Silver Beach and 8 from Washington Park Beach). About 60% (35 samples) of ICC-PCR analyses agreed with the real-time PCR results; 6 samples that tested negative by the ICC-PCR were positive by the real-time PCR, and 17 samples were positive by ICC-PCR but were negative by real-time PCR (Table 3). Silver Beach was more contaminated than Washington Park Beach when evaluated by both ICC-PCR and real-time PCR.
TABLE 3.
Comparison of real-time PCR and ICC-PCR results for HAdV occurrences at the two recreational beaches on Lake Michigan
| Sample date (mo/day/yr) | Time of Day | Silver Beach
|
Washington Parka
|
||
|---|---|---|---|---|---|
| qPCR | ICC-PCR | qPCR | ICC-PCR | ||
| 7/3/2004 | Morning | + | − | − | + |
| Evening | − | − | − | − | |
| 7/4/2004 | Morning | − | + | − | + |
| Evening | − | − | − | − | |
| 7/5/2004 | Morning | − | − | − | + |
| Evening | + | + | + | − | |
| 7/17/2004 | Morning | + | + | − | + |
| Evening | − | − | − | − | |
| 7/18/2004 | Morning | − | + | − | − |
| Evening | + | + | − | − | |
| 7/31/2004 | Morning | − | + | − | − |
| Evening | + | + | − | − | |
| 8/1/2004 | Morning | − | + | NA | NA |
| Evening | − | − | NA | NA | |
| 8/14/2004 | Morning | − | − | − | − |
| Evening | − | − | − | − | |
| 8/15/2004 | Morning | − | + | − | − |
| Evening | − | + | − | − | |
| 8/21/2004 | Morning | + | − | − | − |
| Evening | − | − | + | − | |
| 8/22/2004 | Morning | + | + | − | − |
| Evening | − | + | − | − | |
| 9/3/2004 | Morning | − | + | − | − |
| Evening | + | − | − | + | |
| 9/4/2004 | Morning | − | + | + | − |
| Evening | − | + | − | + | |
| 9/5/2004 | Morning | − | + | − | − |
| Evening | − | − | − | − | |
| 9/6/2004 | Morning | − | − | + | + |
| Evening | − | − | + | + | |
NA, sample not available.
DISCUSSION
Inhibitory problems were noticed during the real-time PCR runs in this study. It has been reported that complex environmental samples like lake water may contain diverse PCR inhibitors including humic and fulvic acids (56). Frequently, these PCR inhibitors cannot be completely removed through classical nucleic acid extraction procedures and often remain as contaminants in the final DNA and/or RNA preparations (36). A decrease in amplification efficiency caused by environmental inhibitors had been documented for a real-time PCR assay used for detecting HAdVs seeded in secondary sewage effluent and creek waters (24). Accordingly, it was deemed that the lake extracts in this study possibly contained environmental inhibitors. To eliminate such adverse components, we tried several approaches including application of LMP agarose during nucleic acid extraction (36), extra ethanol washes prior to nucleic acid elution, further treatment for extracts using ethanol precipitation (45), the InhibitEX tablet (Qiagen, Valencia, CA), and a nucleic acid binding matrix (Qbiogene Inc., Morgan Irvine, CA). However, none of these approaches consistently facilitated the real-time PCR amplification efficiency (data not shown). Other approaches such as optimizing the PCR conditions, diluting the lake extracts, and adding bovine serum albumin in the reaction mixture were also carried out in this study but still did not overcome the inhibitory problems. Alternatively, the real-time PCR standard solutions with matrix conditions resembling the lake extracts were used to quantify the target HAdV hexon gene. Seeking suitable resolution to surmount such inhibitory problems is still essential. An approach that can eliminate environmental inhibitors during the sample collection, concentration, elution steps, and/or nucleic acid extraction processes would improve PCR amplification efficiency.
In addition, reduced amplification for the HAdV generic real-time PCR assay may also be caused by sequence deficiencies in the JTVXF/JTVXP/JTVXR primers and probe. Based on sequence comparison, these primers and probe have numerous mismatches with the hexon genes of various HAdV species (Table 4). This defect may lead to different degrees of reduction in amplification efficiency even though Jothikumar et al. reported that the real-time PCR assay was able to detect all of the representative HAdVs (belonging to species A to F) under various detection limits (26).
TABLE 4.
