Abstract
Insight into the mechanism of lipid transport to the outer membrane of gram-negative bacteria has been hampered by the lack of an effective genetic screen for defective mutants. This work demonstrates an enrichment of conditional mutants defective in lipopolysaccharide export by Ludox density gradient centrifugation and selection for detergent resistance. New temperature-sensitive mutants with lipid export defects were isolated with single missense mutations in msbA. The results demonstrate the power of this approach for the study of lipid export in Escherichia coli.
The envelope of Escherichia coli and other gram-negative bacteria contains two distinct lipid bilayers. These are the inner membrane (IM) and the outer membrane (OM), which are separated by the periplasmic space. The OM is an asymmetric lipid bilayer consisting of an inner face of phospholipids and an outer face of lipopolysaccharide (LPS) (26). LPS is a complex glycolipid unique to gram-negative bacteria that is required for growth of most strains and is a powerful activator of the mammalian innate immune response via its activation of Toll-like receptors (23). The OM also contains a unique set of lipoproteins and β-barrel integral membrane proteins.
The components of the OM, both lipids and proteins, are synthesized in the cytoplasm by use of cytoplasmic precursors and enzymes. These molecules are then transported across the inner membrane: proteins via the SecYEG machinery (27, 35) and lipids by an essential ABC transporter protein called MsbA (5, 26). A genetic approach undertaken to study the role of MsbA in lipid transport resulted in the isolation of an msbA temperature-sensitive mutant (WD2) that carries the single missense mutation A270T. WD2 accumulates newly synthesized LPS and phospholipids at the cytoplasmic leaflet of the inner membrane after short growth at the nonpermissive temperature, resulting in the formation of inner membrane invaginations visible by electron microscopy (6, 9). These results suggest that MsbA is a general lipid transporter and transports both LPS and phospholipids across the IM of E. coli. Acceptance of this interpretation, however, is far from settled, as results obtained using Neisseria meningitidis (a strain that does not require LPS for viability) (31) suggest that MsbA is strictly an LPS transporter (32).
A number of recent reports have focused on the identification of the factors required for assembly of the components of the gram-negative OM (reviewed in reference 29). A major player required for the delivery of LPS to the outer surface of the OM is Imp, an essential OM protein (3, 33). Imp forms an OM complex with the lipoprotein RlpB required for the display of LPS on the surface of the cell (36). LptA, a periplasmic protein, and LptB, a cytoplasmic ATPase, have recently been identified as being necessary for the transport of LPS from the inner membrane to the OM of E. coli (30). In spite of this recent progress, much remains to be learned regarding the mechanisms of lipid trafficking to the OM in gram-negative bacteria. Insights into this process will broaden our understanding of the biology of the gram-negative envelope and could result in the identification of new drug targets. LPS biosynthesis and transport are both essential processes in E. coli (26). LPS biosynthesis is a promising potential drug target, as broad-range LpxC inhibitors are currently being screened and identified as potential therapeutics (1, 13, 14, 20). A genetic screening technique termed chemical conditionality for isolation new mutants with defects in OM protein assembly has been described previously (28). A genetic screen for mutants in the essential pathway of lipid transport to the OM will therefore be very valuable.
Conditional mutations in the eukaryotic protein export pathway often result in alterations in cell density under nonpermissive conditions. This observation led to an effective screen for so-called Sec (secretion) mutants in the organism Saccharomyces cerevisiae by use of the colloidal silica Ludox (19). Ludox forms a density gradient spontaneously during exposure to moderate centrifugal forces (24), and viable cells can be recovered from the gradient. This work demonstrates the use of density gradient centrifugation with Ludox to enrich for temperature-sensitive E. coli mutants with defects in lipid export. This method may be useful in isolating novel conditional mutants with defects in membrane biogenesis.
Description of methodology.
