Abstract
One hundred forty-one Campylobacter jejuni isolates from humans with diarrhea and 100 isolates from retailed poultry meat were differentiated by flaA typing. The bacteria were isolated in a specific geographical area (Dunedin) in New Zealand over a common time period. Twenty nine flaA types were detected, one of which (flaA restriction fragment length polymorphism type 15 [flaA-15]) predominated among isolates from humans (∼30% of isolates). This strain was of low prevalence (5% of isolates) among poultry isolates. flaA-15 strains were five to six times more invasive of HEp2 cells in an in vitro assay than a flaA type (flaA-3) that was commonly encountered on poultry meat (23% of isolates) but was seldom associated with human illness (5%). Competitive-exclusion experiments with chickens, utilizing real-time quantitative PCR to measure the population sizes of specific strains representing flaA-15 (T1016) and flaA-3 (Pstau) in digesta, were carried out. These experiments showed that T1016 always outcompeted Pstau in the chicken intestine. Genomic comparisons of T1016 and Pstau were made using DNA microarrays representing the genome of C. jejuni NCTC 11168. These comparisons revealed differences between the strains in the gene content of the Cj1417c-to-Cj1442c region of the genome, which is associated with the formation of capsular polysaccharide. The strains differed in Penner type (T1016, O42; Pstau, O53). It was concluded that poultry meat was at least one source of human infection with C. jejuni, that some Campylobacter strains detected in poultry meat are of higher virulence for humans than others, and that bacterial attributes affecting strain virulence and commensal colonization ability may be linked.
Campylobacter jejuni is the most common cause of bacterial gastroenteritis in humans in the developed world (1). Domestic poultry are considered to be one of the most important reservoirs of human infection since C. jejuni is a common inhabitant of the avian bowel. Human infections are associated with the poor handling of raw meats in the home after purchase and consumption of contaminated poultry products (10, 19, 24). The New Zealand situation with respect to C. jejuni infections is similar to those of the United Kingdom and the United States, although the notification rate is higher (36). In most countries, little progress has been made in reducing contamination of poultry meat with C. jejuni, and new intervention strategies need to be developed to achieve a reduction in the colonization of broiler flocks by this pathogen and to promote resistance to infection in humans (43). The development of new strategies would be aided by knowledge of the bacterial factors that influence colonization of the avian gut and the pathogenesis of infections (17). It can be postulated that useful molecular knowledge would best be gained from comparisons of C. jejuni strains whose behavior in relation to human and poultry hosts is known. Therefore, we have carried out comparative studies of two C. jejuni strains, T1016 and Pstau, chosen on the basis of epidemiological data (flaA gene typing) in relation to their prevalence in human infections and on poultry meat in a specific geographical area of New Zealand. The strain comparisons utilized an in vitro cell invasion assay, microarray-based comparative genomic hybridization, flagellin A (FlaA) characteristics inferred from deduced amino acid sequences, and competitive-exclusion experiments with chickens.
MATERIALS AND METHODS
Isolation of Campylobacter from chicken portions.
Prepackaged chicken portions, which included thighs, breast, and legs, with skin intact, were purchased from six retail food outlets within Dunedin city in 1998. Each portion was placed in a plastic bag to which was added 100 ml of 1% (wt/vol) peptone water buffered with phosphate-buffered saline (PBS) (pH 7.2; Sigma Chemical Company, St. Louis, MO). The chicken portion was massaged by hand through the plastic bag for 2 min. One milliliter of the resulting liquid was added to 10 ml of Preston's broth supplemented with Campylobacter growth supplement (Oxoid, Unipath Ltd., Basingstoke, England) and selective supplement (Oxoid). The broth was incubated microaerobically (BBl anaerobic jar; BBL CampyPak microaerophilic envelopes) at 42°C for 24 h. The enrichment broth was subcultured on Campylobacter blood-free agar base supplemented with CCDA selective supplement (Oxoid) and incubated at 37°C for 48 h microaerobically. Campylobacter colonies were subcultured to ensure purity and then stored in brain heart infusion broth (Difco Laboratories, Detroit, MI) containing a final concentration of 20% glycerol at −70°C. One hundred Campylobacter isolates were obtained during 1998.
Campylobacter isolates obtained from humans.
The clinical isolates were cultured from human feces submitted to Southern Community Laboratories, Dunedin, as diagnostic specimens for investigation of cases of diarrhea. Colonies were subcultured and stored as described above. One hundred isolates were obtained during 1996 and 1997 and a further 41 in 1998.
Identification of Campylobacter isolates.
Campylobacter isolates were tested for oxidase and catalase activity, hippurate hydrolysis (16), and multiplex PCR (see below). C. jejuni strains ATCC 33560 and NZRM 2397/NCTC 11351 and Campylobacter coli NZRM 3607/NCTC served as controls.
DNA extraction from Campylobacter cultures.
Campylobacter colonies picked from an agar plate were suspended in 600 μl of sterile deionized water and boiled for 10 min to lyse the cells. After centrifugation (13,000 × g for 10 min), the clear supernatant was transferred to another microcentrifuge tube. An equal volume (600 μl) of phenol (BDH Laboratory Supplies) saturated with TES buffer (10 mM Tris, 100 mM EDTA, 20 mM sodium chloride) was added and centrifuged (13,000 × g for 10 min). Five hundred microliters of the upper aqueous layer was transferred to another microcentrifuge tube, an equal volume of chloroform-isoamyl alcohol (24:1) was added, and the mixture was centrifuged (6,000 × g for 5 min). Four hundred microliters of the upper layer was retained. Forty microliters of 3 M sodium acetate and 400 μl of cold (−20°C) ethanol were added and placed at −20°C overnight. The sample was centrifuged (10 min at 13,000 × g). The ethanol was decanted, and the pellet was washed once with cold (−20°C) 70% ethanol. Pellets were dried at 37°C and then dissolved in 45 μl of sterile deionized water and stored at −20°C until required.
Multiplex PCR.