The number of mismatches in hexon genes between sequences of the JTVXF/JTVXR primers and JTVXP probe and those of distinct HAdV species
| HAdV species | No. of mismatches with distinct HAdV species
|
||
|---|---|---|---|
| Forward primer JTVX | TaqMan probe JTVXP | Reverse primer JTVXR | |
| A | 0-2 | 1-2 | 1-3 |
| B | 2-3 | 0-1 | 1-4 |
| C | 0 | 1-2 | 1 |
| D | 0 | 1-3 | 1-2 |
| E | 1-2 | 1 | 1-2 |
| F | 0-1 | 0-3 | 0-1 |
The HAdV concentrations at the two recreational beaches were at the level of 10 to 103 virus particles/liter. These concentrations may be underestimated because of the previously mentioned reduced amplification efficiency possibly resulting from mismatches at the primer and probe sites, especially if high-mismatch HAdV serotypes were dominant in the studied lake samples. In He and Jiang's study conducted on the primary and secondary treated sewage effluents (22), the HAdV concentrations were at a level of 105 virus particles/liter based on a real-time PCR quantification targeting 14 different serotypes of HAdVs (22) (Table 5). Another study found that the HAdV concentrations in the urban river of southern California ranged from 102 to 104 virus particles/liter (8). It is not surprising that the HAdV concentrations at the studied recreational lake beaches were lower than those in sewage and river water given the dilution that is apparent at Lake Michigan shorelines.
TABLE 5.
Literature values of HAdV concentrations and occurrence frequencies in various water sources
| Type of sample | Concentration (no. of viruses particles/liter) | Frequency of occurrence (no. of positive sample/no. of samples tested)a | Reference or source |
|---|---|---|---|
| Primary effluent | 6.6 × 105-7.4 × 105 | NA | 22 |
| Secondary effluent | 6.1 × 105-7.4 × 105 | NA | 22 |
| Secondary effluent with Cl | 8.5 × 105-14 × 105 | NA | 22 |
| River | 102-104 | 6/114 | 8 |
| Drinking water | 1.40 × 10−4-2.45 × 10−4 | 9/204 | 54 |
| River | 5.46 × 10−3 | 4/51 | 55 |
| Dam | 9.97 × 10−4 | 9/51 | 55 |
| Silver Beach | 17-341 | 8/30 | This study |
| Washington Park Beach | 7-3,808 | 6/28 | This study |
NA, not available.
HAdVs were more frequently detected in early summer at Silver Beach but were found more often in the late summer at Washington Park Beach (Fig. 2 and 3). Additionally, the HAdV occurrences during afternoon were not significantly higher (P value of >0.05) in general than those during morning (exclusive of the 6 September 2004 Washington Park Beach samples). This suggests that the HAdVs detected at the two lake beaches were less likely to have originated from an on-site contamination event related to infected humans during high-peak recreational activities. Instead, other sources such as municipal discharges from nearby cities or metropolitan areas (e.g., St. Joseph, Michigan City, or Chicago) may have introduced HAdVs at these two recreational lake beaches.
The outcomes of ICC-PCR testing did not correspond with the real-time PCR results in 40% of the samples (Table 3). More samples tested positive using the ICC-PCR (24 samples) than when tested by HAdV generic real-time PCR (14 samples). This is likely due to the multiplication of infectious viruses in host cells that consequently increased the sensitivity of the PCR in the ICC-PCR tests. However, six samples tested negative by ICC-PCR but were positive by real-time PCR. This result could be attributed to two main reasons. First, the primers used in the ICC-PCR assay (hexAA1885/hexAA1913 and nehexAA1893/nehexAA1905) might be unable to amplify all 51 serotypes of HAdVs because of the mismatches (data not shown), and, second, the ICC-PCR only measured the infectious HAdVs whereas the real-time PCR assay might detect both infectious and noninfectious HAdVs in the lake samples.
Interestingly, both ICC-PCR and real-time PCR results showed that Silver Beach was more polluted than Washington Park Beach (Table 3). Silver Beach is located at the mouth of the St. Joseph River, and HAdVs are likely able to survive in effluents of wastewater treatment plants discharged upstream in the river, survive sunlight inactivation, and be transported to the beach area. Washington Park Beach is located at the mouth of a creek, and the watershed of this creek is much smaller than the St. Joseph River's.
In conclusion, this study revealed the occurrence of HAdVs at recreational Great Lakes beaches using both real-time PCR and ICC-PCR analytical approaches. Application of the real-time PCR was able to quickly quantify the low levels of HAdVs (approximately 10 to 103 virus particles/liter) found at these two lake beaches. Cell culture PCR methods require several weeks to obtain results and make it much more difficult to process large numbers of samples simultaneously. The F species HAdVs (serotypes 40 and 41) detected in three lake samples should raise safety awareness in regard to utilizing recreational beaches in the Great Lakes basin since the HAdV40 and HAdV41 are critical etiological agents of viral gastroenteritis in children.
