Abstract
Polyhydroxyalkanoates (PHAs) are accumulated as intracellular granules by many bacteria under unfavorable conditions, enhancing their fitness and stress resistance. Poly(3-hydroxybutyrate) (PHB) is the most widespread and best-known PHA. Apart from the genes that catalyze polymer biosynthesis, natural PHA producers have several genes for proteins involved in granule formation and/or with regulatory functions, such as phasins, that have been shown to affect polymer synthesis. This study evaluates the effect of PhaP, a phasin, on bacterial growth and PHB accumulation from glycerol in bioreactor cultures of recombinant Escherichia coli carrying phaBAC from Azotobacter sp. strain FA8. Cells expressing phaP grew more, and accumulated more PHB, both using glucose and using glycerol as carbon sources. When cultures were grown in a bioreactor using glycerol, PhaP-bearing cells produced more polymer (2.6 times) and more biomass (1.9 times) than did those without the phasin. The effect of this protein on growth promotion and polymer accumulation is expected to be even greater in high-density cultures, such as those used in the industrial production of the polymer. The recombinant strain presented in this work has been successfully used for the production of PHB from glycerol in bioreactor studies, allowing the production of 7.9 g/liter of the polymer in a semisynthetic medium in 48-h batch cultures. The development of bacterial strains that can efficiently use this substrate can help to make the industrial production of PHAs economically feasible.
Polyhydroxyalkanoates (PHAs) are accumulated as intracellular granules by many bacteria under unfavorable conditions (6). PHAs are carbon and energy reserves and also act as electron sinks, enhancing the fitness and stress resistance of bacteria and contributing to redox balance (11, 23). These thermoplastic polyesters were produced industrially by bacterial fermentation from the late 1980s through the 1990s, first by Imperial Chemical Industries and then by other companies (12). Growing concern about environmental pollution and dwindling petroleum supplies has renewed in the last decade the interest in PHAs, which are totally biodegradable by microorganisms present in most environments and can be produced from different renewable carbon sources (8).
Poly(3-hydroxybutyrate) (PHB) is the best-known PHA and is often used as a model product in the development of fermentation strategies. Accumulation of PHB in recombinant Escherichia coli from several carbon sources, including agroindustrial by-products such as whey, has been studied (1, 16). In the last years, there has been a very important increase in the production of glycerol as a by-product in the synthesis of biodiesel (26). This trend has caused a sharp fall in the cost of glycerol, which is now considered a waste product that must be disposed of. As a result, glycerol has become a very attractive substrate for bacterial fermentations, and its potential as a substrate for PHB production is addressed in this work. The use of glycerol for microbial PHA synthesis has been studied in natural PHA producers, such as Methylobacterium rhodesianum and several Pseudomonas strains (26), and also in recombinant E. coli carrying the PHB synthesis genes from Streptomyces aureofaciens (14).
In previous work we have identified and cloned the genes involved in the synthesis of PHB in Azotobacter sp. strain FA8 (18, 19). The three pha structural genes, phaBAC, were introduced in expression plasmids and used for the construction of recombinant E. coli strains that accumulate the polymer from different carbon sources (16).
Apart from the genes that catalyze polymer biosynthesis, natural PHA producers have several genes for proteins involved in granule formation and/or with regulatory functions. Among this group of gene products, phasins, such as PhaP1 from Cupriavidus necator (formerly called Ralstonia eutropha), are granule-associated proteins that have been shown to affect polymer synthesis and the number and size of PHA granules (20). When the pha region of Azotobacter sp. strain FA8 was analyzed, three genes, phaR, phaP, and phaF, similar to previously described PHA-associated genes, were found (18). Studies by other authors have demonstrated increased growth and PHB production in recombinant E. coli carrying genes coding for PhaP from Paracoccus denitrificans (13) and C. necator (30).
The present work studies the effect of phaP on bacterial growth and PHB accumulation from glycerol in bioreactor cultures of recombinant E. coli carrying phaBAC and phaP of Azotobacter sp. strain FA8. Insight into the behavior of recombinant E. coli strains carrying phasin genes in a bioreactor will be extremely useful for the design of bacterial strains adequate for the efficient production of PHAs from glycerol.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains, plasmids, and primers used in this work are summarized in Table 1.
TABLE 1.