Isolates were tested using the method reported by Harmon et al. (15). A total PCR mix of 50 μl contained the following: 1 μl of Campylobacter DNA, 20 pmol each of primers C1 and C4 and 40 pmol each of primers pg3 and pg50 (Table 1), 2.5 U of Taq DNA polymerase, 5 μl of 10× PCR buffer containing 15 mM MgCl2, 1 μl of 10 mM PCR nucleotide mix, 8 μl of 25 mM MgCl2 solution, and 30.5 μl of deionized water. The PCR program was as follows: 94°C for 4 min, followed by 25 cycles of 94°C for 1 min, 45°C for 1 min, and 72°C for 1 min, and then 72°C for 7 min. PCR products were electrophoresed in 1.5% agarose gels which were then stained in ethidium bromide solution (5 μg/ml) before being viewed by UV transillumination. C. jejuni gave two DNA fragments of 460 bp and 160 bp, whereas C. coli gave a single fragment of 460 bp.
TABLE 1.
PCR and sequencing primers
| Purpose | Primer name | Primer sequence (5′ to 3′) |
|---|---|---|
| flaA gene (multiplex PCR) | pg3 | GAACTTGAACCGATT |
| pg50 | ATGGGATTTCGTAAC | |
| C1 | CAAATAAAGTTAGAGGTAGAATGT | |
| C4 | GGATAAGCACTAGCTACCTGAT | |
| flaA gene (gene amplification) | flaA-F | GGATTTCGTATTAACACAAATGGT |
| flaA-R | CTGTAGTAATCTTAAACAATTTTG | |
| flaA gene sequencing | flaA-F | GGATTTCGTATTAACACAAATGGTGC |
| flaA gene sequencing | flaA-R | CTGTAGTAATCTTAAACAATTTTG |
| flaA gene sequencing | fla Internal SEQ For | CTTTGAAACAGGAGGAAG |
| flaA gene sequencing | Internal-flaA F | TRTTTCTYTAAGAGAATCAAAAGG |
| flaA gene sequencing | Internal-flaA R | ATATCCATCACAGCCATAGCGCC |
flaA gene typing of C. jejuni isolates.
PCR was used to amplify the flaA gene from the Campylobacter isolates (25). A total volume of 50 μl contained the following: 1 μl of Campylobacter DNA, 1 μM each of primers flaA-F and flaA-R (Table 1), 2.5 U of Taq DNA polymerase, 5 μl of 10× PCR buffer containing 15 mM MgCl2 solution, 1 μl of 10 mM PCR nucleotide mix, and 40.5 μl of deionized water. The PCR program was as follows: 94°C for 1 min, followed by 35 cycles of 94°C for 15 s, 53°C for 45 s, and 72°C for 1 min 45 s, and then 72°C for 5 min. The presence of a 1.7-kbp amplicon was confirmed by 0.7% agarose gel electrophoresis, ethidium bromide staining, and UV transillumination.
The flaA gene was digested with the restriction enzyme DdeI (25). Each restriction digest contained the following: 1.5 μl of 10× DdeI buffer, 5 μl of deionized water, 5 U of the DdeI enzyme, and 8 μl of the flaA PCR product. The mixture was incubated at 37°C for 18 h. The restriction fragment length polymorphism (RFLP) profile was detected by 2% agarose gel electrophoresis of the digest, ethidium bromide staining, and UV transillumination.
Penner typing.
T1016 and Pstau were serotyped using the method of Penner and Hennessy (29).
Invasion assay.
Eleven strains differentiated by flaA gene typing were tested for their ability to invade HEp-2 cells (human laryngeal epidermoid carcinoma; ATCC CCL 23) as described by Konkel and Joens (21) but using RPMI medium 1640 (Invitrogen; Carlsbad, CA) instead of Eagle minimal essential medium. HEp-2 (ATCC CCL 23) cells were cultured in 25-cm2 tissue culture flasks and maintained in RPMI medium supplemented with sodium bicarbonate (0.37% [wt/vol]), 1% (vol/vol) penicillin-streptomycin (Invitrogen) (100-ml stock solution containing 5,000 U of penicillin and 5,000 μg of streptomycin). and 10% fetal calf serum (Gibco) at 37°C in an atmosphere containing 5% carbon dioxide. Confluent monolayers were washed three times with RPMI medium without antibiotics, detached by adding 1 ml of trypsin-EDTA solution (0.05% [wt/vol] trypsin and 0.5 mM EDTA in PBS) to the flask, incubated (3 min at 37°C), and then suspended in RPMI medium. The cell density was determined microscopically and adjusted to 2 × 104 cells per ml. One milliliter of HEp-2 cell suspension was added to each well of a 24-well tissue culture tray and incubated at 37°C in an atmosphere containing 5% carbon dioxide until approximately 90% confluence was reached.
Campylobacter cells were harvested from blood agar plates (blood agar base no. 2 [Difco], 5% horse blood [Gibco], CCDA selective supplement [Oxoid]) after 24 h of incubation, washed three times in PBS, and suspended in RPMI medium without antibiotics to give a cell density of approximately 3 × 109 bacteria per ml. The RPMI medium was removed from the HEp-2 cell cultures, and 1 ml of Campylobacter suspension was added to each well and incubated (3 h at 37°C) in an atmosphere containing 5% carbon dioxide. The fluid was removed from each well, and the HEp-2 cell monolayer was washed three times with RPMI medium (without antibiotics). One milliliter of RPMI medium containing 250 μg of gentamicin (Sigma) was added to each well to kill extracellular Campylobacter cells during incubation (37°C for 2 h) in an atmosphere containing 5% carbon dioxide. The HEp-2 monolayers were then washed three times with PBS to remove gentamicin. Intracellular Campylobacter cells were released by lysis of HEp-2 cells by the addition of 1 ml of 0.5% (wt/vol) sodium deoxycholate solution. Numbers of Campylobacter CFU per ml were determined 5 min later by preparing serial 10-fold dilutions of the lysate in PBS buffer which were then spread-plated on Campylobacter blood-free agar base supplemented with CCDA selective supplement. The plates were incubated at 37°C for 48 h in a microaerobic atmosphere. The C. jejuni colonies were counted and used to compare the invasive ability of each strain. Invasion assays were performed in five wells for each strain on three separate occasions, giving 15 data points per strain. The relative invasion ability was calculated by dividing the mean invasion level of each flaA group by the mean invasion level of strains of flaA RFLP type 15 (flaA-15) and multiplying by 100.
Microarray-based comparative genomic hybridization.