In order to determine whether loss of lipid export in the msbA conditional mutant WD2 is accompanied by changes in cell density, parent strain W3110A (W3110 aroA::Tn10; msbA+) and WD2 (W3110A; msbA2) (9) were grown briefly under nonpermissive conditions (44°C for 30 min) in LB containing tetracycline (12.5 μg/ml). Importantly, during this time frame, WD2 cells are growing logarithmically and can be recovered with little loss of viability (9). Cells were harvested and washed, and migration of live cells was analyzed on a Ludox density gradient following moderate centrifugation. Density medium was made as described by Poole (24) and contained 3.75% (wt/vol) polyvinylpyrrolidone (PVP) and 16% (wt/vol) Ludox HS-40 in phosphate-buffered saline (PBS; pH 7.4). Both PVP (catalog no. PVP-40) and Ludox (catalog no. 420816) were purchased from the Sigma Chemical Company, St. Louis, MO. To make 500 ml of media, 50 ml of 10× PBS was added to 18.75 g of PVP and enough water was added to adjust the volume to 300 ml. Once the PVP was dissolved, 200 ml of 40% (wt/vol) Ludox HS-40 was added with stirring. The pH of the Ludox was adjusted to ∼7.5 with concentrated HCl before (but not after) adding to the dissolved PVP. The density was adjusted to 1.11 g/ml with 3.75% PVP in water. The final concentration of PVP was therefore 3.75%, and the final concentration of Ludox was approximately 16% to give a solution of a density of 1.11 g/ml. This solution was used to fill an ultracentrifuge tube (SW41; Beckman), and gradient formation was carried out by centrifugation at 50,000 × g (20,000 rpm) for 1 h at 4°C prior to addition of cells to the top of the tubes. Approximately 1010 E. coli cells were added to the tubes in 0.2 ml PBS, and the gradients were spun at 50,000 × g for 45 min. Cells were recovered from the tube in 0.6-ml aliquots using an 18-gauge needle to puncture the side of the tube approximately 2.5 cm from the bottom.
WD2 migrates distinctly on Ludox gradients.
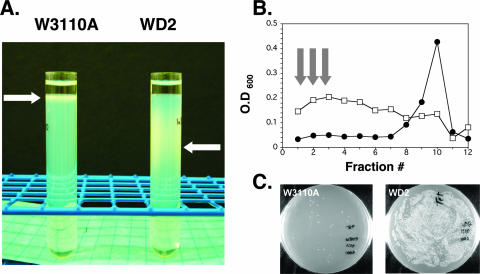
W3110A E. coli is found very close to the top of the gradient under these conditions. As judged by optical density measurement and plating of diluted fractions, very few cells are found more than a few centimeters from the top of the centrifuge tube (Fig. 1). In contrast, WD2 that had been temperature shifted migrated to denser regions of the gradient (Fig. 1). Cells were found broadly distributed on the gradient, with a peak of cells migrating halfway from the bottom of the tube. It is likely that growing W3110A cells also migrate at a range of densities; however, these results suggest those densities are not within the resolvable range of these Ludox gradients. Exponential-phase E. coli has been reported to migrate at a range of densities of between 1.03 and 1.10 on Percoll gradients (17). That study also found that the cell density of E. coli increases upon prolonged growth in stationary phase (17). All cells described in the current report were grown only in exponential phase. WD2 therefore displays an apparent density more like that of stationary-phase cells during growth in exponential phase. By performing experiments using density marker beads on identical gradients, it was found that W3110A and WD2 mutant cells migrated at densities of <1.10 and 1.12, respectively, under these conditions. Ludox resolves in a density range of 1.098 to 1.142 under these conditions based upon marker bead migration. While Percoll (22; GE Healthcare Biosciences, Uppsala, Sweden) is very similar to Ludox in its density-forming properties (resolving in a density range of 1.098 to 1.121 under these conditions) and is somewhat easier to use, Ludox provides a steeper gradient and gave better enrichment of msbA mutant cells in our hands. Cells isolated from the Ludox density gradient can be plated and grown at the permissive temperature with only a small loss of viability (Fig. 1C). In this manner, Ludox gradients can provide an at least 10- to 25-fold increase in enrichment of WD2 compared to W3110A based upon the average results obtained with several gradients. The conditional lpxA mutant SM101 (11) migrated similarly to the parent strain on this gradient following growth at the nonpermissive temperature for 30 min (data not shown), indicating that loss of LPS transport, but not synthesis, results in altered mobility of cells on Ludox gradients.