E. coli strains, plasmids, and primers used in this study
| E. coli strain, plasmid, or primer | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| DH5α | F− λ−endA1 hsdR17 hsdM+supE44 thi-1 recA1 gyrA96 relA1 Δ(argF lacZYA)U169 φ80d Δ(lacZ)M15 | Life Technologies |
| M15 | Strain used to express and purify proteins; derived from K-12 | Qiagen |
| M15/pQP | M15 carrying plasmid pQP | This work |
| K1060 | F−fadE62 lacI60 tyrT58(AS) fabB5 mel-1 supF58 | E. coli Genetic Stock Center |
| K24vk | As K1060, carrying pJP24 and pBBR1MCS-2 | This work |
| K24Kvc | As K1060, carrying pJP24K and pBBR1MCS | This work |
| K24PF | As K1060, carrying pJP24 and pAD-PF | This work |
| K24KP | As K24K, carrying pJP24K and pAD-P | This work |
| Plasmids | ||
| pQE31 | Expression vector, Apr | Qiagen |
| pBBR1MCS | Broad host range; lacPOZ′; mob RP4 Cmr | 9 |
| pBBR1MCS-2 | Broad host range; lacPOZ′; mob RP4 Kmr | 9 |
| pBlueScript SK− | AprlacPOZ; T7 and T3 promoters | Stratagene |
| pJP24 | pQE32 derivative carrying a 4.3-kb BamHI HindIII insert containing phaBAC from Azotobacter sp. strain FA8, Apr | 16 |
| pJP24K | pJP24 derivative; Apr Kmr | 16 |
| pRX23 | pBlueScript carrying a 5-kb XhoI genomic fragment from Azotobacter sp. strain FA8 containing ISr′, phaF, phaP, and phaR; Apr | This work |
| pBSK-PF | pBlueScript carrying phaF and phaP from Azotobacter sp. strain FA8; Apr | This work |
| pQ-P | pQE31 carrying phaP under lac control; Apr | This work |
| pAD-PF | pBBR1MCS-2 carrying phaF and phaP from Azotobacter sp. strain FA8; Kmr | This work |
| pAD-P | pBBR1MCS carrying phaP from Azotobacter sp. strain FA8; Cmr | This work |
| Primers | ||
| phaPup | CATGgGATcCCGTAATGGCTTTTTTTGATCa,b | This work |
| phaPLow | TCGaaGcTTGCCGTCAGGCAGTCTTb,c | This work |
Restriction site for BamHI is underlined.
Letters in lowercase differ from the original gene sequence.
Restriction site for HindIII is underlined.
Culture conditions and growth media.
For DNA manipulations and strain construction, cultures were grown at 37°C in LB. Ampicillin (100 μg/ml), kanamycin (50 μg/ml), and chloramphenicol (20 μg/ml) were added when indicated. For growth and PHB accumulation analysis bacteria were grown in one of the following media: M9-lactose medium, containing (per liter of deionized water) 1 g NH4Cl, 6 g Na2HPO4, 3 g KH2PO4, 0.5 g NaCl, 0.2 g MgSO4·7H2O, 10 mg CaCl2, and 30 g lactose, or MYA medium, containing (per liter of deionized water) 6 g Na2HPO4, 3 g KH2PO4, 1.4 g (NH4)2SO4, 0.5 g NaCl, 0.2 g MgSO4·7H2O, 10 g yeast extract, and 5 g casein amino acids. MYA was supplemented with either glucose (MYA-Glu) or glycerol (MYA-Gly) at 30 g/liter.
DNA manipulations.
Plasmid and genomic DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, and DNA ligations were performed by standard procedures (15) and following specific instructions from the manufacturers. Transformations were carried out as previously described (7). E. coli strain DH5α was routinely used as a host for most of the recombinant plasmids.
Cloning of phaP and phaF from Azotobacter sp. strain FA8.
A 5.1-kb XhoI genomic fragment from Azotobacter sp. strain FA8 containing genes phaF, phaP, and phaR, together with parts of two flanking insertion sequences (18), was cloned into pBlueScript, giving rise to plasmid pRX23. This plasmid was cut with SmaI and HindIII, and the 1.8-kb fragment containing phaF and phaP was cloned into pBlueScript cut with the same enzymes, resulting in plasmid pBSK-PF. To obtain plasmid pAD-PF, the insert from pBSK-PF was subcloned into the Kmr vector pBBR1MCS-2 using the BamHI and HindIII restriction sites. A 1.1-kb fragment containing phaP was cut from pBSK-PF with SalI and cloned into the Cmr vector pBBR1MCS, resulting in plasmid pAD-P.