Details of the C. jejuni NCTC 11168 open reading frame DNA microarray and its use, including primer selection, the parameters for primer synthesis, selection of amplicons, and the purification and printing of DNA on slides, have been described previously (40). Additional information is available at http://ibs-isb.nrc-cnrc.gc.ca/glycobiology/campychips_e.html.
Genomic DNA from strains T1016 and Pstau was sheared into fragments ranging between 0.5 and 5 kb (mean size, ∼1.5 kb) using the method of Bodenteich et al. (5). Briefly, genomic DNA was suspended in 35% glycerol and nebulized in an aerosol nebulizer (Medex, Carlsbad, CA) for 45 s at 15 lb/in2. Five micrograms of sheared DNA was fluorescently labeled using direct chemical coupling with the Label-IT (Mirus Corp., Madison, WI) cyanine dyes Cy3 and Cy5 as recommended by the manufacturer. Probes were purified from unincorporated dyes by sequentially passing samples through SigmaSpin (Sigma, Oakville, Canada) and Qiaquick (Qiagen, Mississauga, Canada) columns. Labeled DNA sample yields and dye incorporation efficiencies were calculated using the Nanodrop ND-1000 spectrophotometer (Nanodrop, Rockland, DE).
The hybridization profile for each strain was obtained by cohybridizing labeled DNA from the tester (T1016 or Pstau) strain and from the NCTC 11168 (reference) strain to the microarray. Equivalent amounts (1 to 2 μg) of labeled tester and control samples with similar dye incorporation efficiencies were pooled, lyophilized, and hybridized to microarrays as previously described (40).
Microarrays were scanned using a Chipreader laser scanner (Bio-Rad, Mississauga, Canada) according to the manufacturer's recommendations. Spot quantification, visual inspection of potential outliers, and flagging of anomalous spots were performed using the software program ArrayPro Analyzer (version 4.5; Media Cybernetics). The microarray data exported from ArrayPro were imported into the BioArray Software Environment (BASE version 1.2) (34). Spots flagged due to poor spot morphology or low signal intensity (less than 5× local background) were filtered out and removed from further analysis. After print-tip Loess normalization, data were used to calculate the average log ratio [log2(Signal Tester/Signal Control)] from the two replicates for each gene represented on the microarray. Log ratio data were visualized and analyzed using The Institute for Genomic Research's MultiExperiment Viewer (TMEV) (MEV version 3.0) software (35). Gene conservation profiles were assessed based on empirically determined log ratio values (41). Clustering of samples based on log ratio profile similarities was performed by the average linkage hierarchical clustering method of Eisen et al. (11), as implemented in TMEV, using the Pearson correlation coefficient as a distance metric. The support tree method of bootstrapping implemented in TMEV was used to test the reliability of the clustering patterns (500 bootstrap resamplings). Percentages of resampled trees supporting a given tree node were determined. To facilitate tree topology visualization, tree information was coded into the Newick format and the trees were visualized using TreeView (version 1.6.6) (26).
Competitive performance experiments with chickens.
Ethical approval for animal experimentation was obtained from the Otago University Animal Ethics Committee (application 36/02). One-day-old “standard hybrid” chicks were obtained from a local supplier. The chicks were housed in a plastic-lined, 1-m3, autoclavable, open-topped metal container. Autoclaved and dried wood shavings were used as bedding on the floor of the container. The chicks were fed standard chick starter mash (Reliance Stock Food, Dunedin, New Zealand), and water, which had been sterilized prior to use, was in constant supply. A 200-W heat lamp was used to keep a constant temperature within the enclosure.
Two C. jejuni strains were used in the experiments: T1016 and Pstau. For each experiment, 10 1-day-old chickens were gavaged with 3 × 105 C. jejuni cells. Five experimental protocols (chick colonization groups A to G) were used, in which the strains were tested alone or in combination. The chicks were euthanized 7 days after inoculation, and the ceca were removed from each bird. Half a gram of cecum contents was placed in separate containers containing 4.5 ml of PBS. Approximately 3.5 g of 3-mm-diameter glass beads were added, and the preparations were homogenized using a vortex mixer. The homogenate was serially diluted in 10-fold steps, and aliquots of each dilution were used to inoculate blood agar plates containing 10 μg of vancomycin per ml. The plates were incubated at 42°C for 48 h microaerobically. Campylobacter colonies were enumerated, expressed as CFU per g of cecum contents. Ten colonies were subcultured, tested for oxidase and catalase activity, and examined in gram-stained smears.
Quantification of C. jejuni from chicken gut contents using real-time TaqMan quantitative PCR.
To determine population levels of specific Campylobacter strains (T1016 and Pstau) in cecum contents, two sets of PCR probes and primers were designed. The strain-specific primers and probes were designed on the basis of the flaA gene sequences of the two strains (NCBI accession numbers AY751740 [T1016] and AY751741 [Pstau]). The flaA genes of the strains were sequenced by the Centre for Gene Research, University of Otago, using primers given in Table 1. TaqMan MGB probes and their primers were designed using Primer Express software, version 1.5, and the guidelines provided by PE Applied Biosystems (Foster City, CA).
To construct a standard curve by which Campylobacter cells could be enumerated, two cecum samples were randomly selected from two chicks, one of which had been inoculated with Pstau and the other of which had been inoculated with T1016. DNA was extracted from the cecum samples using the QIAamp DNA minikit (Qiagen GmbH, Hilden, Germany). The T1016 DNA extract corresponded to 1 × 1010 C. jejuni cells per gram of cecum contents. The DNA was serially diluted to create the standard curve, with Campylobacter concentrations ranging from 1 × 1010 CFU per g to 105 CFU per g. The Pstau DNA corresponded to 1 × 109 C. jejuni cells per gram of cecum contents. This was used to prepare a standard curve of 1 × 109 CFU per g to 1 × 105 CFU per g.
Taqman real-time quantitative PCR was performed with the following reagents in a total volume of 25 μl: 12.5 μl 2× TaqMan Universal PCR master mix (PE Applied Biosystems), 300 nM of each primer (PE Applied Biosystems), 200 nM of the strain-specific probe (PE Applied Biosystems), 2.5 μl of template, and 5 μl of deionized water.