FIG. 1.
Sedimentation of msbA conditional mutant WD2 cells on a Ludox density gradient. Mutant WD2 and parent E. coli W3110A cells were grown and analyzed using a Ludox density gradient as described in the text. (A) Photograph of the centrifuge tubes following centrifugation. Arrows show the area where most cells are found. (B) Light scattering of each fraction was measured by recording the A600 of individual diluted samples. Fractions from an identical blank gradient were used to subtract background absorbance. Black circles refer to A600 of fractions from gradient containing W3110A cells, and squares refer to WD2. O.D 600, optical density at 600 nm. (C) Equal volumes of pooled fractions 1, 2, and 3 (arrows in panel B) from W3110A (left plate) or WD2 (right plate) tubes were plated to assess relative CFU levels recovered from the gradient and to demonstrate cell viability. Plates were incubated at 30°C overnight.
Ludox enrichment of temperature-sensitive mutants.
In order to isolate a collection of temperature-sensitive mutants with density properties similar to those of WD2, W3110A cells were mutagenized for 5 min at 37°C with nitrosoguanidine (50 μg/ml), washed several times with ice-cold PBS, and allowed to recover at 30°C for 2 h (9). Mutagenized W3110A and WD2 cells were then diluted into fresh LB and grown at 42°C for 30 min in log phase. Cells were washed, resuspended in 0.2 ml PBS, layered on top of a Ludox density gradient, and centrifuged as described above. Most of the mutagenized cells were recovered near the top of the gradient where unmutagenized cells were located (data not shown). Using the migration of WD2 cells as a guide, fractions from the gradient with mutagenized cells near the density of 1.12 were pooled, diluted to 25 ml with LB containing 20% glycerol, and stored at −80°C. Aliquots of cells were plated on LB-tetracycline plates to give approximately 100 to 200 colonies per plate following an overnight incubation at 30°C. Temperature-sensitive colonies were isolated at a frequency of ∼1% by replica plating at 30 and 42°C.
Coisolation of deep rough mutants.
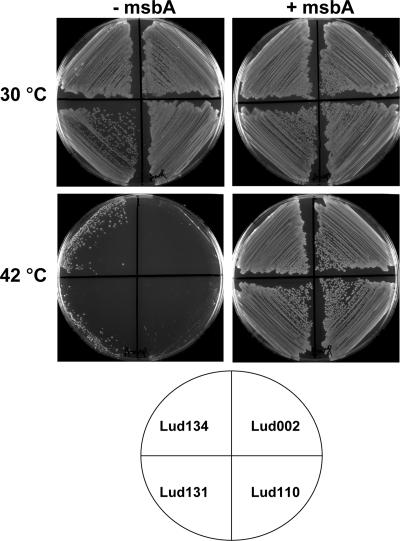
Many temperature-sensitive mutants from this first round of screening were found to contain truncated LPS (not required for growth at 42°C) in addition to mutations causing temperature sensitivity. This is commonly referred to as a deep rough phenotype. Analysis of the phenotype of these cells was based upon the solubility of their LPS (25) in a single-phase acidic Bligh-Dyer solution (2, 18) and by their sensitivity to detergents (reference 26 and data not shown). The reason for the apparent enrichment of deep rough mutants by the density gradient is not clear at this time but may be related to the absence of surface-exposed carbohydrate in both deep rough and lipid export mutants. In order to remove deep rough mutants from the collection of dense mutants, mutagenized Ludox-enriched cells were plated at 30°C on LB containing 0.1% sodium dodecyl sulfate (SDS) and appropriate antibiotics and grown overnight at 30°C prior to replica plating. These conditions are lethal for deep rough mutants but do not kill W3110A E. coli or WD2 at 30°C (W. T. Doerrler, unpublished observations). Plating on 0.1% SDS removed an additional 75 to 90% of CFU from the density-enriched cells. SDS-resistant temperature-sensitive mutants isolated from the density gradient were dramatically enriched with msbA or msbA-like mutants, as determined by the ability of a plasmid copy of msbA to restore their growth at 42°C (Fig. 2). Eleven out of 23 mutants in an initial screening were complemented with a plasmid containing the msbA gene. Not all these mutants, however, harbored msbA mutations (see below). The combined use of density enrichment (10- to 25-fold) and detergent resistance (4- to 10-fold) therefore produces a total predicted 40- to 2,500-fold enrichment for this method.