Construction of phaP expression vector.
Primers phaPup and phaPlow (Table 1) were used to obtain a 591-bp amplification fragment using plasmid pRX23 as a template. The resulting amplification fragment was cut with HindIII and BamHI and ligated into vector pQE31. The resulting plasmid, pQP, confers resistance to ampicillin and expresses phaP from a promoter operator element consisting of the phage T5 promoter and two lac operator sequences. The insert was cloned in such a way that the amino-terminal end of the phaP gene product is fused to the first six amino acids of the lacZ gene, followed by six histidine residues, resulting in a fusion protein. The phasin protein was expressed in strain M15/pQP and purified by affinity chromatography as recommended by the supplier, using the QIAexpress system kit (Qiagen). The purified protein was used to obtain an anti-PhaP antiserum in mice.
Western analysis.
For immunoblotting experiments, cells were pelleted and sonicated in 1 ml of 100 mM phosphate buffer, pH 7.5, plus 1 mM phenylmethylsulfonyl fluoride. Protein concentration was measured according to the Bradford method (3). Equal amounts (30 μg) of total protein were used for a Western immunoblotting assay. Proteins were separated by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (BA85; Schleicher & Schuell). For Western blotting, the membrane was probed with mouse anti-PhaP polyclonal serum diluted 1:2,500, followed by alkaline phosphatase-conjugated coat anti-mouse immunoglobulin G (NEN) diluted 1:5,000. The blots were developed with the Phototope-Star chemiluminescent detection kit (New England Biolabs).
Bioreactor cultivation.
Batch cultures were carried out at 37.0 ± 0.2°C in a 5.6-liter stirred tank reactor equipped with six flat-bladed disk turbines (BioFlo110; New Brunswick Scientific Co., Edison, NJ) with a working volume of 3 liters MYA-Gly, containing 50 μg/ml kanamycin and 20 μg/ml chloramphenicol for plasmid maintenance. Culture pH was controlled at 7.20 by automatic addition of either 3 N KOH or 5 N H2SO4. Dissolved oxygen concentration was maintained above 30% of air saturation throughout the fermentation by the automatic control of the agitation speed (up to 1,000 rpm) while sparging the fermentor with 1 vessel volume of air per minute. Dissolved oxygen was measured using an Ag/AgCl polarometric oxygen probe (Mettler Toledo, Greifensee, Switzerland). Foam was suppressed by adding 30 μl/liter Antifoam 289 (Sigma-Aldrich). Residual glycerol concentration was determined using an enzymatic kit (Roche Diagnostics, Germany) at the end of the fermentation.
Determination of plasmid stability.
The amount of plasmid-containing cells was determined by plating samples from the corresponding cultures on LB with or without the appropriate antibiotics. Plasmid stability was recorded as the percentage of antibiotic-resistant cells.
Biomass determination.
Samples taken from the bioreactor were immediately chilled at 0°C by placement in an ice bath. Cells from 10-ml samples were washed twice with deionized water, recovered by centrifugation, dried at 85°C for 36 h, and weighed. Biomass content was defined as grams (dry weight) of cells (CDW) per liter. All results are indicative of replicate experiments.
Analysis of PHB production.
For qualitative detection of PHB inclusion bodies, cells were observed by fluorescence microscopy after staining with the basic oxazine Nile Blue A as previously described (17). PHB content in flask cultures was determined gravimetrically after alkaline treatment with 0.2 N NaOH as described previously (5). PHB content in bioreactor cultures was quantitatively determined by gas chromatography using a slight modification of the method described by Braunegg et al. (4, 16). Pure PHB from C. necator was used as a standard. PHB concentration was defined as g polymer per liter of culture broth. PHB content was defined as a percentage of CDW. All results are indicative of duplicate or triplicate experiments.
Purification of PHB.