Amplification was carried out using an ABI Prism 7700 sequence detection system instrument (PE Applied Biosystems). The PCR program was as follows: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min. Numerical values (CFU per g) were assigned to the standard curve amplification plot values. The ABI Prism 7700 SDS version 1.9 software calculated numerical values for the test samples.
Comparison of predicted amino acid sequences of FlaA.
Hydropathic profiles of the FlaA proteins of T1016 and Pstau were obtained by the Kyte-Doolittle method (22) (http://fasta.bioch.virginia.edu). To identify candidate amino acid motifs that indicate sites of glycosylation, the predicted amino acid sequences of FlaA proteins from C. jejuni strains T1016, Pstau, and 81-176 and C. coli strain VC167 were compared using ClustalW (www.ebi.ac.uk).
RESULTS
Prevalence of C. jejuni strains on chicken portions and in human feces.
C. jejuni was detected in 76.9% of chicken portions. All of the isolates from human feces were C. jejuni. The RFLP profiles (flaA types) generated by digestion of the flaA gene with DdeI had three to six fragments, ranging from 150 bp to 1,000 bp in size. Strain T1016 (flaA-15) was Penner serotype O42, and Pstau (flaA-3) was O53.
Prevalence of C. jejuni strains in chicken portions and in human infections.
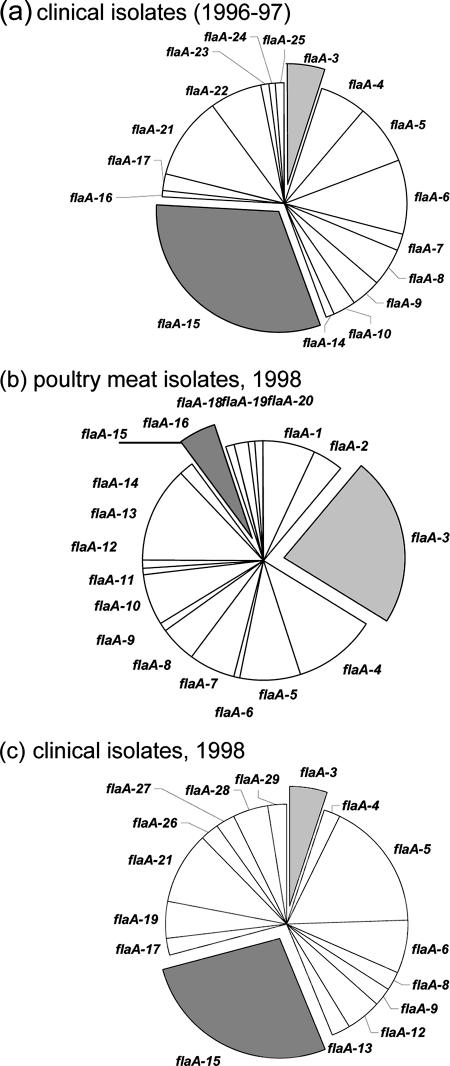
flaA-15 strains were most prevalent among human fecal isolates in both years in which isolates were obtained from the diagnostic laboratory (Fig. 1). In contrast, flaA-3 and flaA-13 strains were the most common in chicken portions (23% and 13%, respectively). flaA-15 strains were detected in chicken portions but at a low level of prevalence (Fig. 1). Thus, flaA-15 strains were a consistent cause of human gastroenteritis over 2 years of observation, yet Campylobacter strains of this type were relatively uncommon in chicken portions sold by food outlets in the patient catchment. Conversely, consumers were commonly exposed to flaA-3 and flaA-13 strains, yet few human infections resulted from this exposure (5% and 0%, respectively).
FIG. 1.
Comparison of flaA types detected among clinical isolates obtained in 1996 and 1997 (a), poultry meat isolates obtained in 1998 (b), or clinical isolates obtained in 1998 (c). The prevalence of each strain is shown as wedges of pie graphs.
Comparison of invasive abilities of C. jejuni strains.
Table 2 records the abilities of representative strains of the flaA-3, -13, and -15 types to invade HEp-2 cells. flaA-15 strains were five to six times more invasive than flaA-3 and flaA-13 strains (Mann-Whitney nonparametric test; P < 0.0001). Therefore, the flaA-15 strains were considered to be potentially more virulent than strains of the other two flaA RFLP types (12, 33).
TABLE 2.
In vitro invasive abilities of C. jejuni strains
| Gene and RFLP type | C. jejuni strain | No. of C. jejuni cells within HEp-2 cell lysate (CFU/ml)c | Mean invasion levela | Relative invasive abilityb |
|---|---|---|---|---|
| flaA-15 | T1018 | 2.69 × 104 (2.40 × 103) | 5.58 × 104 | 100.0 |
| T157 | 2.01 × 104 (7.76 × 102) | |||
| T1012 | 9.02 × 104 (6.17 × 103) | |||
| T1016 | 8.62 × 104 (8.01 × 103) | |||
| flaA-13 | T3 | 1.30 × 104 (7.86 × 102) | 1.18 × 104 | 21.1 |
| WCW | 1.20 × 104 (1.74 × 103) | |||
| T4 | 1.04 × 104 (1.24 × 103) | |||
| flaA-3 | PSTT2 | 1.31 × 104 (6.81 × 103) | 0.93 × 104 | 16.7 |
| Pst1 | 8.03 × 103 (8.54 × 102) | |||
| Tau1 | 1.30 × 104 (5.16 × 102) | |||
| PStau | 3.25 × 103 (2.46 × 102) |
Average number of CFU/ml of strains tested; applies to all strains in RFLP group.
(Mean invasion level of each flaA group/mean invasion level of flaA-15 strains) × 100; applies to all strains in RFLP group.
Results are means (standard errors of the means).
Colonization of the chicken gut.
Preliminary experiments showed that batches of 1-day-old chickens obtained from the local supplier were free of C. jejuni. Inoculation of chickens with either Campylobacter strain T1016 (flaA-15) or Pstau (flaA-3) resulted in colonization of the birds (Table 3, colonization groups A and B). Similar Campylobacter population levels were present in gut samples regardless of whether the birds were examined 7 or 14 days after inoculation (data not shown; Mann-Whitney test, P > 0.05). In general, selective agar plate enumerations gave total Campylobacter population sizes similar to those obtained using Taqman real-time quantitative PCR (Table 3).