FIG. 2.
Restoration of growth at 42°C to Ludox msbA mutants harboring a plasmid copy of wild-type msbA. Ludox msbA mutants were transformed with pWSK29 (−msbA) or pWTD10 (+msbA), and ampicillin-resistant colonies were streaked on LB-ampicillin (100 μg/ml) plates and grown overnight at 30 and 42°C. pWTD10 was constructed by cloning the XbaI/BamHI fragment from pWTD1 (7) into a similarly digested and dephosphorylated vector (pWSK29) (34). IPTG (isopropyl-β-d-thiogalactopyranoside) was not included in the growth media due to toxicity caused by overexpression of MsbA.
New msbA mutants isolated.
The msbA structural gene was sequenced from mutants whose growth was complemented by a plasmid copy of msbA (Fig. 2), and a new collection of temperature-sensitive msbA missense mutants was identified (Table 1). Among these mutants were Lud131 and Lud134, each harboring the mutation A270T, also found in WD2, a mutant previously isolated using localized mutagenesis (9). This mutation was found independently two times in this screening, perhaps indicating that this is a mutational “hotspot” for conditional msbA mutations. Additional mutants isolated by this screen were Lud002 and Lud110, harboring missense mutations in msbA resulting in the amino acid changes S168L and T285I, respectively (Fig. 2, Table 1). The temperature-sensitive phenotype could be cotransduced with the msbA-linked marker aroA::Tn10 present in these strains. It was observed that both Lud002 (S168L) and Lud110 (T285I) had spontaneous reversion rates that were lower by at least 1 order of magnitude (i.e., reversion rates were below 10−5; Fig. 2) than were previously reported for WD2 (9) or for Lud131 or Lud134 (all with the A270T mutation). Interestingly, each of these MsbA mutations maps to the predicted periplasmic surface of the membrane domain of MsbA (Table 1). This location may compose an important surface for formation of effective MsbA dimers in the inner membrane (10) or may play an important role in the transport mechanism. This report therefore adds to the structural information available for MsbA and may be helpful in determining important domains and surfaces of other ABC transporters.
TABLE 1.
MsbA temperature-sensitive mutants isolated following Ludox density gradient enrichment of chemically mutagenized E. coli
| Mutant | Mutation | Topological locationa |
|---|---|---|
| Lud002 | S168L (TCG→TTG) | TM4, near periplasm |
| Lud131 | A270T (GCG→ACG) | TM5, near periplasm |
| Lud134 | A270T (GCG→ACG) | |
| Lud110 | T285I (ACC→ATC) | TM6, near periplasm |
Locations of mutations were determined using the SOSUI prediction program (12). TM, transmembrane domain.
Altered lipid export in new msbA mutants.
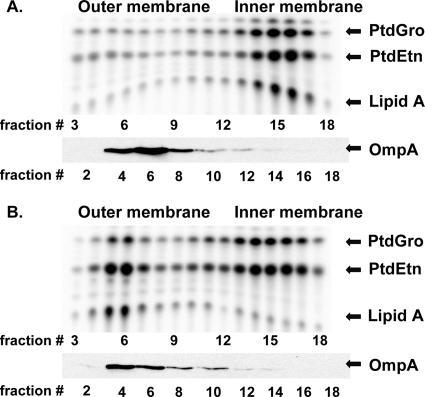
Lipid export was analyzed using Lud002 (harboring the amino acid substitution S168L; Table 1) to confirm that the temperature-sensitive phenotype accompanies a defect in transport of newly synthesized lipids to the OM in this mutant. Prior analysis of mutant WD2 revealed that LPS and phospholipids accumulate at the inner surface of the IM upon shifting to the nonpermissive temperature of 44°C (6, 9). Lud002 displays a similar accumulation of LPS and phospholipids in the IM (Fig. 3). Due to their lower rates of spontaneous reversion, Lud002 and Lud110 may be more useful than WD2 in carrying out genetic analyses such as isolation of multicopy and second site suppressors.