PHB produced at the end of the fermentation experiments was extracted from lyophilized cells with hot CHCl3 using a Soxhlet apparatus, ethanol precipitated, and recovered by filtration. The precipitate was dried, dissolved in CHCl3, filtered to remove contaminating particles, and dried on a glass petri dish to obtain a thin film. The resulting polymer was characterized by gas chromatography as described above, and used for differential scanning calorimetry (DSC) measurements.
DSC measurements.
Glass transition temperature (Tg), melting temperature (Tm), and crystallinity of purified PHB samples were determined by DSC using Mettler 822 and STARe thermal analysis system version 6.1 software (Mettler Toledo AG, Switzerland) as previously described (16). Crystallinity of PHB was estimated from the enthalpy of fusion obtained by DSC. The fusion enthalpy of a theoretical 100% crystalline sample was assumed to be 146 J/g (2). All results are indicative of duplicate or triplicate experiments.
Transmission electron microscopy.
Cells were fixed by adding 2.5% (vol/vol) glutaraldehyde and embedded in agar. One-millimeter pieces of the agar were fixed for 30 min in phosphate-buffered 2.5% (vol/vol) glutaraldehyde, rinsed three times with phosphate buffer, and postfixed in phosphate-buffered 1% (wt/vol) osmium tetroxide for 1 h. The agar pieces were rinsed with water again and fixed for 1 h in 1% (wt/vol) aqueous uranyl acetate. All fixations were carried out at room temperature. After dehydration in a graded series of ethanol and two changes in propylene oxide, the agar pieces containing bacterial cells were embedded in Epon 812 resin (Spi Supplies, New Chester, PA). Thin sections stained with uranyl acetate and lead citrate were examined in a Philips EM 301 transmission electron microscope.
RESULTS
Effect of phaP and phaF on PHB synthesis in E. coli carrying phaBAC from Azotobacter sp. strain FA8.
K24 is an E. coli strain carrying the PHB biosynthetic genes from Azotobacter sp. strain FA8 in Apr plasmid pJP24 (16). As a first approach to analyze the possible effects of putative PHA regulatory genes found in Azotobacter sp. strain FA8 on recombinant E. coli, pAD-PF, a Kmr plasmid containing phaP and phaF, was introduced into K24. Both recombinant plasmids have compatible replication origins and different selection markers, so they can coexist in the recombinants, allowing the combined expression of the heterologous genes. Plasmid pAD-PF contains phaP and phaF, situated in opposite (converging) orientations and approximately 300 bases upstream from each gene. phaP is situated colinearly with the lac promoter. The growth behavior and ability to accumulate the polymer in M9-lactose medium were analyzed in K24 containing pAD-PF (K24PF) or the Kmr vector without an insert (K24vk). Results obtained in three independent experiments (in which each parameter was measured in duplicate) showed that although CDW concentration at the end of the cultivation was similar (1.1 ± 0.1 g/liter and 1.1 ± 0.2 g/liter for K24vk and K24PF, respectively), the PHB content was higher for the strain bearing pAD-PF (24.6% ± 1.1% and 31.8% ± 3.8% for K24vk and K24PF, respectively). To verify if the effects observed were due to the expression of phaP, a plasmid containing only this gene was constructed.
Construction of a stable PHA-accumulating recombinant strain that expresses PhaP.
Strain K24 was not able to accumulate high concentrations of PHB due to the instability of its Apr recombinant plasmid, but pJP24K, a Kmr derivative of pJP24 analyzed in a previous study (16), was efficiently maintained in bioreactor cultures of the corresponding strain, K24K. In view of this, we decided to study the effects of PhaP in strain K24K. Plasmid pAD-P, a Cmr plasmid that expresses phaP under lac control, was constructed and introduced into strain K24K. This plasmid contains the coding region of phaP including 392 bases from its upstream region.
phaP expression in the recombinants was verified by Western blot experiments (Fig. 1). The phaP-containing insert was cloned in both orientations, but only the orientation colinear with the lac promoter allowed the expression of PhaP, as detected in Western blot assays (data not shown).
FIG. 1.
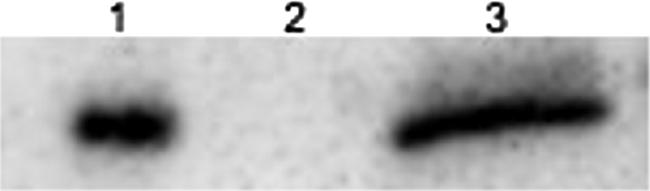
Immunoblot assays of PhaP in E. coli strains containing different plasmids. Lane 1, M15/pQP; lane 2, K24Kvc cultured in MYA-Gly in flask cultures, 24 h; lane 3, K24KP cultured in MYA-Gly in flask cultures, 24 h. Equal amounts (30 μg) of total protein were used for each lane.