TABLE 3.
Campylobacter populations in cecum samples from chickens inoculated with T1016 and/or Pstau
| Chick colonization group, description | C. jejuni plate count (CFU per g of cecum contents)a | C. jejuni Taqman real-time qPCR count (CFU per g of cecum contents)b | flaA-15 strain population size as % of C. jejuni Taqman real-time qPCR count |
|---|---|---|---|
| A, T1016 (flaA-15) | 1.04 × 109 (3.1 × 108) | 1.74 × 109 (3.3 × 108) | 100 |
| B, PStau (flaA-3) | 1.25 × 109 (2.5 × 108) | 5.50 × 109 (9.2 × 107) | Not applicable |
| C, T1016 (flaA-15)/PStau (flaA-3), mixed 1:1 | 1.16 × 109 (2.3 × 108) | 1.80 × 109 (3.9 × 108) | 99 |
| D, T1016 (flaA-15) and then PStau (flaA-3) | 1.89 ×109 (6.5 × 108) | 1.05 × 1010 (2.5 × 108) | 100 |
| E, T1016 (flaA-15) then PStau (flaA-3) | 1.66 × 109 (3.8 × 108) | 8.16 × 109 (2.6 × 109) | 100 |
| F, PStau (flaA-3) and then T1016 (flaA-15) | 1.84 × 109 (4.8 × 108) | 3.40 × 109 (1.7 × 109) | 100 |
| G, Pstau (flaA-3) and then T1016 (flaA-15) | ND | 1.83 × 109 (2.86 × 108) | 85 |
Results are means (standard errors of the means). ND, not done.
Results are means (standard errors of the means). qPCR, quantitative PCR.
Competitive performance of Campylobacter strains in the chicken gut.
Regardless of the order of inoculation, strain T1016 formed the greatest proportion of the total Campylobacter population relative to Pstau (Table 3, colonization groups C to G)). Therefore, in addition to being potentially more virulent for humans (HEp-2 cell invasion), the flaA-15 strain was more competitive in the chicken gut than the flaA-3 strain.
Characterization of genetic diversity of strains T1016 and Pstau using C. jejuni NCTC 11168 microarray.
Gene conservation assessments were made by comparing microarray data using empirical rules described previously for Campylobacter strain genomic analyses (41). Using the format “T1016 call-Pstau call,” decisions were made in relation to microarray scores (log ratios) as to whether the gene in question was absent, likely to be conserved, or likely to be divergent with respect to the reference genome. Four clusters of genes in which the genomes of T1016 and Pstau differed were noted (Table 4). Neither strain harbored genes carried by the virulence plasmid pVir (3) (Table 4). The cluster of six genes (Cj1321 to Cj1326) considered to be characteristic of strains originating in livestock (6), including chickens, was absent from T1016 and Pstau, with the exception of Cj1325, which was detected in Pstau.
TABLE 4.
Genomic comparison of strains T1016 and Pstau relative to NCTC 11168
| GeneIDa | Putative protein | Microarray differential | Gene conservation assessment (T1016, Pstau)b |
|---|---|---|---|
| Cj0030 | Hypothetical protein | −2.31 | A, C |
| Cj0057 | Periplasmic protein | −2.27 | D, C |
| Cj0177 | Outer membrane siderophore | −2.77 | A, C |
| Cj0178 | Outer membrane siderophore | −3.45 | A, C |
| Cj0179 | Biopolymer transport protein | −2.48 | D, C |
| Cj0180 | Biopolymer transport protein | −2.56 | D, C |
| Cj0181 | TonB transport protein | −2.52 | A, C |
| Cj0481 | Lyase | −3.03 | A, C |
| Cj0483 | Altronate hydrolase C-terminus | −2.81 | A, C |
| Cj0484 | Transmembrane transport protein | −2.53 | D, C |
| Cj0486 | Sugar transport protein | −2.33 | D, C |
| Cj0490 | Aldehyde dehydrogenase C-terminus | −2.85 | A, C |
| Cj0690c | Restriction/modification enzyme | −3.16 | A, C |
| Cj0727 | Periplasmic solute-binding protein | −2.46 | A, C |
| Cj0728 | Periplasmic protein | −2.90 | A, C |
| Cj0730 | ABC transport system permease | −2.57 | A, C |
| Cj0731 | ABC transport system permease | −3.00 | A, C |
| Cj0732 | ABC transport system ATP-binding protein | −2.53 | A, C |
| Cj0733 | Hypothetical protein | −2.72 | A, C |
| Cj0736 | Hypothetical protein | −4.18 | A, C |
| Cj0741 | Hypothetical protein | −3.02 | A, C |
| Cj0755 | Iron uptake protein | −3.11 | A, C |
| Cj0860 | Integral membrane protein | 2.60 | C, A |
| Cj1055c | Integral membrane protein | 2.73 | C, A |
| Cj1141 (neuB1) | N-Acetylneuraminic acid synthetase | −1.78 | A, D |
| Cj1307 | Amino acid activating enzyme | −2.57 | A, C |
| Cj1417c | Hypothetical protein | 2.00 | C, D |
| Cj1420c | Hypothetical protein | 3.16 | C, A |
| Cj1439c | UDP-galactopyranose mutase | −1.20 | A, D |
| Cj1442c | Hypothetical protein | 2.36 | C, A |
| Cj1555c | Hypothetical protein | −2.29 | A, D |
| Cj1585c | Oxidoreductase | −3.13 | C, A |
| Cj1668c | Periplasmic protein | −1.64 | A, C |
| Cj1678 | Lipoprotein | 2.33 | A, C |
| Cj1727c | MetY, O-acetylhomoserine (thiol)-lyase | −2.80 | D, C |
| LIO87_ORF18d | 3.00 | D, A | |
| CjPO3_Cj1135 | Two-domain glycosyltransferase | −1.46 | A, D |
| RM1221_ORF0432 | Virulence plasmid | −1.94 | A, D |
| RM1221_ORF0433 | Virulence plasmid | −2.05 | A, D |
| RM1221_ORF0434 | Virulence plasmid | −1.35 | A, D |
| RM1221_ORF1270 | Virulence plasmid | −2.88 | A, D |
| RM1221_ORF1280 | Virulence plasmid | −3.46 | A, C |
| RM1221_ORF1704 | Virulence plasmid | −3.17 | A, C |
| RM1221_ORF1956 | Virulence plasmid | 1.56 | D, A |
Clusters of genes that differ between strains are in bold font. ID, identifier.