Acknowledgments
This study was funded by a research grant from the Center for Water Sciences at Michigan State University and by NOAA Center of Excellence for Great Lakes and Human Health, Great Lakes Environmental Research Laboratory.
We thank Rebecca Ives, Stephanie Molloy, Theng Theng Fong, and Ai Masago for assisting with sample collection and processing.
Footnotes
Published ahead of print on 12 October 2007.
REFERENCES
- 1.Allard, A., B. Albinsson, and G. Wadell. 1992. Detection of adenoviruses in stools from healthy persons and patients with diarrhea by two-step polymerase chain reaction. J. Med. Virol. 37:149-157. [DOI] [PubMed] [Google Scholar]
- 2.Allard, A., B. Albinsson, and G. Wadell. 2001. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J. Clin. Microbiol. 39:498-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allard, A., R. Girones, P. Juto, and G. Wadell. 1990. Polymerase chain reaction for detection of adenovirus in stool samples. J. Clin. Microbiol. 28:2659-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Avellón, A., P. Perez, J. C. Aguilar, R. ortiz de Lejarazu, and J. E. Echevarria. 2001. Rapid and sensitive diagnosis of human adenovirus infections by generic polymerase chain reaction. J. Virol. Methods 92:113-120. [DOI] [PubMed] [Google Scholar]
- 6.Castignolles, N., F. Petit, I. Mendel, L. Simon, L. Cattolico, and C. Buffet-Janvresse. 1998. Detection of adenovirus in the waters of the Seine River estuary by nested-PCR. Mol. Cell Probes 12:175-180. [DOI] [PubMed] [Google Scholar]
- 7.Chapron, C. D., N. A. Ballester, J. H. Fontaine, C. N. Frades, and A. B. Margolin. 2000. Detection of astroviruses, enteroviruses, and adenovirus types 40 and 41 in surface waters collected and evaluated by the information collection rule and an integrated cell culture-nested PCR procedure. Appl. Environ. Microbiol. 66:2520-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, S., and S. C. Jiang. 2005. Real-time PCR quantification of human adenoviruses in urban rivers indicates genome prevalence but low infectivity. Appl. Environ. Microbiol. 71:7426-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choo, Y.-J., and S.-J. Kim. 2006. Detection of human adenoviruses and enteroviruses in Korean oysters using cell culture, integrated cell culture-PCR, and direct PCR. J. Microbiol. 44:162-170. [PubMed] [Google Scholar]
- 10.Crabtree, K. D., C. P. Gerba, J. B. Rose, and C. N. Haas. 1997. Waterborne adenovirus: a risk assessment. Water Sci. Technol. 35:1-6. [Google Scholar]
- 11.Cromeans, T. L., J. Narayanan, K. H. Jung, G. Ko, D. Wait, and M. D. Sobsey. 2003. Development of molecular methods for the detection of viruses in water. Report 90995F. American Water Works Association, Denver, CO.
- 12.Cruz, J. R., P. Caceres, F. Cano, J. Flores, A. Bartlett, and B. Torún. 1990. Adenovirus types 40 and 41 and rotaviruses associated with diarrhea in children from Guatemala. J. Clin. Microbiol. 28:1780-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Echavarria, M., M. Forman, J. Ticehurst, J. S. Dumler, and P. Charache. 1998. PCR method for detection of adenovirus in urine of healthy and human immunodeficiency virus-infected individuals. J. Clin. Microbiol. 36:3323-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enriquez, C. E., and C. P. Gerba. 1995. Concentration of enteric adenovirus 40 from tap, sea and waste water. Water Res. 29:2554-2560. [Google Scholar]
- 15.Fong, T. T., D. W. Griffin, and E. K. Lipp. 2005. Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. Appl. Environ. Microbiol. 71:2070-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Formiga-Cruz, M., G. Tofiño-Quesada, S. Bofill-Mas, D. N. Lees, K. Henshilwood, A. K. Allard, A.-C. Condin-Hansson, B. E. Hernroth, A. Vantarakis, A. Tsibouxi, M. Papapetropoulou, D. Furones, and R. Girones. 2002. Distribution of human viral contamination in shellfish from different growing areas in Greece, Spain, Sweden, and the United Kingdom. Appl. Environ. Microbiol. 68:5990-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Formiga-Cruz, M., A. Hundesa, P. Clemente-Casares. N. Albinana-Gimenez, A. Allard, and R. Girones. 2005. Nested multiplex PCR assay for detection of human enteric viruses in shellfish and sewage. J. Virol. Methods 125:111-118. [DOI] [PubMed] [Google Scholar]
- 18.Foy, H. M. 1997. Adenoviruses, p. 119-138. In A. S. Evans and R. A. Karlow (ed.), Viral infection in humans: epidemiology and control, 4th ed. Plenum Press, New York, NY.