The effect of PhaP in K24K was analyzed in agitated flask cultures (50 ml in 500-ml Erlenmeyer flasks) in MYA medium supplemented with glucose or glycerol. Cell growth and PHB content were monitored in the cultures of K24K bearing pAD-P (K24KP) or the Cmr plasmid without insert (K24Kvc) (Table 2). In both carbon sources the PhaP-bearing strains grew more and contained more polymer. Cultures of K24KP grown in glucose achieved higher biomass and PHB accumulation levels than those grown in glycerol, but growth of K24Kvc was similar on the two carbon sources. Plasmid loss was less than 1% for both cultures. Cells were observed by transmission electron microscopy, revealing PHA granules of similar size, present in similar numbers in the two strains, but more cells containing polymer granules were observed for K24KP (data not shown).
TABLE 2.
Effect of PhaP on biomass and PHB accumulation in 48-h flask cultures of K24Kvc and K24KP in MYA-Glu and MYA-Glya
| Insert in plasmid | Carbon source | CDW (g/liter) | % PHBb,c | PHB (g/liter)c | μmax (h−1) |
|---|---|---|---|---|---|
| None | Glucose | 4.9 ± 0.1 | 27.6 ± 1.4 | 1.4 ± 0.1 | 0.9 ± 0.1 |
| phaP | Glucose | 12.0 ± 0.1 | 48.1 ± 1.3 | 5.8 ± 0.2 | 1.1 ± 0.1 |
| None | Glycerol | 5.6 ± 0.1 | 9.8 ± 1.5 | 0.6 ± 0.1 | 0.9 ± 0.1 |
| phaP | Glycerol | 7.4 ± 1.2 | 46.1 ± 6.6 | 3.4 ± 0.1 | 1.0 ± 0.1 |
Means from triplicate experiments are shown.
The amount of PHB is given as an average weight percentage of the CDW.
PHB was determined gravimetrically after alkaline treatment.
As a result of enhanced growth and polymer accumulation, K24KP produced several times more polymer than the strain without PhaP, and this difference was more evident when using glycerol as the carbon source.
Effect of PhaP on cell growth and PHB production from glycerol in bioreactor cultures.
Strain K24K bearing plasmid pBBR1MCS (K24Kvc) or pAD-P (K24KP) was grown in MYA-Gly medium. Cell growth, PHB production, and cell morphology were analyzed. Table 3 shows cell growth and PHB content of a representative bioreactor culture. The strain expressing PhaP consumed more substrate, grew 1.9 times more (15.3 versus 8.0 g/liter), and produced 2.6-times-more PHB (7.9 versus 3.1 g/liter) than the strain with the vector alone. Plasmid loss was less than 1% for both cultures.
TABLE 3.
Biomass and PHB accumulation in 48-h bioreactor cultures of strains K24Kvc and K24KP in MYA-Glya
| Strain | CDW (g/liter) | PHBb (g/liter) | Residual CDWc (g/liter) | % PHBd (wt/wt) | Residual glycerole (g/liter) | PHB productivity (g PHB liter−1 h−1) | YPHB/Sf | μ max (h−1) |
|---|---|---|---|---|---|---|---|---|
| K24Kvc | 8.0 ± 1.2 | 3.1 ± 0.1 | 5.0 ± 0.9 | 38.2 ± 1.2 | 2.2 | 0.1 | 0.1 | 0.6 ± 0.1 |
| K24KP | 15.3 ± 1.3 | 7.9 ± 0.5 | 7.3 ± 1.1 | 51.9 ± 3.3 | 0.5 | 0.2 | 0.3 | 0.6 ± 0.1 |
Means from triplicate determinations of a representative experiment are shown.
PHB was determined by gas chromatography.
Residual CDW was calculated by subtracting the weight of PHB.
The amount of PHB is given as an average weight percentage of the CDW.
Residual glycerol was enzymatically determined at the end of the fermentation.
PHB yield was calculated on the basis of glycerol consumption.