A, likely to be absent; C, likely to be conserved; D, likely to be divergent.
Comparison of FlaA proteins of T1016 and Pstau.
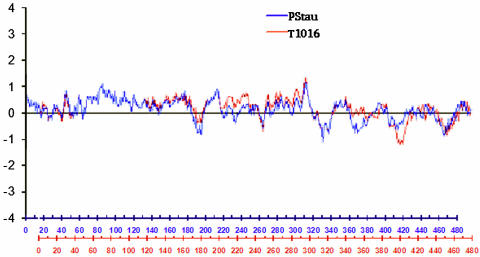
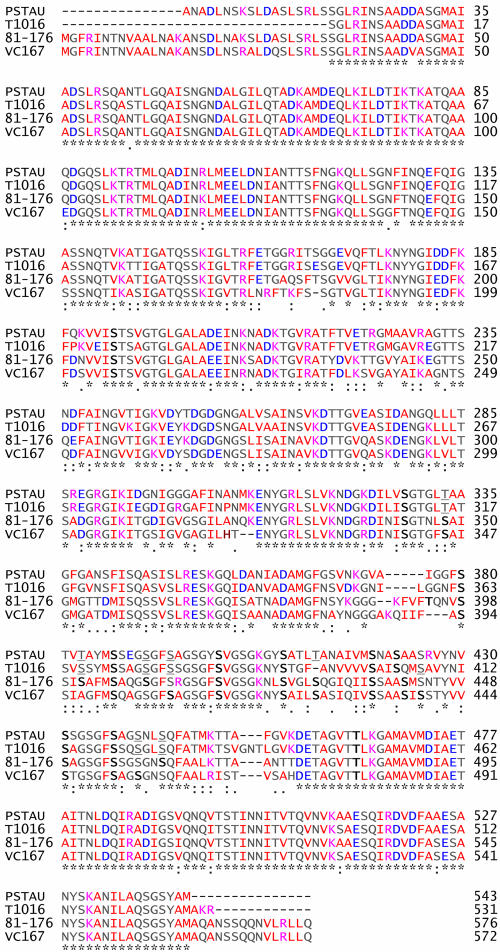
The hydrophobicity (residues 220 to 260) and hydrophilicity (residues 400 to 420) of the FlaA proteins of T1016 and Pstau differed (Fig. 2). The predicted amino acid sequences of the FlaA proteins showed 86% identity, having conserved N- and C-terminal domains and a central variable region as reported by Logan and colleagues (23). Eighteen residues were detected as putative glycosylation sites in T1016 and Pstau (Fig. 3). Eleven serine/threonine residues known to be glycosylated in 81-176 and VC176 (42) were conserved in T1016 and Pstau. FlaA of Pstau had three serine residues, which are glycosylated in 81-176 and VC176, replaced by threonine (T333, T383, and T412). T1016 had only one substitution of this nature (T315). One serine residue (S366) in the FlaA protein of T1016 is likely to be glycosylated because it is near hydrophobic residues and a glycosylated serine residue of 81-176 (42). Additionally, T1016 lacks a conserved residue between F394 and A395 known to be glycosylated in other strains. Therefore, the FlaA proteins of T1016 and Pstau varied in putative sites of glycosylation and in the hydropathy profile.
FIG. 2.
Hydropathic profiles of T1016 and Pstau FlaA proteins (Kyte-Doolittle calculation over a window length of 19).
FIG. 3.
Alignment of predicted amino acid sequences of FlaA proteins. Campylobacter strains are listed on the left. The asterisk indicates identity, whereas the colon and period indicate amino acid similarities. Boldface letters show conserved residues in Pstau and T1016 that have been shown to be glycosylated in 81-176 and VC167. Underlined letters indicate residues that may be modified given the hydrophobic nature of surrounding residues and/or conserved localization with a S/T residue that has been shown to be modified in 81-176 or VC167. Red letters indicate hydrophobic residues, whereas lilac and blue indicate residues that are negatively and positively charged, respectively. Numbers at right facilitate amino acid positioning.
DISCUSSION
Extensive genome-based investigations of C. jejuni strains in recent years, although confounded to some extent by huge differences in gene content between strains, have provided clues as to factors involved in the pathogenesis of human infections and colonization of the chicken intestine but do not yet provide a comprehensive view of the mechanisms of C. jejuni virulence and commensal colonization (9, 18, 27, 28). These studies have generally focused on single isolates of C. jejuni. In contrast, we have compared properties exhibited by two strains isolated from poultry meat and which were chosen on the basis of epidemiological data obtained in a defined geographical area of New Zealand. These data were based on RFLP of the flaA gene, recently validated as a suitable high-throughput alternative to multilocus sequence typing (8). flaA-15 strains were a predominant cause of human diarrhea but had a much lower prevalence in poultry meat. Therefore, consumers in the catchment region had low exposure to, but high infection rates with, flaA-15 strains. Conversely, flaA types commonly encountered in poultry meat, such as flaA-3, were seldom associated with infection. This outcome might reflect the existence of other, currently unknown sources of human infection caused by flaA-15 strains. Alternatively, it might indicate that flaA-15 strains had higher virulence for humans than, for example, flaA-3 strains; indeed, on the basis of the results of the in vitro invasion assay, flaA-15 strains could be considered to be the more virulent (12, 21, 33).
These observations opened two areas of investigation: comparison of flaA-15 and flaA-3 strains for genetic or phenotypic differences that might explain differing virulence and an opportunity to explore competitive exclusion in the chicken intestine. Perhaps the less-virulent strain could be used to reduce colonization of the chicken intestine by the more-virulent strain? If this method were successful and were used commercially, the prevalence of the human virulent strain could be reduced in broiler flocks, thus minimizing exposure of consumers to the virulent strain.