- 19.Girones, R., M. Puig, A. Allard, F. Lucena, G. Wadell, and J. Jofre. 1995. Detection of adenovirus and enterovirus by PCR amplification in polluted waters. Water Sci. Technol. 31:351-357. [Google Scholar]
- 20.Grabow, W. O. K., M. B. Taylor, and J. C. de Villiers. 2001. New methods for the detection of viruses: call for review of drinking water quality guidelines. Water Sci. Technol. 43:1-8. [PubMed] [Google Scholar]
- 21.Greening, G. E., J. Hewitt, and G. D. Lewis. 2002. Evaluation of integrated cell culture-PCR (C-PCR) for virological analysis of environmental samples. J. Appl. Microbiol. 93:745-750. [DOI] [PubMed] [Google Scholar]
- 22.He, J. W., and S. Jiang. 2005. Quantification of enterococci and human adenoviruses in environmental samples by real-time PCR. Appl. Environ. Microbiol. 71:2250-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heim, A., C. Ebnet, G. Harste, P. Pring-Aakerblom. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR, J. Med. Virol. 70:228-239. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, S., H. Dezfulian, and W. Chu. 2005. Real-time quantitative PCR for enteric adenovirus serotype 40 in environmental waters. Can. J. Microbiol. 51:393-398. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, S. C., and W. Chu. 2004. PCR detection of pathogenic viruses in southern California urban rivers. J. Appl. Microbiol. 97:17-28. [DOI] [PubMed] [Google Scholar]
- 26.Jothikumar, N., T. L. Cromeans, V. R. Hill, X. Lu, M. D. Sobsey, and D. D. Erdman. 2005. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl. Environ. Microbiol. 71:3131-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ko, G., N. Jothikumar, V. R. Hill, and M. D. Sobsey. 2005. Rapid detection of infectious adenoviruses by mRNA real-time RT-PCR. J. Virol. Methods 127:148-153. [DOI] [PubMed] [Google Scholar]
- 28.Kukkula, M., P. Arstila, M. L. Klossner, L. Maunula, C. H. Bonsdorff. 1997. Waterborne outbreak of viral gastroenteritis. Scand. J. Infect. Dis. 29:415-418. [DOI] [PubMed] [Google Scholar]
- 29.Lee, C., S. H. Lee, E. Han, and S. J. Kim. 2004. Use of cell culture-PCR assay based on combination of A549 and BGMK cell lines and molecular identification as a tool to monitor infectious adenoviruses and enteroviruses in river water. Appl. Environ. Microbiol. 70:6695-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, H. K., and Y. S. Jeong. 2004. Comparison of total culturable virus assay and multiplex integrated cell culture-PCR for reliability of waterborne virus detection. Appl. Environ. Microbiol. 70:3632-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, S. H., C. Lee, K. W. Lee, H. B. Cho, and S. J. Kim. 2005. The simultaneous detection of both enteroviruses and adenoviruses in environmental water samples including tap water with an integrated cell culture-multiplex-nested PCR procedure. J. Appl. Microbiol. 98:1020-1029. [DOI] [PubMed] [Google Scholar]
- 32.Lin, B., G. J. Vora, D. Thach, E. Walter, D. Metzgar, C. Tibbetts, and D. A. Stenger. 2004. Use of oligonucleotide microarrays for rapid detection and serotyping acute respiratory disease-associated adenoviruses. J. Clin. Microbiol. 42:3232-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Logan, C., J. J. O'Leary, and O. O'Sullivan. 2006. Real-time reverse transcription-PCR for detection of rotavirus and adenovirus as causative agents of acute viral gastroenteritis in children. J. Clin. Microbiol. 44:3189-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, X., and D. D. Erdman. 2006. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch. Virol. 151:1587-1602. [DOI] [PubMed] [Google Scholar]
- 35.McMillian, N. S., S. A. Martin, M. D. Sobsey, D. A. Wait, R. A. Meriwether, and J. N. MacCormack. 1992. Outbreak of pharyngoconjunctival fever in a summer camp-North Carolina, 1991. Morb. Mortal. Wkly. Rep. 41:342-347. [PubMed] [Google Scholar]
- 36.Moreira, D. 1998. Efficient removal of PCR inhibitors using agarose-embedded DNA preparations. Nucleic Acids Res. 26:3309-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papapetropoulou, M., and A. C. Vantarakis. 1998. Detection of adenovirus outbreak at a municipal swimming pool by nested PCR amplification. J. Infect. 36:101-103. [DOI] [PubMed] [Google Scholar]