The cells were microscopically observed to detect changes in cell morphology, as other authors have reported a tendency for some recombinant strains to form filaments when grown in bioreactors (28). Cells expressing PhaP were slightly bigger, probably because of their increased polymer content, but no filamentation was observed in any of the cultures (data not shown). K24KP cells from the 24-h bioreactor cultures were observed in a transmission electron microscope. Large granules occupying a great portion of the cells could be observed (see Fig. S1 in the supplemental material).
Physical properties of PHB obtained from glycerol bioreactor cultures were investigated by DSC. The polymer from K24Kvc had a lower Tg and Tm than the phasin-bearing recombinant (46.3 ± 0.3 and 169.8 ± 0.4 for K24Kvc and 56.9 ± 1.0 and 177.3 ± 0.5 for K24KP, respectively). The PHB obtained from K24Kvc also showed a lower percent crystallinity (52.6 ± 0.4) than the polymer from K24KP (66.4 ± 1.5). These results show that the recombinant strains produce PHB with different physical characteristics.
DISCUSSION
Previous studies (24, 31) that investigated the effect of PhaP on PHB accumulation in recombinant E. coli carrying pha from C. necator have shown an increase in polymer accumulation and a higher number of smaller PHA granules in strains carrying phaP. York et al. (31) reported a 100% increase in polymer production in 105-h flask cultures in LB containing 2% glucose. We analyzed the effect of phaP of Azotobacter sp. strain FA8 on PHB accumulation and bacterial growth in recombinant E. coli carrying the pha-synthesizing genes from this strain. As expected, the cells carrying PhaP accumulated more polymer. When the effect of PhaP on cell growth was first analyzed in flask cultures grown in minimal medium with lactose, no differences were observed, but when cells were grown in conditions in which higher cell densities were achieved, those carrying phaP grew more and accumulated more PHB. When cultures were grown in a bioreactor using glycerol, the differences observed were greater. The increase in biomass production is partly due to higher polymer accumulation, but when the residual biomasses were compared, the value for PhaP-bearing cells was also higher. These results suggest that PhaP exerts a growth-promoting effect in the PHB-synthesizing recombinants, probably by acting as a barrier between the polymer and cytoplasmic components. In a previous study in which the effect of phaP from C. necator in recombinant E. coli expressing the nonnatural BPEC pathway was analyzed (27), PHA- or polythioester-producing cells carrying phaP accumulated more polymer in agitated flask cultures, but no differences were observed in the growth behavior of the cells. The difference of these results from those observed in the present work could be due to the different polymer-synthesizing pathways, but they could also be explained by taking into account the fact that the cultures did not grow to high cell densities.
The enhancing effect of PhaP on growth adds an additional advantage to the use of recombinant strains harboring this protein, because it increases not only PHA content in the cells but also total biomass. Different hypotheses have been proposed to explain the enhancing effect of phasins on PHB accumulation in natural PHA producers or in recombinant bacteria (21). These proteins could increase the activity of the enzymes involved in polymer synthesis or benefit PHA-synthesizing cells by their structural role, forming a barrier between the hydrophobic polymer and cytoplasmic components (29). The first hypothesis is supported by in vitro studies showing that PhaP from C. necator increases the activity of PhaC2 of Pseudomonas aeruginosa (22), but an in vivo study (25) has reported no increase in the activity of the three structural pha genes in PhaP-expressing strains. The results obtained in the present work, in accordance with those obtained by York et al. (31), demonstrate that the PhaP-containing recombinants exhibit both increased polymer accumulation and cell growth, suggesting a protective effect of PhaP, which is more evident in denser cultures. Following this hypothesis, the effect of this protein on growth promotion and polymer accumulation would be expected to be even greater in high-density cultures, such as those used in the industrial production of the polymers.
When the physical properties of PHB purified from the two recombinants were compared, it was found that the strain carrying phaP produces PHB with different physical characteristics. A recent study (10) that analyzed the characteristics of PHB produced by mutants of phasin genes in C. necator has found no effect on the molecular weight of the polymers. Further analysis would be needed in order to establish whether the characteristics observed in the present work are due to a direct effect of PhaP on the polymer, or if they are a consequence of the differences in growth observed.