FIG. 3.
Accumulation of newly synthesized lipids in the inner membrane of Lud002 (MsbAS168L). Lud002 cells harboring vector pWSK29 (A) or pWTD10 (B) were grown at 30°C to late log phase in LB-ampicillin and then diluted 1:3 into fresh media that had been prewarmed to 44°C. Growth was continued at this temperature for 40 min with 2.5 μCi/ml 32Pi added during the final 10 min of growth. Cells were recovered by centrifugation, and spheroplasts were prepared by treatment with EDTA-lysozyme and lysed by sonication. Washed membranes were separated into IM and OM fractions by use of a 30-to-60% sucrose gradient (6, 21). Portions of gradient fractions were subjected to mild acid hydrolysis, and lipids were extracted with organic solvents as described previously (6, 37). This treatment has no effect on phospholipids and allows the LPS and phospholipids to be analyzed simultaneously. Lipid species were resolved by thin-layer chromatography using the solvent chloroform:pyridine:88% formic acid:water (50:50:16:5) and analyzed using a PhosphorImager equipped with IQMac software. Fractions were analyzed by Western blotting using an anti-proOmpA polyclonal antibody to identify OM fractions as described previously (8). Abbreviations: PtdGro, phosphatidylglycerol; PtdEtn, phosphatidylethanolamine.
Isolation of htrB mutants.
Seven mutants that were complemented by wild-type msbA did not contain mutations in the msbA structural gene. These mutants (Lud111, -112, -119, -124, -129, -130, and -133) were also complemented by a plasmid copy of htrB and were assumed to contain mutations in the gene htrB. The temperature sensitivity of these strains was not cotransduced with the msbA-linked marker aroA::Tn10. HtrB is a lipid A lauroyl transferase that is required for the production of pentaacylated Kdo2-lipid A from tetraacylated Kdo2-lipid A (4, 26). HtrB deletion mutants do not grow above 33°C and exhibit lipid export defects with accumulation of tetraacylated LPS in the inner membrane during growth at elevated temperatures (15, 37). MsbA was first identified as a multicopy suppressor of an htrB deletion mutant (16). Indeed, these putative htrB mutants were found to produce lipid A with four, rather than six, acyl chains that accumulated in the inner membrane at 42°C (data not shown). These mutants were not characterized further, but their coisolation in this screening speaks to the specificity of this procedure.
Conclusions.
The molecular details of lipid trafficking to the outer membrane of gram-negative bacteria remain unclear despite exciting recent progress (29, 30). While several new players have recently been identified, including MsbA and LptB in the inner membrane and cytoplasm, the Imp/RlpB complex in the OM, and LptA in the periplasm, the interplay between these molecules and the identity of other proteins that take part in this process remain unknown. Identification of components of the machinery involved in lipid trafficking to OM could result in the identification of new antibacterial and anti-inflammatory drugs. We are currently screening larger collections of Ludox-enriched conditional mutants for novel lipid export mutants.
In summary, density enrichment was used to isolate conditional mutants with lipid export defects, including msbA and likely htrB mutants (9, 37). This approach is based on the altered mobility of an msbA temperature-sensitive mutant on density gradients composed of the colloidal silica Ludox. Ludox is easy to use and nontoxic to bacterial cells. It has been used for decades in the study of the bacterial cell cycle (24) and has also been used successfully to isolate some of the yeast Sec mutants that are conditionally defective in secretion of proteins (19). This method provides an important new genetic tool to isolate novel mutants in lipid export.
Acknowledgments
W.T.D. thanks Christian R. H. Raetz for past and continuing support of his work. In addition, W.T.D. thanks Deanna Carrick (Duke University, Department of Cell Biology) for her technical contributions to this work and Gregg Pettis (Louisiana State University Department of Biological Sciences) for his critical reading of the manuscript.