Genetic comparisons between strains T1016 (flaA-15) and Pstau (flaA-3) were conducted by performing microarray-based comparative genomic hybridization, despite this approach being limited by what genes are represented in the genome of the standard strain (NCTC 11168, Penner O2) and that in vivo gene expression may be a more valid comparison than the presence or absence of genes in the genome (37, 43). Nevertheless, our comparison revealed that the two strains differed in the content of three gene clusters. Genes within regions encoding Cj0177 to Cj0181, Cj0481 to Cj0490, and Cj0727 to Cj0736 were absent or divergent in T1016 but conserved with respect to the standard strain in Pstau. In contrast, some genes within the cluster Cj1417c to Cj1442c were conserved in T1016 but absent or divergent in Pstau. These genes, specifically Cj1417c, Cj1420c, and Cj1442c, are therefore of potential interest for further research because they were absent or divergent in the strain of lesser virulence and because they are within the genomic region encoding capsule formation (Penner typing) (9, 20). Mutation of these genes in T1016 and other similar strains might impact on virulence and colonization of the chicken intestine (3) and assist in obtaining detailed mechanistic knowledge of these phenomena.
Glycosylation of Campylobacter flagellin is associated with virulence and colonization of the chicken intestine (17, 38). In silico comparison of the glycosylation sites in the FlaA molecules of the two strains showed marked differences. These variations in glycosylation of FlaA might influence bacterial adherence (13, 42) and interactions with the immune system (39). Moreover, differences in the hydropathy profile of the FlaA proteins from T1096 and Pstau were observed, located mainly in the central portion of the protein, which, in other Campylobacter flagellins, is known to be surface exposed (31).
T1016 always outcompeted Pstau in the chicken intestine, no matter the order in which the strains were used to inoculate the birds. The detection and measurement of the Campylobacter populations utilized culture-based and nucleic acid-based analytical methods because C. jejuni can form viable but nonculturable cells (32) which would nevertheless be detectable by PCR. Real-time, quantitative PCR also permitted differential counts of the two strains to be made with a lower detection limit than that obtained in previous research (7) and avoided the use of mutant bacteria whose colonization potential might have been altered from the wild type (4). These assays showed that the more virulent strain (T1016) was the best adapted of the two strains for life in the chicken intestine because it was always numerically dominant. It seems, admittedly based on circumstantial observations in the case of our study, that virulence and commensal colonization properties may be linked, therefore holding little promise for competitive exclusion of higher-virulence strains by those of lesser virulence.
The two strains differed in Penner type, a phenotypic reflection of capsular material on the bacterial cell surface (9, 20). Whereas T1016 was of Penner type O42, Pstau belonged to O53. This difference in serotype was presumably associated with altered gene content that we detected within the genomic regions encoding capsule formation (9, 20), a phenotypic trait that is clearly linked to virulence (2, 30).
We concluded from our study that poultry meat was at least one source of human infection with C. jejuni in the Dunedin region, that some Campylobacter strains detected in poultry meat are of higher virulence for humans than others, and that bacterial attributes affecting strain virulence and commensal colonization may be linked. This last observation is in keeping with the views of other researchers (14, 44). Overall, the results of our strain comparisons support the view that the success of C. jejuni as a commensal and as a pathogen is related to its potential to remodel its cell surface properties (37), and they help to explain the epidemiological results that we obtained concerning this pathogen, which is a considerable hazard to public health in New Zealand. Our study provides a basis for future research in which the cell surface properties of T1016 and Pstau could be modified and examined experimentally in order to better understand the determinants of Campylobacter virulence and the mediators of ecological behavior.
Acknowledgments
Development of the real-time, quantitative PCR assay was supported by the Poultry Industry Association of New Zealand.
Footnotes
Published ahead of print on 5 October 2007.
REFERENCES
- 1.Allos, B. M. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201-1206. [DOI] [PubMed] [Google Scholar]
- 2.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacon, D. J., C. M. Szymanski, D. H. Burr, R. P. Silver, R. A. Alm, and P. Guerry. 2001. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40:769-777. [DOI] [PubMed] [Google Scholar]
- 4.Barrow, P., and K. Page. 2000. Inhibition of colonisation of the alimentary tract of young chickens with Campylobacter jejuni by pre-colonisation with strains of C. jejuni. FEMS Microbiol. Lett. 182:87-91. [DOI] [PubMed] [Google Scholar]
- 5.Bodenteich, A. S., Y. Chisoe, F. Wang, and B. A. Roe. 1994. Shotgun cloning as the strategy of choice to generate templates for high throughput dideoxynucleotide sequencing, p. 42-50. In M. D. Adams, C. Fields, and C. Venter (ed.), Automated DNA sequencing and analysis techniques. Academic Press, London, United Kingdom.