- 38.Payment, P., S. Fortin, and M. Trudel. 1984. Ferric chloride flocculation for nonflocculating beef extract preparations. Appl. Environ. Microbiol. 47:591-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pina, S., M. Puig, F. Lucena, J. Jofre, and R. Girones. 1998. Viral pollution in the environment and shellfish: human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 64:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piranha, J., A. Pacheco, R. C. Gamba, D. U. Mehnert, P. Garrafa, and K. Barrella. 2006. Fecal contamination (viral and bacteria) detection in groundwater used for drinking purposes in São Paulo, Brazil. Geomicrobiol. J. 23:279-283. [Google Scholar]
- 41.Puig, M., J. Jofre, F. Lucena, A. Allard, W. Wadell, and R. Girones. 1994. Detection of adenoviruses and enteroviruses in polluted waters by nested PCR amplification. Appl. Environ. Microbiol. 60:2963-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pusch, D., D. Y. Oh, S. Wolf, R. Dumke, U. Schröter-Bobsin, M. Hohne, I. Roske, and E. Schreier. 2005. Detection of enteric viruses and bacterial indicators in German environmental waters. Arch. Virol. 150:929-947. [DOI] [PubMed] [Google Scholar]
- 43.Rigotto, C., T. C. Sincero, C. M. Simoes, and C. R. Barardi. 2005. Detection of adenoviruses in shellfish by means of conventional-PCR, nested-PCR, and integrated cell culture PCR (ICC/PCR). Water Res. 39:297-304. [DOI] [PubMed] [Google Scholar]
- 44.Rohayem, J., S. Berger, T. Juretzek, O. Herchenröder, M. Mogel, M. Poppe, J. Henker, and A. Rethwilm. 2004. A simple and rapid single-step multiplex RT-PCR to detect norovirus, astrovirus and adenovirus in clinical stool samples. J. Virol. Methods 118:49-59. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Sarantis, H., G. Johnson, M. Brown, M. Petric, and R. Tellier. 2004. Comprehensive detection and serotyping of human adenoviruses by PCR and sequencing. J. Clin. Microbiol. 42:3963-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sprengel, J., B. Schmitz, D. Huess-Neitzel, C. Zock, and W. Doerfler. 1994. Nucleotide sequence of human adenovirus type 12 DNA: comparative functional analysis. J. Virol. 68:379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swenson, P. D., G. Wadell, A. Allard, and J. C. Hierholzer. 2003. Adenoviruses, p. 1404-1417. In R. H. Yolken, M. L. Landry, T. F. Smith, and J. L. Waner (ed.), Manual of clinical microbiology, vol. II, 8th ed. ASM Press, Washington, DC. [Google Scholar]
- 49.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgens. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turner, M., G. R. Istre, H. Beauchamp, M. Baum, and S. Arnold. 1987. Community outbreak of adenovirus type 7a infections associated with a swimming pool. South. Med. J. 80:712-715. [DOI] [PubMed] [Google Scholar]
- 51.Uhnoo, I., G. Wadell, L. Svensson, E. Olding-Stenkvist, and R. Molby. 1986. Etiology and epidemiology of acute gastroenteritis in Swedish children. J. Infect. 13:73-89. [DOI] [PubMed] [Google Scholar]
- 52.U.S. Environmental Protection Agency. 1995. Virus monitoring protocol for the Information Collection Requirements rule. Publication EPA/814-B-95-002. U.S. Environmental Protection Agency, Washington, DC.
- 53.Van Heerden, J., M. M. Ehlers, W. B. Van Zyl, and W. O. Grabow. 2003. Incidence of adenoviruses in raw and treated water. Water Res. 37:3704-3708. [DOI] [PubMed] [Google Scholar]
- 54.Van Heerden, J., M. M. Ehlers, and W. O. Grabow. 2005. Detection and risk assessment of adenoviruses in swimming pool water. J. Appl. Microbiol. 99:1256-1264. [DOI] [PubMed] [Google Scholar]
- 55.Van Heerden, J., M. M. Ehlers, W. B. Van Zyl, and W. O. Grabow. 2004. Prevalence of human adenoviruses in raw and treated water. Water Sci. Technol. 50:39-43. [PubMed] [Google Scholar]
- 56.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, W., M. C. McDonough, and D. D. Erdman. 2000. Species-specific identification of human adenoviruses by a multiplex PCR assay. J. Clin. Microbiol. 38:4114-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]