This work had its beginnings in the laboratory of C. R. H. Raetz and was supported by his National Institutes of Health (NIH) grant (GM-51310). Additional financial support has come from the NIH (grant 1 F32 AI-10613-01) and the Louisiana Board of Regents Louisiana Education Quality Support Fund (grant 2005-08-RD-A-04).
Footnotes
Published ahead of print on 26 October 2007.
REFERENCES
- 1.Barb, A. W., A. L. McClerren, K. Snehelatha, C. M. Reynolds, P. Zhou, and C. R. H. Raetz. 2007. Inhibition of lipid A biosynthesis as the primary mechanism of CHIR-090 antibiotic activity in Escherichia coli. Biochemistry 46:3793-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 3.Braun, M., and T. J. Silhavy. 2002. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol. 45:1289-1302. [DOI] [PubMed] [Google Scholar]
- 4.Clementz, T., J. J. Bednarski, and C. R. H. Raetz. 1996. Function of the htrB high temperature requirement gene of Escherchia coli in the acylation of lipid A: HtrB catalyzed incorporation of laurate. J. Biol. Chem. 271:12095-12102. [DOI] [PubMed] [Google Scholar]
- 5.Doerrler, W. T. 2006. Lipid trafficking to the outer membrane of Gram-negative bacteria. Mol. Microbiol. 60:542-552. [DOI] [PubMed] [Google Scholar]
- 6.Doerrler, W. T., H. S. Gibbons, and C. R. H. Raetz. 2004. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 279:45102-45109. [DOI] [PubMed] [Google Scholar]
- 7.Doerrler, W. T., and C. R. H. Raetz. 2002. ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 277:36697-36705. [DOI] [PubMed] [Google Scholar]
- 8.Doerrler, W. T., and C. R. H. Raetz. 2005. Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli YaeT mutant. J. Biol. Chem. 280:27679-27687. [DOI] [PubMed] [Google Scholar]
- 9.Doerrler, W. T., M. C. Reedy, and C. R. H. Raetz. 2001. An Escherichia coli mutant defective in lipid export. J. Biol. Chem. 276:11461-11464. [DOI] [PubMed] [Google Scholar]
- 10.Dong, J., G. Yang, and H. S. McHaourab. 2005. Structural basis of energy transduction in the transport cycle of MsbA. Science 308:1023-1028. [DOI] [PubMed] [Google Scholar]
- 11.Galloway, S. M., and C. R. H. Raetz. 1990. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J. Biol. Chem. 265:6394-6402. [PubMed] [Google Scholar]
- 12.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 13.Kadam, R. U., D. Garg, A. Chavan, and N. Roy. 2007. Evaluation of Pseudomonas aeruginosa deacetylase LpxC inhibitory activity of dual PDE4-TNFalpha inhibitors: a multiscreening approach. J. Chem. Inf. Model. 47:1188-1195. [DOI] [PubMed] [Google Scholar]
- 14.Kadam, R. U., A. V. Shivange, and N. Roy. 2007. Escherichia coli versus Pseudomonas aeruginosa deacetylase LpxC inhibitors selectivity: surface and cavity-depth-based analysis. J. Chem. Inf. Model. 47:1215-1224. [DOI] [PubMed] [Google Scholar]
- 15.Karow, M., O. Fayet, A. Cegielska, T. Ziegelhoffer, and C. Georgopoulos. 1991. Isolation and characterization of the Escherichia coli htrB gene, whose product is essential for bacterial viability above 33°C in rich media. J. Bacteriol. 173:741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karow, M., and C. Georgopoulos. 1993. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutations in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol. Microbiol. 7:69-79. [DOI] [PubMed] [Google Scholar]
- 17.Makinoshima, H., A. Nishimura, and A. Ishihama. 2002. Fractionation of Escherichia coli cell populations at different stages during growth transition to stationary phase. Mol. Microbiol. 43:269-279. [DOI] [PubMed] [Google Scholar]
- 18.Nishijima, M., and C. R. H. Raetz. 1979. Membrane lipid biogenesis in Escherichia coli: identification of genetic loci for phosphatidylglycerophosphate synthetase and construction of mutants lacking phosphatidylglycerol. J. Biol. Chem. 254:7837-7844. [PubMed] [Google Scholar]