A recombinant E. coli strain carrying a DNA fragment from Streptomyces aureofaciens (14), in contrast with results observed in the present work, accumulated more polymer when growing on glycerol than when growing on glucose, probably due to the presence of other genes from S. aureofaciens or to different growth conditions. On the other hand, the relative amount of polymer accumulated by recombinant E. coli from these carbon sources has been observed to depend on aeration conditions (unpublished results). There are, to our knowledge, no other publications describing the synthesis of PHB from glycerol using recombinant E. coli.
The recombinant strain presented in this work has been successfully used for the production of PHB from glycerol in bioreactor studies, allowing the production of 7.9 g/liter of the polymer in a semisynthetic medium in 48-h batch cultures. The development of bacterial strains that can efficiently use this substrate can help in making the industrial production of PHAs economically feasible.
Supplementary Material
Acknowledgments
We thank Beatriz S. Méndez for helpful discussions, Patricio R. Santagapita and M. del Pilar Buera for DSC analysis, and Bernd Rehm for kindly giving us plasmid pBBR1MCS.
This work was supported by grants from UBA (project no. X134) and CABBIO (project no. 8). M.J.P. is a career investigator from CONICET. A.D.A. and P.I.N. have graduate student fellowships from CONICET.
Footnotes
Published ahead of print on 26 October 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ahn, W. S., S. J. Park, and S. Y. Lee. 2000. Production of poly(3-hydroxybutyrate) by fed-batch culture of recombinant Escherichia coli with a highly concentrated whey solution. Appl. Environ. Microbiol. 66:3624-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barham, P. G., A. Keller, E. L. Otum, and P. A. Holmes. 1984. Crystallization and morphology of a bacterial thermoplastic: poly-β-hydroxybutyrate. J. Mater. Sci. 19:2781-2794. [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Braunegg, G., B. Sonnleitner, and R. M. Lafferty. 1978. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in bacterial biomass. Eur. J. Appl. Microbiol. Biotechnol. 6:29-37. [Google Scholar]
- 5.Choi, J., and S. Y. Lee. 1999. Efficient and economical recovery of poly(3-hydroxybutyrate) from recombinant Escherichia coli by simple digestion with chemicals. Biotechnol. Bioeng. 62:546-553. [DOI] [PubMed] [Google Scholar]
- 6.Dawes, E. A., and P. J. Senior. 1973. The role and regulation of energy reserve polymers in micro-organisms. Adv. Microb. Physiol. 10:135-266. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan, D. 1985. Techniques for transformation of Escherichia coli, p. 109-135. In D. M. Glover (ed.), DNA cloning: a practical approach, vol. 1. IRL Press, Oxford, England. [Google Scholar]
- 8.Khanna, S., and A. K. Srivastava. 2005. Recent advances in microbial polyhydroxyalkanoates. Process Biochem. 40:607-619. [Google Scholar]
- 9.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 10.Kuchta, K., L. Chi, H. Fuchs, M. Pötter, and A. Steinbüchel. 2007. Studies on the influence of phasins on accumulation and degradation of PHB and nanostructure of PHB granules in Ralstonia eutropha H16. Biomacromolecules 8:657-662. [DOI] [PubMed] [Google Scholar]
- 11.López, N. I., M. E. Floccari, A. Steinbüchel, A. F. García, and B. S. Méndez. 1995. Effect of poly(3-hydroxybutyrate) (PHB) content on the starvation-survival of bacteria in natural waters. FEMS Microbiol. Ecol. 16:95-112. [Google Scholar]
- 12.Madison, L. L., and G. W. Huisman. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63:21-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maehara, A., S. Ueda, H. Nakano, and T. Yamane. 1999. Analyses of a polyhydroxyalkanoic acid granule-associated 16-kilodalton protein and its putative regulator in the pha locus of Paracoccus denitrificans. J. Bacteriol. 181:2914-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahishi, L. H., G. Tripathi, and S. K. Rawal. 2003. Poly(3-hydroxybutyrate) (PHB) synthesis by recombinant Escherichia coli harbouring Streptomyces aureofaciens PHB biosynthesis genes: effect of various carbon and nitrogen sources. Microbiol. Res. 158:19-27. [DOI] [PubMed] [Google Scholar]