- 6.Champion, O. L., M. W. Gaunt, O. Gundogdu, A. Elmi, A. A. Witney, J. Hinds, N. Dorrell, and B. W. Wren. 2005. Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc. Natl. Acad. Sci. USA 102:16043-16048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., and N. Stern. 2001. Competitive exclusion of heterologous Campylobacter spp. in chicks. Appl. Environ. Microbiol. 67:848-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Djordjevic, S. P., L. E. Unicomb, P. J. Adamson, L. Mickan, R. Rios, and the Australian Campylobacter Study Group. 2007. Clonal complexes of Campylobacter jejuni identified by multilocus sequence typing are reliably predicted by restriction fragment length polymorphism analyses of the flaA gene. J. Clin. Microbiol. 45:102-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberhart-Phillips, J., N. Walker, N. Garrett, D. Bell, D. Sinclair, W. Rainger, and M. Bates. 1997. Campylobacteriosis in New Zealand: results of a case-control study. J. Epidemiol. Commun. Health 51:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everest, P. H., H. Goosens, J. P. Butzler, D. Lloyd, S. Knutton, J. M. Ketley, and P. H. Williams. 1992. Differentiated Caco-2 cells as a model for enteric invasion by Campylobacter jejuni and C. coli. J. Med. Microbiol. 37:319-325. [DOI] [PubMed] [Google Scholar]
- 13.Grant, C. C., M. E. Konkel, W. Cieplak, and L. S. Tompkins. 1993. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect. Immun. 61:1764-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanel, I., J. Muller, W. Muller, and F. Schulze. 2004. Correlation between invasion of Caco-2 eukaryotic cells and colonization ability in the chick gut in Campylobacter jejuni. Vet. Microbiol. 101:75-82. [DOI] [PubMed] [Google Scholar]
- 15.Harmon, K. M., G. M. Ransom, and I. V. Wesley. 1997. Differentiation of Campylobacter jejuni and Campylobacter coli by polymerase chain reaction. Mol. Cell. Probes 11:195-200. [DOI] [PubMed] [Google Scholar]
- 16.Harvey, S. M. 1980. Hippurate hydrolysis by Campylobacter fetus. J. Clin. Microbiol. 11:435-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471-484. [DOI] [PubMed] [Google Scholar]
- 18.Hofreuter, D., J. Tsai, R. O. Watson, V. Novik, B. Altman, M. Benitez, C. Clark, C. Perbost, T. Jarvie, L. Du, and J. E. Galan. 2006. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74:4694-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jorgensen, F., R. Bailey, S. Willimas, P. Henderson, D. R. Wareing, F. J. Bolton, J. A. Frost, L. Ward, and T. J. Humphrey. 2002. Prevalence and numbers of Salmonella and Campylobacter spp. on raw, whole chickens in relation to sampling methods. Int. J. Food Microbiol. 76:151-164. [DOI] [PubMed] [Google Scholar]
- 20.Karlyshev, A. V., D. Linton, N. A. Gregson, A. J. Lastovica, and B. W. Wren. 2000. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Microbiol. 35:529-541. [DOI] [PubMed] [Google Scholar]
- 21.Konkel, M. E., and L. A. Joens. 1989. Adhesion to and invasion of HEp-2 cells by Campylobacter spp. Infect. Immun. 57:2984-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 23.Logan, S. M., J. F. Kelly, P. Thibault, C. P. Ewing, and P. Guerry. 2002. Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol. Microbiol. 46:587-597. [DOI] [PubMed] [Google Scholar]
- 24.Mattick, K., K. Durham, G. Domingue, F. Jorgenson, M. Sen, D. W. Schaffner, and T. Humphrey. 2003. The survival of foodborne pathogens during domestic washing-up and subsequent transfer onto washing-up sponges, kitchen surfaces and food. Int. J. Food Microbiol. 85:213-226. [DOI] [PubMed] [Google Scholar]
- 25.Nachamkin, I., K. Bohachick, and C. M. Patton. 1993. Flagellin gene typing of Campylobacter jejuni by restriction fragment length polymorphism analysis. J. Clin. Microbiol. 31:1531-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 27.Parker, C. T., B. Quinines, W. G. Miller, S. T. Horn, and R. E. Mandrell. 2006. Comparative genomic analysis of Campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in C. jejuni strain RM1221. J. Clin. Microbiol. 44:4125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson, B. M., C. Pin, J. Wright, K. I'Anson, T. Humphrey, and J. M. Wells. 2003. Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays. FEBS Lett. 554:224-230. [DOI] [PubMed] [Google Scholar]
- 29.Penner, J. L., and J. N. Hennessy. 1980. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J. Clin. Microbiol. 12:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poly, F., D. Threadgill, and A. Stintzi. 2004. Identification of Campylobacter jejuni ATCC 43431-specific genes by whole microbial genome comparisons. J. Bacteriol. 186:4781-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Power, M. P., P. Guerry, W. D. McCubbin, C. M. Kay, and T. J. Trust. 1994. Structural and antigenic characteristics of Campylobacter coli FlaA flagellin. J. Bacteriol. 176:3303-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rollins, D. M., and R. R. Colwell. 1986. Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl. Environ. Microbiol. 52:531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz-Palacios, G. M., L. E. Cervantes, D. S. Newburg, Y. Lopez-Vidal, and J. J. Calva. 1992. In vitro models for studying Campylobacter infections, p. 176-183. In I. Nachamkin, M. J. Blaser, and L. S. Tompkins (ed.), Campylobacter jejuni: current status and future trends. American Society for Microbiology, Washington, DC.
- 34.Saal, L. H., C. Troein, J. Vallon-Christersson, S. Gruvberger, A. Borg, and C. Peterson. 2002. BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biol. 3:SOFTWARE0003. [DOI] [PMC free article] [PubMed]
- 35.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 36.Savill, M., A. Hudson, M. Devane, N. Garrett, B. Gilpin, and A. Ball. 2003. Elucidation of potential transmission routes of Campylobacter in New Zealand. Water Sci. Technol. 47:33-38. [PubMed] [Google Scholar]
- 37.Stintzi, A., D. Marlow, K. Palyada, H. Naikare, R. Panciera, L. Whitworth, and C. Clarke. 2005. Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect. Immun. 73:1797-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szymanski, C. M., D. H. Burr, and P. Guerry. 2002. Campylobacter protein glycosylation affects host cell interactions. Infect. Immun. 70:2242-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szymanski, C. M., S. M. Logan, D. Linton, and B. W. Wren. 2003. Campylobacter—a tale of two protein glycosylation systems. Trends Microbiol. 11:233-238. [DOI] [PubMed] [Google Scholar]
- 40.Taboada, E. N., R. R. Acedillo, C. D. Carrillo, W. A. Findlay, D. T. Medeiros, O. L. Mykytczuk, M. J. Roberts, C. A. Valencia, J. M. Farber, and J. H. Nash. 2004. Large-scale comparative genomics meta-analysis of Campylobacter jejuni isolates reveals low level of genome plasticity. J. Clin. Microbiol. 42:4566-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taboada, E. N., R. R. Acedillo, C. C. Luebbert, W. A. Findlay, and J. H. Nash. 2005. A new approach for the analysis of bacterial microarray-based comparative genomic hybridization: insights from an empirical study. BMC Genomics 6:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thibault, P., S. M. Logan, J. F. Kelly, J.-R. Brisson, C. P. Ewing, T. J. Trust, and P. Guerry. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276:34862-34870. [DOI] [PubMed] [Google Scholar]
- 43.Woodall, C. A., M. A. Jones, P. A. Barrow, J. Hinds, G. L. Marsden, D. J. Kelly, N. Dorrell, B. W. Wren, and D. J. Maskell. 2005. Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infect. Immun. 73:5278-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziprin, R., C. Young, J. Byrd, L. Stanker, M. Hume, S. Gray, B. Kim, and M. Konkel. 2001. Role of Campylobacter jejuni potential virulence genes in cecal colonization. Avian Dis. 45:549-557. [PubMed] [Google Scholar]