- 19.Novick, P., C. Field, and R. Schekman. 1980. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21:205-215. [DOI] [PubMed] [Google Scholar]
- 20.Onishi, H. R., B. A. Pelak, L. S. Gerckens, L. L. Silver, F. M. Kahan, M. H. Chen, A. A. Patchett, S. M. Galloway, S. A. Hyland, M. S. Anderson, and C. R. H. Raetz. 1996. Antibacterial agents that inhibit lipid A biosynthesis. Science 274:980-982. [DOI] [PubMed] [Google Scholar]
- 21.Osborn, M. J., and R. Munson. 1974. Separation of the inner (cytoplasmic) and outer membranes of Gram-negative bacteria. Methods Enzymol. 31:642-653. [DOI] [PubMed] [Google Scholar]
- 22.Pertoft, H., K. Rubin, L. Kjellen, T. C. Laurent, and B. Klingeborn. 1977. The viability of cells grown or centrifuged in a new density gradient medium, Percoll. Exp. Cell Res. 110:449-457. [DOI] [PubMed] [Google Scholar]
- 23.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. Van Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 24.Poole, R. K. 1977. Fluctuations in buoyant density during the cell cycle of Escherichia coli K12: significance for the preparation of synchronous cultures by age selection. J. Gen. Microbiol. 98:177-186. [DOI] [PubMed] [Google Scholar]
- 25.Raetz, C. R. H., T. A. Garrett, C. M. Reynolds, W. A. Shaw, J. D. Moore, D. C. Smith, Jr., A. A. Ribeiro, R. C. Murphy, R. J. Ulevitch, C. Fearns, D. Reichart, C. K. Glass, C. Benner, S. Subramaniam, R. Harkewicz, R. C. Bowers-Gentry, M. W. Buczynski, J. A. Cooper, R. A. Deems, and E. A. Dennis. 2006. Kdo2-lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J. Lipid Res. 47:1097-1111. [DOI] [PubMed] [Google Scholar]
- 26.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robson, A., and I. Collinson. 2006. The structure of the Sec complex and the problem of protein translocation. EMBO Rep. 7:1099-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruiz, N., B. Falcone, D. Kahne, and T. J. Silhavy. 2005. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121:307-317. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz, N., D. Kahne, and T. J. Silhavy. 2006. Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 4:57-66. [DOI] [PubMed] [Google Scholar]
- 30.Sperandeo, P., R. Cescutti, R. Villa, C. Di Benedetto, D. Candia, G. Deho, and A. Polissi. 2007. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J. Bacteriol. 189:244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steeghs, L., R. den Hartog, A. den Boer, B. Zomer, P. Roholl, and P. van der Ley. 1998. Meningitis bacterium is viable without endotoxin. Nature 392:449-450. [DOI] [PubMed] [Google Scholar]
- 32.Tefsen, B., M. P. Bos, F. Beckers, J. Tommassen, and H. de Cock. 2005. MsbA is not required for phospholipid transport in Neisseria meningitidis. J. Biol. Chem. 280:35961-35966. [DOI] [PubMed] [Google Scholar]
- 33.Tefsen, B., J. Geurtsen, F. Beckers, J. Tommassen, and H. de Cock. 2005. Lipopolysaccharide transport to the bacterial outer membrane in spheroplasts. J. Biol. Chem. 280:4504-4509. [DOI] [PubMed] [Google Scholar]
- 34.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 35.Wickner, W., and R. Schekman. 2005. Protein translocation across biological membranes. Science 310:1452-1456. [DOI] [PubMed] [Google Scholar]
- 36.Wu, T., A. C. McCandlish, L. S. Gronenberg, S. S. Chng, T. J. Silhavy, and D. Kahne. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 103:11754-11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou, Z., K. A. White, A. Polissi, C. Georgopoulos, and C. R. H. Raetz. 1998. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 273:12466-12475. [DOI] [PubMed] [Google Scholar]