- 15.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1988. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16.Nikel, P. I., A. de Almeida, E. C. Melillo, M. A. Galvagno, and M. J. Pettinari. 2006. New recombinant Escherichia coli strain tailored for the production of poly(3-hydroxybutyrate) from agroindustrial by-products. Appl. Environ. Microbiol. 72:3949-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostle, A. G., and J. G. Holt. 1982. Nile blue A as a fluorescent stain for poly-β-hydroxybutyrate. Appl. Environ. Microbiol. 44:238-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pettinari, J. M., L. Chaneton, G. Vázquez, A. Steinbüchel, and B. S. Méndez. 2003. Insertion sequence-like elements associated with putative polyhydroxybutyrate regulatory genes in Azotobacter sp. FA8. Plasmid 50:36-44. [DOI] [PubMed] [Google Scholar]
- 19.Pettinari, M. J., G. J. Vazquez, D. Silberschmidt, B. Rehm, A. Steinbüchel, and B. S. Méndez. 2001. Poly(3-hydroxybutyrate) synthesis genes in Azotobacter sp. strain FA8. Appl. Environ. Microbiol. 67:5331-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pötter, M., H. Muller, F. Reinecke, R. Wieczorek, F. Fricke, B. Bowien, B. Friedrich, and A. Steinbüchel. 2004. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology 150:2301-2311. [DOI] [PubMed] [Google Scholar]
- 21.Pötter, M., and A. Steinbüchel. 2005. Poly(3-hydroxybutyrate) granule-associated proteins: impacts on poly(3-hydroxybutyrate) synthesis and degradation. Biomacromolecules 6:552-560. [DOI] [PubMed] [Google Scholar]
- 22.Qi, Q., A. Steinbüchel, and B. H. Rehm. 2000. In vitro synthesis of poly(3-hydroxydecanoate): purification and enzymatic characterization of type II polyhydroxyalkanoate synthases PhaC1 and PhaC2 from Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 54:37-43. [DOI] [PubMed] [Google Scholar]
- 23.Ruiz, J. A., R. O. Fernández, P. I. Nikel, B. S. Méndez, and M. J. Pettinari. 2006. dye (arc) mutants: insights into an unexplained phenotype and its suppression by the synthesis of poly(3-hydroxybutyrate) in Escherichia coli recombinants. FEMS Microbiol. Lett. 258:55-60. [DOI] [PubMed] [Google Scholar]
- 24.Seo, M. C., H. D. Shin, and Y. H. Lee. 2003. Functional role of granule-associated genes, phaP and phaR, in poly-beta-hydroxybutyrate biosynthesis in recombinant E. coli harboring phbCAB operon. Biotechnol. Lett. 25:1243-1249. [DOI] [PubMed] [Google Scholar]
- 25.Seo, M. C., H. D. Shin, and Y. H. Lee. 2004. Transcription level of granule-associated phaP and phaR genes and granular morphogenesis of poly-beta-hydroxyalkanoate granules in Ralstonia eutropha. Biotechnol. Lett. 26:617-622. [DOI] [PubMed] [Google Scholar]
- 26.Solaiman, D. K., R. D. Ashby, T. A. Foglia, and W. N. Marmer. 2006. Conversion of agricultural feedstock and coproducts into poly(hydroxyalkanoates). Appl. Microbiol. Biotechnol. 71:783-789. [DOI] [PubMed] [Google Scholar]
- 27.Tessmer, N., S. Konig, U. Malkus, R. Reichelt, M. Pötter, and A. Steinbüchel. 2007. Heat-shock protein HspA mimics the function of phasins sensu stricto in recombinant strains of Escherichia coli accumulating polythioesters or polyhydroxyalkanoates. Microbiology 153:366-374. [DOI] [PubMed] [Google Scholar]
- 28.Wang, F., and S. Y. Lee. 1997. Production of poly(3-hydroxybutyrate) by fed-batch culture of filamentation-suppressed recombinant Escherichia coli. Appl. Environ. Microbiol. 63:4765-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wieczorek, R., A. Pries, A. Steinbüchel, and F. Mayer. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 177:2425-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.York, G. M., J. Stubbe, and A. J. Sinskey. 2001. New insight into the role of the PhaP phasin of Ralstonia eutropha in promoting synthesis of polyhydroxybutyrate. J. Bacteriol. 183:2394-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.York, G. M., J. Stubbe, and A. J. Sinskey. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J. Bacteriol. 184:59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


